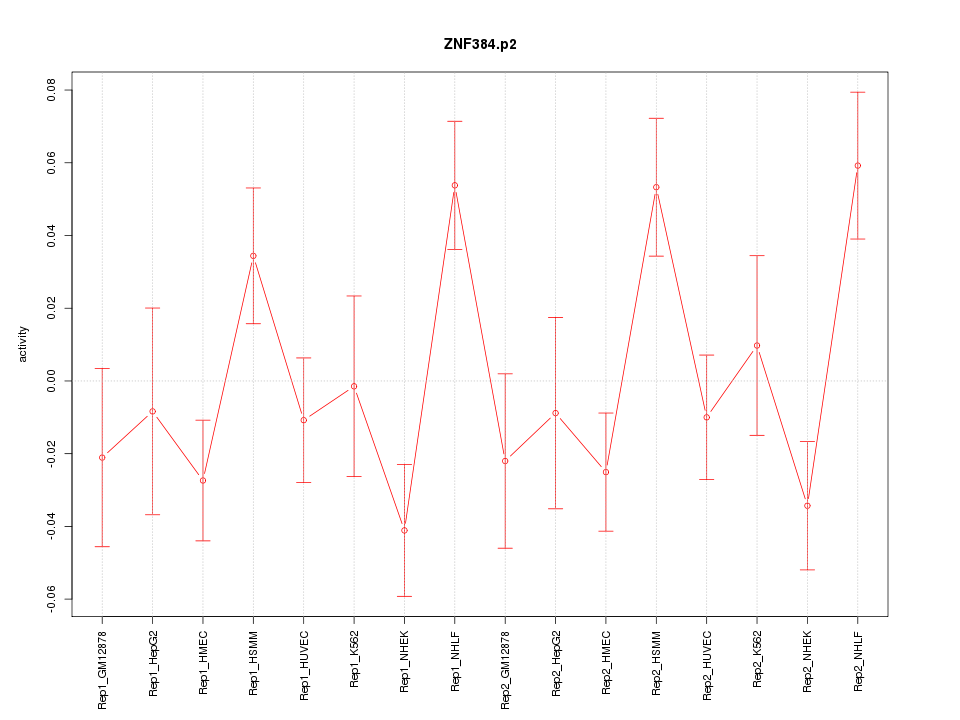
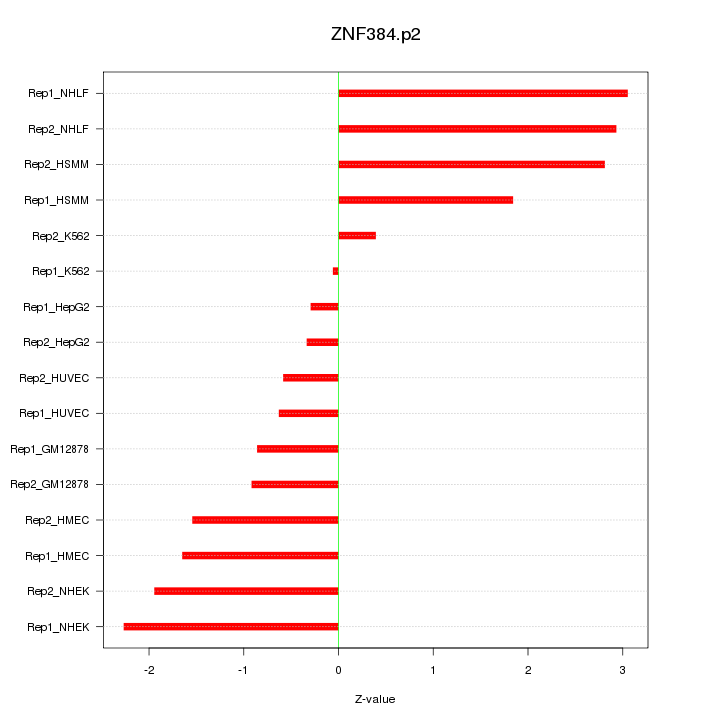
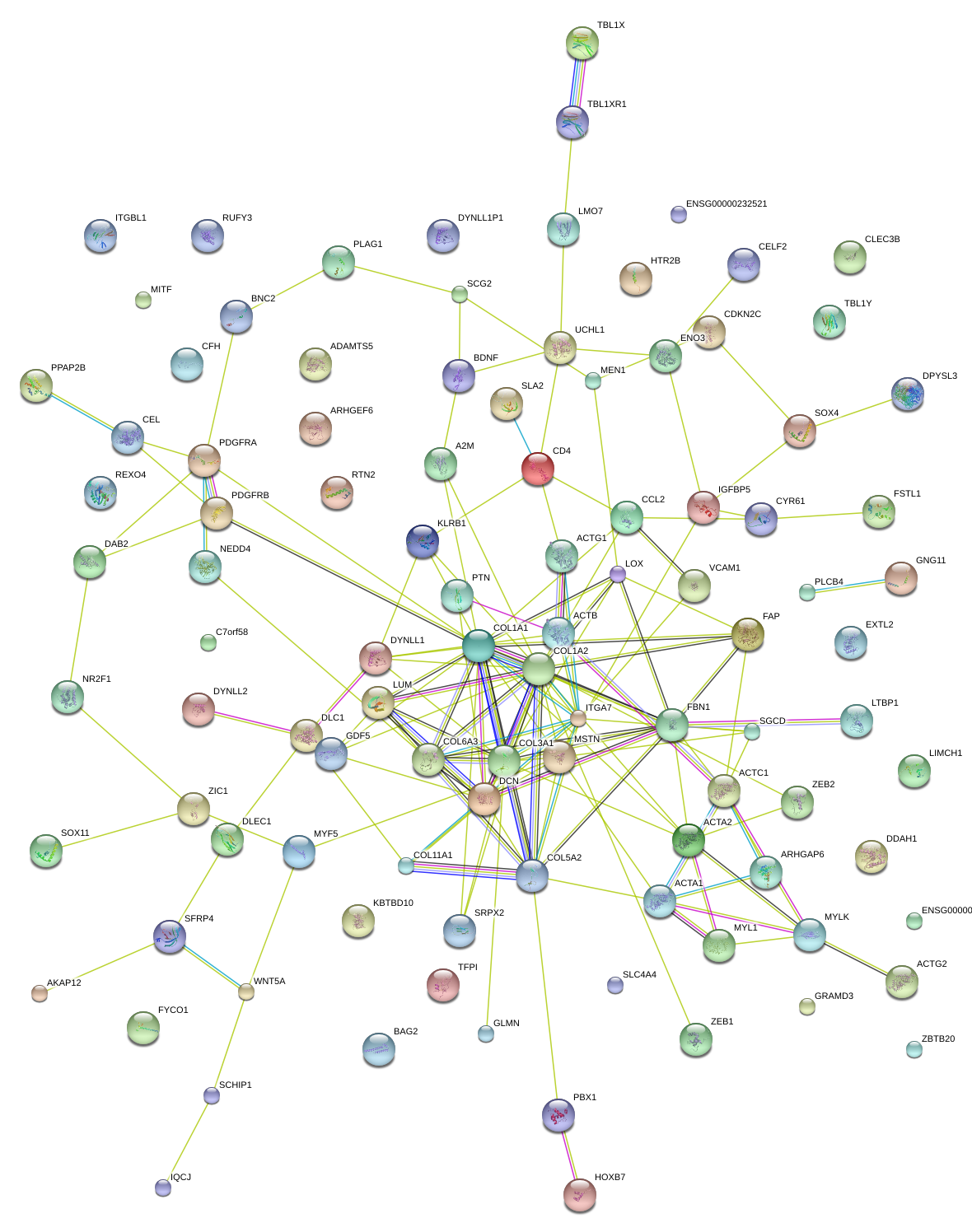
|
chr2_+_189547358
|
7.832
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189547323
|
7.560
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189547377
|
6.128
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_-_189752575
|
5.666
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr2_-_188127430
|
4.476
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr4_+_54790020
|
3.846
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr2_-_237987509
|
3.735
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_237987579
|
3.725
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr12_-_90029672
|
3.473
|
NM_002345
|
LUM
|
lumican
|
|
chr7_+_93862137
|
3.250
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr2_+_170074457
|
3.016
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr2_-_190635566
|
3.008
|
NM_005259
|
MSTN
|
myostatin
|
|
chr2_-_188127294
|
2.936
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr13_+_100902942
|
2.893
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr12_-_90100733
|
2.850
|
NM_001920
|
DCN
|
decorin
|
|
chr2_-_237987554
|
2.825
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_144994349
|
2.607
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_144994038
|
2.488
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_210888010
|
2.286
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr2_-_217268492
|
2.253
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr2_+_5750229
|
2.223
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr8_-_13018123
|
2.121
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_+_162795328
|
2.098
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_-_116272911
|
2.093
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr1_+_194887630
|
2.075
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chrX_-_135691168
|
2.064
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr1_-_103346639
|
1.987
|
NM_001190709
NM_001854
NM_080629
NM_080630
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr5_-_42847716
|
1.982
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr12_-_90100679
|
1.931
|
|
DCN
|
decorin
|
|
chr11_-_27637771
|
1.884
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr12_-_90100710
|
1.880
|
|
DCN
|
decorin
|
|
chr4_-_81213207
|
1.867
|
|
|
|
|
chr15_-_46725087
|
1.817
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr4_+_40953698
|
1.718
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr4_+_41395493
|
1.631
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr20_+_9236446
|
1.520
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr3_-_55496607
|
1.490
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr2_-_224175293
|
1.466
|
|
SCG2
|
secretogranin II
|
|
chr3_-_55496370
|
1.452
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr5_+_155686333
|
1.406
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr2_-_162808123
|
1.400
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr3_+_70068440
|
1.397
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr5_-_149515331
|
1.356
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr4_+_40953666
|
1.340
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr4_+_40953652
|
1.326
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr1_+_163084707
|
1.297
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_-_124822113
|
1.295
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr2_-_224175386
|
1.291
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr7_+_93388951
|
1.289
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr5_-_146813343
|
1.281
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr1_-_56817823
|
1.281
