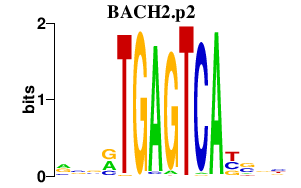
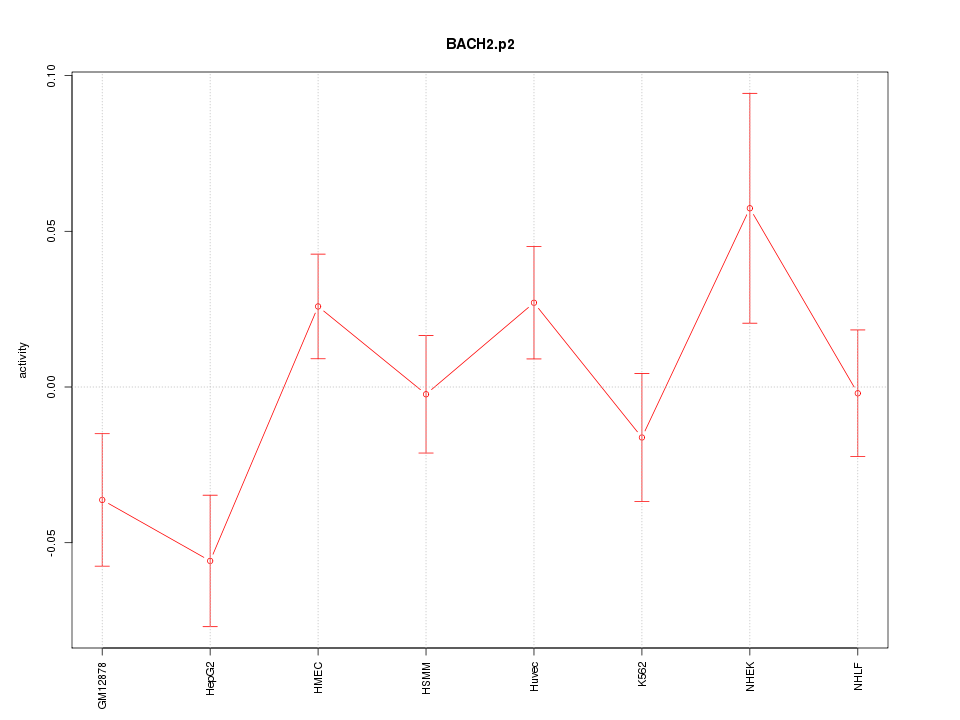
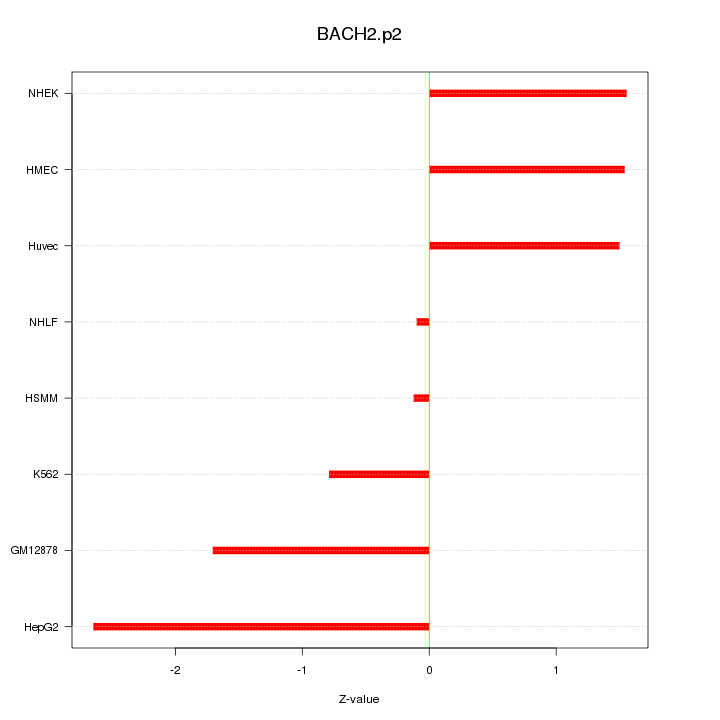
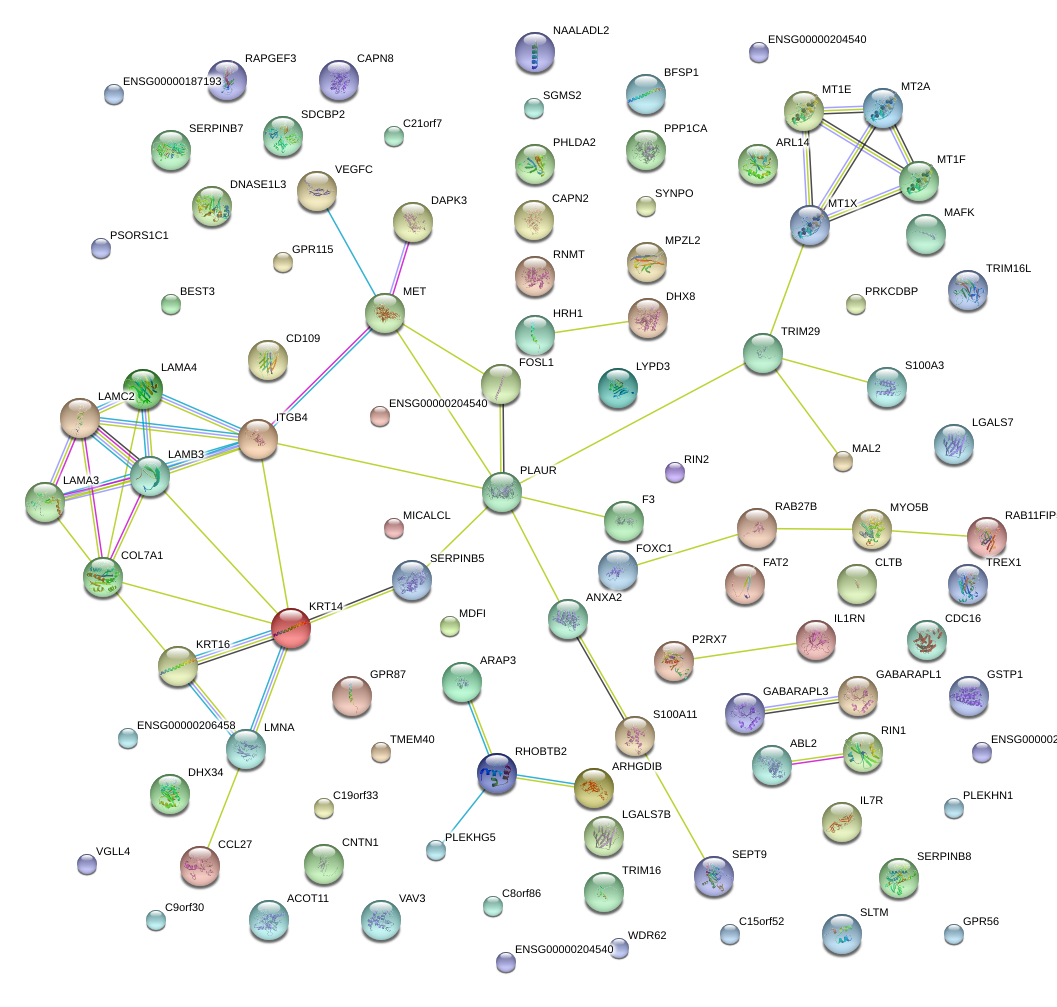
|
chr18_+_19706979
|
7.057
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr1_+_221955876
|
6.456
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr3_+_11153778
|
5.225
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr10_-_88720908
|
4.549
|
|
LOC100133190
|
hypothetical LOC100133190
|
|
chr7_+_116099648
|
4.487
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr4_+_108965167
|
4.137
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr11_-_6298284
|
3.975
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr1_-_6474346
|
3.753
|
NM_020631
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr19_+_43486640
|
3.735
|
NM_033520
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr1_-_177378801
|
3.671
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr12_-_68369322
|
3.606
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr14_+_104218487
|
3.579
|
|
|
|
|
chr2_+_113591940
|
3.574
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr6_+_31190505
|
3.573
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr6_+_74462226
|
3.550
|
NM_001159587
NM_001159588
NM_133493
|
CD109
|
CD109 molecule
|
|
chr1_-_207891264
|
3.418
|
NM_001017402
|
LAMB3
|
laminin, beta 3
|
|
chr21_+_29439768
|
3.411
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr8_-_38505331
|
3.194
|
NM_207412
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr17_-_36996611
|
3.180
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr6_+_74462654
|
3.164
|
|
CD109
|
CD109 molecule
|
|
chr6_+_74462630
|
3.155
|
|
CD109
|
CD109 molecule
|
|
chr6_+_74462485
|
3.149
|
|
CD109
|
CD109 molecule
|
|
chr9_+_102231616
|
3.003
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr6_+_74462577
|
2.975
|
|
CD109
|
CD109 molecule
|
|
chr9_+_676640
|
2.962
|
|
|
|
|
chr3_-_11585264
|
2.908
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr11_+_12265022
|
2.810
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr8_+_22900863
|
2.741
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr18_+_59593591
|
2.718
|
NM_003784
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr16_+_56219802
|
2.712
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr11_-_119499188
|
2.672
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr17_+_18566060
|
2.638
|
NM_001037330
|
TRIM16L
|
tripartite motif containing 16-like
|
|
chr11_-_2907169
|
2.594
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chr3_-_48607596
|
2.587
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr11_-_117640166
|
2.499
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr5_-_141041982
|
2.486
|
NM_022481
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr15_-_58477407
|
2.467
