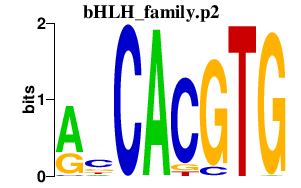
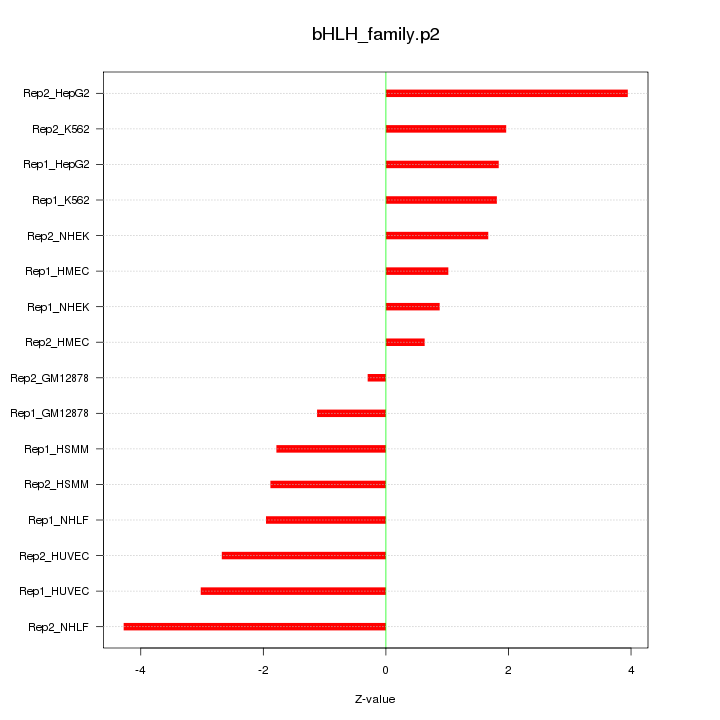
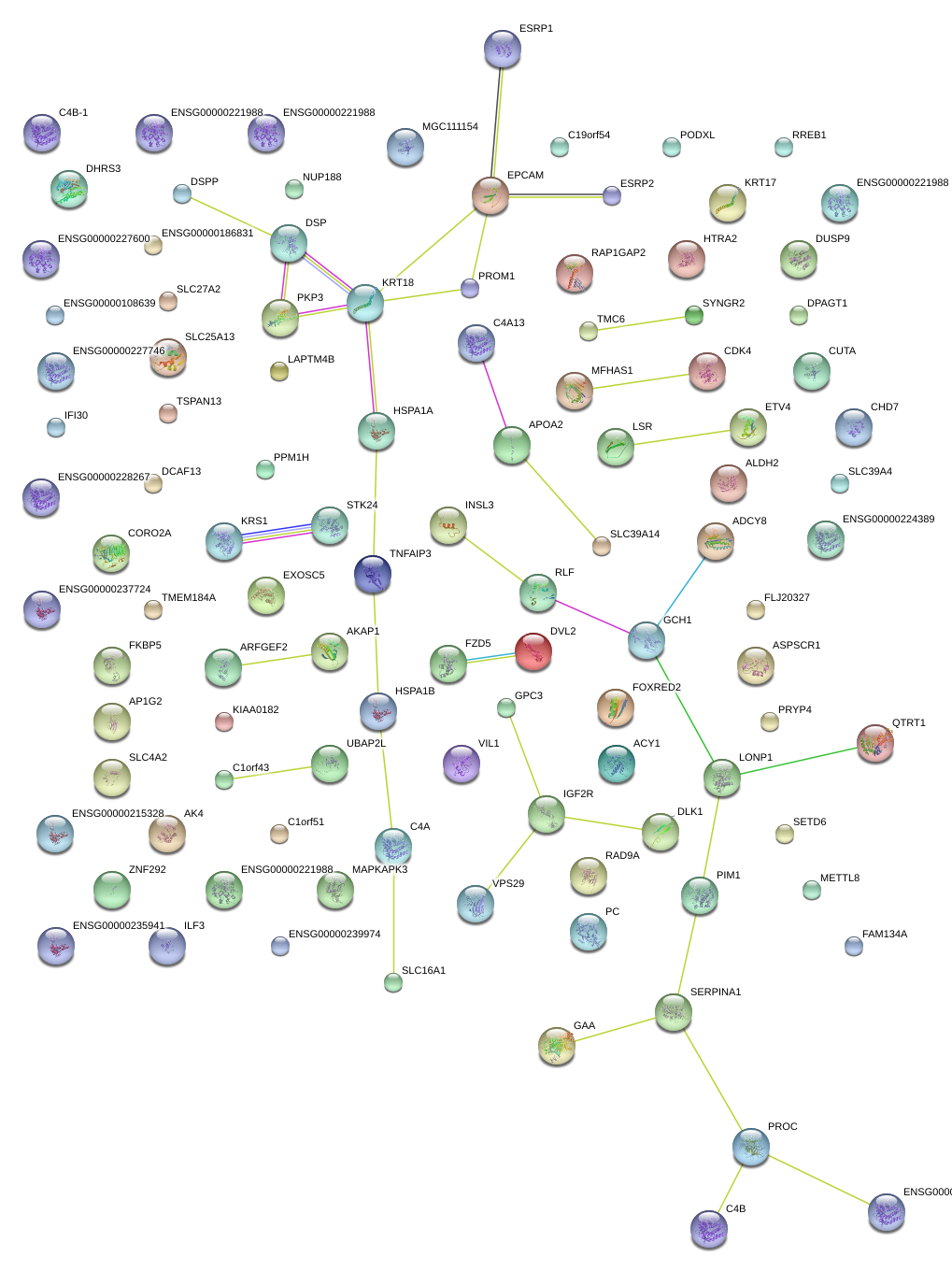
|
chr12_-_61615124
|
4.332
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr12_-_61614930
|
3.751
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chrX_+_152562013
|
3.659
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr1_-_113300138
|
3.285
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr6_+_160310118
|
3.261
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr1_-_159459999
|
3.253
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chrX_+_130985316
|
3.185
|
NM_001042452
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr19_+_10625987
|
3.042
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr6_+_37245860
|
3.035
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr19_+_10626106
|
2.936
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr19_-_10625449
|
2.893
|
|
LOC147727
|
hypothetical LOC147727
|
|
chr2_-_208342296
|
2.855
|
NM_003468
|
FZD5
|
frizzled homolog 5 (Drosophila)
|
|
chr17_-_73636362
|
2.722
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr14_-_54438913
|
2.632
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr16_-_66827449
|
2.628
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chrX_-_132947141
|
2.594
|
|
GPC3
|
glypican 3
|
|
chr2_+_218992053
|
2.536
|
NM_007127
|
VIL1
|
villin 1
|
|
chr7_+_16759845
|
2.528
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr14_-_93919305
|
2.484
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr17_+_77529070
|
2.475
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chrX_-_132947290
|
2.443
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr11_-_66482366
|
2.362
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr19_+_40431122
|
2.337
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr19_+_10625896
|
2.288
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr1_-_152459667
|
2.270
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr1_-_113300351
|
2.264
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr12_-_56432292
|
2.236
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr4_-_15694407
|
2.211
|
|
PROM1
|
prominin 1
|
|
chr3_+_50629604
|
2.180
|
NM_004635
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr8_-_145609737
|
2.111
|
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr1_-_152459506
|
2.075
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chrX_+_130984912
|
2.058
|
NM_001042453
NM_016542
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr15_+_48261649
|
2.056
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr14_+_100270222
|
2.054
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr17_+_52518098
|
2.053
|
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr12_-_56432377
|
2.021
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr6_+_32090516
|
1.978
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr17_+_73676219
|
1.976
|
NM_004710
|
SYNGR2
|
synaptogyrin 2
|
|
chr3_+_51992339
|
1.938
|
NM_000666
NM_001198895
NM_001198896
NM_001198897
NM_001198898
|
ACY1
|
aminoacylase 1
|
|
chr12_-_56432385
|
1.935
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr17_+_77528782
|
1.916
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr11_+_384199
|
1.903
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr6_-_33493792
|
1.851
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr12_-_56432375
|
1.825
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr11_+_66915997
|
1.818
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr8_-_8788149
|
1.813
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr9_-_99994704
|
1.809
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr4_-_15694691
|
1.789
|
NM_001145847
NM_001145848
|
PROM1
|
prominin 1
|
|
chr19_-_46595053
|
1.767
|
NM_020158
|
EXOSC5
|
exosome component 5
|
|
chr1_+_40399618
|
1.757