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_-_224175339
|
1.271
|
|
SCG2
|
secretogranin II
|
|
chr3_-_124821868
|
1.249
|
|
MYLK
|
myosin light chain kinase
|
|
chr12_-_54392328
|
1.243
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr20_-_34707971
|
1.241
|
NM_032214
NM_175077
|
SLA2
|
Src-like-adaptor 2
|
|
chr7_-_136678973
|
1.232
|
|
PTN
|
pleiotrophin
|
|
chr5_+_92944680
|
1.232
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr17_-_45617899
|
1.230
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr2_+_73973600
|
1.228
|
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr12_+_79634820
|
1.221
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr5_+_155686750
|
1.209
|
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr10_+_11246934
|
1.204
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_+_73973640
|
1.197
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr1_+_51207242
|
1.197
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr5_-_39460687
|
1.193
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr2_+_33213111
|
1.179
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr13_+_75232797
|
1.172
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr1_-_103346482
|
1.146
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr3_-_55490401
|
1.140
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr5_-_149515613
|
1.128
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr1_+_163084934
|
1.127
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr5_+_125786975
|
1.121
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr2_-_231697999
|
1.108
|
NM_000867
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B
|
|
chr1_+_100957883
|
1.098
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr21_-_27261276
|
1.050
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr3_-_121652183
|
1.047
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr5_+_125786717
|
1.033
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chrX_+_9391314
|
1.015
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr20_-_33489383
|
0.970
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr5_-_149515502
|
0.969
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr17_-_44042924
|
0.957
|
|
HOXB7
|
homeobox B7
|
|
chr7_-_37922902
|
0.950
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr4_+_71806538
|
0.943
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr3_+_45042754
|
0.932
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chrX_+_99785818
|
0.928
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr1_+_51207035
|
0.920
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr3_+_160269734
|
0.876
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr6_+_57145017
|
0.872
|
NM_004282
|
BAG2
|
BCL2-associated athanogene 2
|
|
chr1_-_85816520
|
0.869
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr6_+_151688327
|
0.864
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_-_121440704
|
0.844
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr1_-_101132993
|
0.835
|
NM_001033025
NM_001439
|
EXTL2
|
exostoses (multiple)-like 2
|
|
chrX_-_11194015
|
0.832
|
NM_013423
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr12_-_9159780
|
0.829
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr3_-_115960807
|
0.827
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr4_+_72423633
|
0.823
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr3_+_148609841
|
0.823
|
NM_003412
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr2_-_210876560
|
0.820
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr17_+_29606408
|
0.820
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr17_+_4795128
|
0.815
|
NM_001193503
NM_001976
NM_053013
|
ENO3
|
enolase 3 (beta, muscle)
|
|
chr1_+_85819004
|
0.809