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr3_-_152517204
|
2.456
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr5_-_141041933
|
2.444
|
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr5_+_150000394
|
2.417
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr11_-_117640078
|
2.385
|
|
|
|
|
chr11_-_117640407
|
2.343
|
NM_005797
NM_144765
|
MPZL2
|
myelin protein zero-like 2
|
|
chr1_+_181421984
|
2.323
|
|
LAMC2
|
laminin, gamma 2
|
|
chr3_+_175641340
|
2.305
|
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr3_+_161877640
|
2.218
|
NM_025047
|
ARL14
|
ADP-ribosylation factor-like 14
|
|
chr11_-_117640196
|
2.206
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr1_-_36589744
|
2.200
|
|
|
|
|
chr1_+_221966951
|
2.172
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr1_-_108032645
|
2.171
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr11_-_65860450
|
2.170
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr1_+_154351162
|
2.151
|
|
LMNA
|
lamin A/C
|
|
chr1_+_154351082
|
2.121
|
NM_005572
NM_170707
NM_170708
|
LMNA
|
lamin A/C
|
|
chr1_-_150276114
|
2.108
|
NM_005620
|
S100A11
|
S100 calcium binding protein A11
|
|
chr18_+_59788242
|
2.070
|
NM_001031848
NM_002640
NM_198833
|
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8
|
|
chr19_-_48866111
|
2.065
|
NM_001005376
NM_001005377
NM_002659
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr18_+_50646616
|
2.047
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr17_-_37022465
|
2.028
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr19_-_3920736
|
1.998
|
NM_001348
|
DAPK3
|
death-associated protein kinase 3
|
|
chr6_+_41712907
|
1.996
|
|
MDFI
|
MyoD family inhibitor
|
|
chr6_+_1556543
|
1.988
|
|
FOXC1
|
forkhead box C1
|
|
chr12_-_15005761
|
1.968
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr3_-_12775718
|
1.955
|
NM_018306
|
TMEM40
|
transmembrane protein 40
|
|
chr18_+_59295123
|
1.938
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr7_+_1543644
|
1.856
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr1_+_891739
|
1.789
|
NM_001160184
NM_032129
|
PLEKHN1
|
pleckstrin homology domain containing, family N member 1
|
|
chr12_+_10256682
|
1.761
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr4_-_177950658
|
1.759
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr6_+_47774247
|
1.758
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr1_+_181422016
|
1.746
|
|
LAMC2
|
laminin, gamma 2
|
|
chr19_-_48661563
|
1.738
|
NM_014400
|
LYPD3
|
LY6/PLAUR domain containing 3
|
|
chr15_-_38417564
|
1.736
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr20_-_17487480
|
1.726
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr1_-_94779727
|
1.685
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr17_+_71229110
|
1.682
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr18_-_45630154
|