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr16_+_84204424
|
1.750
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr8_+_95722448
|
1.744
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr14_-_23106372
|
1.724
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr19_+_40431359
|
1.723
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr1_-_159459814
|
1.713
|
|
APOA2
|
apolipoprotein A-II
|
|
chr6_-_35804337
|
1.706
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr1_+_65385799
|
1.700
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr1_+_148521852
|
1.688
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr1_+_40399642
|
1.685
|
|
RLF
|
rearranged L-myc fusion
|
|
chr6_+_87921980
|
1.638
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr17_+_77528699
|
1.635
|
NM_024083
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr1_-_12599934
|
1.576
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr2_-_171999473
|
1.574
|
NM_024770
|
METTL8
|
methyltransferase like 8
|
|
chr12_-_109424260
|
1.570
|
NM_016226
NM_057180
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae)
|
|
chr17_-_38979178
|
1.561
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr1_+_152459918
|
1.555
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr17_-_7078446
|
1.539
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr17_+_75689985
|
1.539
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr19_+_10673105
|
1.535
|
NM_031209
|
QTRT1
|
queuine tRNA-ribosyltransferase 1
|
|
chr11_-_118477739
|
1.514
|
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase)
|
|
chr3_+_50629593
|
1.503
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr19_+_18145578
|
1.499
|
NM_006332
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr22_-_35233032
|
1.495
|
NM_024955
NM_001102371
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr20_+_46971618
|
1.489
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr9_+_130749791
|
1.479
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr12_+_51628921
|
1.465
|
NM_199187
NM_000224
|
KRT18
|
keratin 18
|
|
chr11_+_384238
|
1.460
|
|
PKP3
|
plakophilin 3
|
|
chr6_+_31891509
|
1.459
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr2_+_74610039
|
1.452
|
NM_013247
NM_145074
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr7_-_1562272
|
1.452
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr2_+_47449790
|
1.442
|
NM_002354
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr16_+_57106883
|
1.426
|
NM_001160305
NM_024860
|
SETD6
|
SET domain containing 6
|
|
chr2_+_219751230
|
1.405
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr19_-_45948211
|
1.402
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr8_+_22280942
|
1.400
|
NM_001135153
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr17_-_7078399
|
1.352
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr6_+_7486862
|
1.352
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr19_+_40431618
|
1.351
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr6_+_138230045
|
1.343
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr8_+_22280703
|
1.338
|
NM_001128431
NM_001135154
NM_015359
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr7_-_95789204
|
1.332
|
NM_001160210
NM_014251
|
SLC25A13
|
solute carrier family 25, member 13 (citrin)
|
|
chr8_+_98857190
|
1.326
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr6_+_7486836
|
1.316
|
|
DSP
|
desmoplakin
|
|
chr6_+_7052985
|
1.316
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr8_+_22280731
|
1.311
|
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr2_+_219751192
|
1.309
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr6_-_33493898
|
1.302
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr19_+_18145619
|
1.297
|
|
IFI30
|
interferon, gamma-inducible protein 30
|
|
chr7_+_150390566
|
1.296
|
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr8_+_61753793