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr12_-_9159789
|
0.807
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr7_+_120415958
|
0.804
|
NM_024913
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr12_+_6768966
|
0.796
|
|
CD4
|
CD4 molecule
|
|
chr7_+_120416103
|
0.794
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr19_-_50688310
|
0.793
|
NM_206901
|
RTN2
|
reticulon 2
|
|
chr9_-_16860719
|
0.787
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr15_-_53996597
|
0.786
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr12_+_74160779
|
0.785
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr10_-_90740935
|
0.779
|
NM_001141945
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr1_+_160868822
|
0.766
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr16_+_6763810
|
0.761
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr9_-_87904306
|
0.758
|
NM_016548
|
GOLM1
|
golgi membrane protein 1
|
|
chr5_-_88155360
|
0.757
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr9_-_16717887
|
0.755
|
|
BNC2
|
basonuclin 2
|
|
chr9_-_87904280
|
0.753
|
|
GOLM1
|
golgi membrane protein 1
|
|
chr12_+_10256682
|
0.750
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr12_+_79625538
|
0.749
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr6_-_152681171
|
0.735
|
NM_015293
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr1_-_84928473
|
0.732
|
NM_001166295
NM_001166417
NM_001166293
NM_001166294
NM_014021
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr3_-_101316038
|
0.710
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr1_+_51206819
|
0.709
|
NM_001262
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr2_-_152299229
|
0.708
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr5_-_121441910
|
0.705
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr1_+_160868932
|
0.703
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr10_+_11247059
|
0.699
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr15_-_88150823
|
0.693
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr10_+_31648103
|
0.681
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr17_-_61253122
|
0.670
|
NM_001037325
|
CCDC46
|
coiled-coil domain containing 46
|
|
chr2_+_151922350
|
0.669
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr8_-_120720200
|
0.667
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr8_+_22493928
|
0.663
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr4_+_54790190
|
0.651
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr10_+_63331018
|
0.638
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chrX_+_54850832
|
0.637
|
NM_177433
|
MAGED2
|
melanoma antigen family D, 2
|
|
chr3_-_167037861
|
0.637
|
|
BCHE
|
butyrylcholinesterase
|
|
chr7_+_120416680
|
0.636
|
NM_001105533
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr1_-_171442937
|
0.631
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr8_-_122722674
|
0.627
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr11_-_2127368
|
0.619
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr7_-_37923040
|
0.619
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr4_+_169654742
|
0.616
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr11_+_46359781
|
0.615
|
NM_002391
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr7_-_136678999
|
0.610
|
|
PTN
|
pleiotrophin
|
|
chr1_+_154095883
|
0.608
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr15_-_75135496
|
0.601
|
|
TSPAN3
|
tetraspanin 3
|
|
chr2_-_188086612
|
0.600
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr4_-_186934059
|
0.598
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_39868770
|
0.596
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr9_+_101628829
|
0.591
|
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr7_+_22733626