1.671
|
|
MYO5B
|
myosin VB
|
|
chr1_+_221966835
|
1.620
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr11_+_67107641
|
1.614
|
NM_000852
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr20_-_1257809
|
1.594
|
NM_080489
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr1_+_221966684
|
1.594
|
NM_001748
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr11_+_67107852
|
1.586
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr1_+_221966849
|
1.578
|
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr20_-_1257782
|
1.562
|
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr1_-_151788341
|
1.562
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr20_+_19903778
|
1.530
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr17_+_71229151
|
1.519
|
|
ITGB4
|
integrin, beta 4
|
|
chr16_+_55199978
|
1.509
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr9_-_34652629
|
1.463
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr12_-_46439127
|
1.463
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr1_+_54780515
|
1.454
|
|
ACOT11
|
acyl-CoA thioesterase 11
|
|
chr3_-_58175437
|
1.421
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr19_+_43971689
|
1.417
|
NM_001042507
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B
|
|
chr3_+_48482213
|
1.416
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr2_-_73170290
|
1.416
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr5_+_35892747
|
1.414
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr8_+_120289735
|
1.409
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr5_-_175776137
|
1.401
|
NM_001834
NM_007097
|
CLTB
|
clathrin, light chain B
|
|
chr1_+_154351200
|
1.386
|
|
LMNA
|
lamin A/C
|
|
chr11_-_65424434
|
1.372
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr12_-_46438447
|
1.361
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr1_-_221919982
|
1.359
|
|
CAPN8
|
calpain 8
|
|
chr11_-_33701073
|
1.354
|
NM_001127227
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr7_-_158304062
|
1.327
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr11_-_65424385
|
1.327
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr1_-_221920023
|
1.317
|
NM_001143962
|
CAPN8
|
calpain 8
|
|
chr3_+_48482625
|
1.302
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr19_-_43955984
|
1.291
|
NM_002307
|
LGALS7
LGALS7B
|
lectin, galactoside-binding, soluble, 7
lectin, galactoside-binding, soluble, 7B
|
|
chr15_+_38849450
|
1.291
|
NM_001130448
|
C15orf62
|
chromosome 15 open reading frame 62
|
|
chr1_+_181421796
|
1.287
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr11_+_1812115
|
1.285
|
NM_138567
|
SYT8
|
synaptotagmin VIII
|
|
chr2_-_85494639
|
1.282
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr12_-_10767115
|
1.263
|
NM_001145426
NM_003651
|
CSDA
|
cold shock domain protein A
|
|
chr3_-_123995216