|
1.290
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr17_+_2627099
|
1.280
|
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr6_+_32230172
|
1.273
|
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr17_-_73636386
|
1.272
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr6_+_32229771
|
1.271
|
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr17_+_46692908
|
1.267
|
|
UTP18
|
UTP18, small subunit (SSU) processome component, homolog (yeast)
|
|
chr1_+_1971762
|
1.266
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr6_-_47385313
|
1.266
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr2_+_169021039
|
1.262
|
NM_203463
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr4_+_74491272
|
1.261
|
|
ALB
|
albumin
|
|
chr9_+_696744
|
1.258
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr11_+_118463906
|
1.257
|
NM_001024382
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr6_+_31891293
|
1.254
|
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr6_-_53517466
|
1.251
|
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr3_-_4483939
|
1.237
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr1_-_27113018
|
1.235
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr17_+_70256397
|
1.231
|
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr16_-_2037819
|
1.228
|
NM_002528
|
NTHL1
|
nth endonuclease III-like 1 (E. coli)
|
|
chr1_-_21868380
|
1.227
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr7_-_23476497
|
1.227
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr17_-_7078304
|
1.224
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr19_+_7608010
|
1.220
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr8_+_95722516
|
1.219
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr4_-_56996400
|
1.218
|
NM_002703
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase
|
|
chr17_-_2561676
|
1.215
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr6_-_47385205
|
1.211
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr16_+_67236699
|
1.210
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr2_+_215885063
|
1.208
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr14_-_23727826
|
1.199
|
|
IPO4
|
importin 4
|
|
chr12_-_101835026
|
1.189
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr1_-_159257567
|
1.187
|
|
F11R
|
F11 receptor
|
|
chr11_+_120828058
|
1.181
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr10_+_69314347
|
1.173
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr2_+_215885053
|
1.169
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr8_+_95722575
|
1.166
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr19_-_38485269
|
1.165
|
NM_004364
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr9_+_130749812
|
1.161
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr19_+_7608015
|
1.160
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr12_+_51629164
|
1.158
|
|
KRT18
|
keratin 18
|
|
chrX_-_100549561
|
1.155
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr5_-_131160541
|
1.151
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr7_-_103417198
|
1.140
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr2_+_17585462
|
1.139
|
|
VSNL1
|
visinin-like 1
|
|
chr1_-_27689129
|
1.135
|
|
WASF2
|
WAS protein family, member 2
|
|
chr6_-_47385605
|
1.133
|
NM_014452
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr19_+_7607976
|
1.133
|
NM_001127396
NM_006949
|
STXBP2
|
syntaxin binding protein 2
|
|
chr2_+_17585287
|
1.129
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr12_-_101835183
|
1.129
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr17_-_31964723
|
1.128
|
|
MYO19
|
myosin XIX
|
|
chr6_+_32057779
|
1.128
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr2_+_215884923
|
1.126
|
NM_004044
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr16_+_29726962
|
1.124