|
0.589
|
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr11_+_46359836
|
0.588
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr1_+_51206963
|
0.580
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr4_-_91979113
|
0.578
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr12_+_13240868
|
0.577
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr6_+_151603346
|
0.573
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_-_111119846
|
0.572
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr17_-_44037332
|
0.571
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr20_+_9146036
|
0.560
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr4_+_78172966
|
0.559
|
|
SEPT11
|
septin 11
|
|
chr14_+_95792299
|
0.556
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr5_+_35892747
|
0.555
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr9_+_109085643
|
0.549
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr12_+_6768934
|
0.547
|
NM_001195014
NM_001195015
NM_001195016
NM_001195017
|
CD4
|
CD4 molecule
|
|
chr11_+_46359821
|
0.546
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr12_+_6768866
|
0.546
|
NM_000616
|
CD4
|
CD4 molecule
|
|
chrX_+_135079120
|
0.545
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr12_-_9123126
|
0.544
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr10_+_124310170
|
0.541
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr5_+_125828685
|
0.536
|
NM_001146322
|
GRAMD3
|
GRAM domain containing 3
|
|
chr3_+_69895651
|
0.535
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chrX_-_37916138
|
0.532
|
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr1_-_92124201
|
0.531
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr6_+_121798443
|
0.520
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr17_-_64462976
|
0.519
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr3_-_15814678
|
0.519
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr7_+_134226690
|
0.518
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chr2_-_158439868
|
0.518
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr16_+_6009095
|
0.517
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr1_-_199608941
|
0.517
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr17_-_10362583
|
0.509
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr11_-_27679617
|
0.507
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr7_-_76667045
|
0.507
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr3_-_167037946
|
0.506
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr10_-_69422391
|
0.503
|
|
HERC4
|
hect domain and RLD 4
|
|
chrX_-_135690699
|
0.502
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_+_13240987
|
0.500
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr11_+_46359855
|
0.499
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr17_+_37222731
|
0.491
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr1_+_144152864
|
0.490
|
|
TXNIP
|
thioredoxin interacting protein
|
|
chr15_+_65217412
|
0.490
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|
|
chr2_-_105421661
|
0.488
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr1_-_171441234
|
0.488
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr12_-_24993260
|
0.488
|
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr13_-_109600709
|
0.488
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr14_-_73250094
|
0.483
|
NM_006029
|
PNMA1
|
paraneoplastic antigen MA1
|
|
chr2_-_192420168
|
0.483
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr17_+_61729306
|
0.483
|
NM_002737
|
PRKCA
|
protein kinase C, alpha
|
|
chr3_+_161040439
|
0.483
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_-_24993190
|
0.483
|
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|