|
1.252
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr9_-_129381086
|
1.249
|
NM_001035534
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr18_+_59405513
|
1.246
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr1_-_17180659
|
1.238
|
NM_001135247
NM_017459
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr18_+_54039818
|
1.222
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_-_84197370
|
1.220
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr17_+_71229191
|
1.203
|
|
ITGB4
|
integrin, beta 4
|
|
chr6_-_84197477
|
1.201
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr1_+_27062215
|
1.195
|
NM_006142
|
SFN
|
stratifin
|
|
chr12_-_105057940
|
1.188
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr6_-_84197463
|
1.181
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr3_+_11242666
|
1.148
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr18_+_54039779
|
1.144
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_-_155374756
|
1.124
|
|
ETV3
|
ets variant 3
|
|
chr22_-_35914200
|
1.124
|
NM_031910
NM_182486
|
C1QTNF6
|
C1q and tumor necrosis factor related protein 6
|
|
chr6_-_159159186
|
1.124
|
|
EZR
|
ezrin
|
|
chr6_-_84197573
|
1.121
|
NM_002395
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr2_+_27705018
|
1.108
|
NM_001145047
NM_001145048
NM_001145049
|
GPN1
|
GPN-loop GTPase 1
|
|
chrX_-_15198093
|
1.102
|
NM_001031739
NM_001168530
NM_024087
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr16_-_62573
|
1.087
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr17_-_3407780
|
1.076
|
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr6_-_159159242
|
1.075
|
|
EZR
|
ezrin
|
|
chr4_+_76700281
|
1.072
|
NM_178497
|
C4orf26
|
chromosome 4 open reading frame 26
|
|
chrX_+_12903145
|
1.069
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr3_-_51912376
|
1.064
|
NM_152397
|
IQCF1
|
IQ motif containing F1
|
|
chr11_+_384238
|
1.061
|
|
PKP3
|
plakophilin 3
|
|
chr5_-_175775966
|
1.042
|
|
CLTB
|
clathrin, light chain B
|
|
chr11_-_65424436
|
1.039
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr3_+_69216747
|
1.033
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr18_+_54039730
|
1.032
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr3_+_185499679
|
1.031
|
NM_002808
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2
|
|
chr4_+_74825138
|
1.029
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr6_-_138470160
|
1.021
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr5_+_145297490
|
1.016
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr11_+_10433241
|
1.016
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr12_-_56445653
|
0.998
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr11_-_66898223
|
0.990
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr9_-_90456824
|
0.989
|
NM_001100111
|
LOC286238
|