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr17_+_70256389
|
1.120
|
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr20_+_60744231
|
1.117
|
NM_016354
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr11_-_118477994
|
1.114
|
NM_001382
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase)
|
|
chr11_-_107234733
|
1.112
|
|
SLC35F2
|
solute carrier family 35, member F2
|
|
chr6_+_7486891
|
1.110
|
|
DSP
|
desmoplakin
|
|
chr3_-_12983959
|
1.108
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr1_-_159257486
|
1.102
|
|
F11R
|
F11 receptor
|
|
chr11_-_118757477
|
1.098
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr12_-_121316907
|
1.095
|
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr16_-_28410580
|
1.090
|
NM_000086
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr17_+_70256341
|
1.071
|
NM_004252
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr6_-_47385470
|
1.068
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr12_-_121316997
|
1.067
|
NM_022916
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr17_-_44058612
|
1.065
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr22_-_35232979
|
1.059
|
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr17_-_77478835
|
1.058
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr20_+_60744268
|
1.056
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr9_-_132444639
|
1.051
|
|
LOC100272217
|
hypothetical LOC100272217
|
|
chr5_-_133734616
|
1.050
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr14_-_23727850
|
1.049
|
NM_024658
|
IPO4
|
importin 4
|
|
chr5_+_149717489
|
1.049
|
|
TCOF1
|
Treacher Collins-Franceschetti syndrome 1
|
|
chr1_+_11255831
|
1.048
|
NM_013319
|
UBIAD1
|
UbiA prenyltransferase domain containing 1
|
|
chr16_+_67236730
|
1.048
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr2_-_172675480
|
1.048
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr17_-_32730804
|
1.044
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr3_+_53855603
|
1.041
|
NM_018725
|
IL17RB
|
interleukin 17 receptor B
|
|
chr6_-_33494042
|
1.040
|
NM_001014433
NM_001014837
NM_001014838
NM_001014840
NM_015921
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr2_-_219750975
|
1.031
|
|
C2orf24
|
chromosome 2 open reading frame 24
|
|
chrX_+_152608159
|
1.028
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr1_+_44213229
|
1.028
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chrX_-_131401319
|
1.027
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chrX_-_100549444
|
1.016
|
|
GLA
|
galactosidase, alpha
|
|
chr1_+_116717552
|
1.009
|
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chrX_-_100490329
|
1.009
|
NM_001145951
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr1_+_10015577
|
1.002
|
NM_001105562
NM_006048
|
UBE4B
|
ubiquitination factor E4B (UFD2 homolog, yeast)
|
|
chr1_+_44213200
|
0.998
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr20_+_47986342
|
0.996
|
|
RNF114
|
ring finger protein 114
|
|
chr6_-_43305156
|
0.996
|
NM_006443
NM_199184
|
C6orf108
|
chromosome 6 open reading frame 108
|
|
chr20_+_37024383
|
0.995
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr2_+_28971007
|
0.993
|
NM_015131
|
WDR43
|
WD repeat domain 43
|
|
chrX_+_105823723
|
0.991
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr2_-_220116508
|
0.985
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr11_+_34083784
|
0.984
|
|
NAT10
|
N-acetyltransferase 10 (GCN5-related)
|
|
chr16_-_1465007
|
0.983
|
|
CLCN7
|
chloride channel 7
|
|
chr3_+_185515013
|
0.983
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr6_-_35996915
|
0.983
|
NM_003137
|
SRPK1
|
SRSF protein kinase 1
|
|
chr17_+_70256370
|
0.983
|
|
SLC9A3R1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1
|
|
chr7_-_99536966
|
0.980
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr22_-_17798998
|
0.979
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|