hypothetical protein LOC286238
|
|
chr11_-_47502115
|
0.985
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr6_-_159159257
|
0.980
|
NM_003379
|
EZR
|
ezrin
|
|
chr11_-_92570626
|
0.952
|
|
|
|
|
chr17_-_24068759
|
0.947
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr17_-_36792161
|
0.944
|
NM_021013
|
KRT34
|
keratin 34
|
|
chr6_-_112033775
|
0.938
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr17_+_7196151
|
0.937
|
|
KCTD11
|
potassium channel tetramerisation domain containing 11
|
|
chr19_-_60573552
|
0.930
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr5_+_40877053
|
0.909
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr16_+_66121040
|
0.905
|
NM_001193523
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr4_+_152549776
|
0.899
|
NM_001109977
|
FAM160A1
|
family with sequence similarity 160, member A1
|
|
chr11_-_92570693
|
0.895
|
NM_152313
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4
|
|
chr3_+_185499732
|
0.890
|
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2
|
|
chr19_-_56215084
|
0.881
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr17_-_18102767
|
0.878
|
NM_002018
|
FLII
|
flightless I homolog (Drosophila)
|
|
chr3_+_64645819
|
0.878
|
|
LOC100507098
|
hypothetical LOC100507098
|
|
chr17_+_72827227
|
0.876
|
|
SEPT9
|
septin 9
|
|
chr21_-_35343065
|
0.876
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr16_-_2848106
|
0.873
|
NM_022119
|
PRSS22
|
protease, serine, 22
|
|
chr17_+_7195931
|
0.871
|
NM_001002914
|
KCTD11
|
potassium channel tetramerisation domain containing 11
|
|
chr6_+_107066422
|
0.871
|
NM_001624
|
AIM1
|
absent in melanoma 1
|
|
chr17_-_24069299
|
0.857
|
NM_001142624
NM_001142625
NM_001144943
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr17_-_7064092
|
0.855
|
NM_001365
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr7_-_1465487
|
0.853
|
|
MICALL2
|
MICAL-like 2
|
|
chr7_+_48094750
|
0.840
|
NM_181597
|
UPP1
|
uridine phosphorylase 1
|
|
chr3_-_48576204
|
0.838
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr12_+_55018925
|
0.837
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr17_-_18102594
|
0.826
|
|
FLII
|
flightless I homolog (Drosophila)
|
|
chr11_-_102174102
|
0.826
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr6_-_35804337
|
0.821
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chrX_+_99785818
|
0.818
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr17_+_77261457
|
0.817
|
|
|
|
|
chr1_-_155374793
|
0.814
|
NM_005240
NM_001145312
|
ETV3
|
ets variant 3
|
|
chr6_-_112033752
|
0.809
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr11_+_384199
|
0.806
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr3_-_197549640
|
0.799
|
NM_138461
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr1_+_151270301
|
0.799
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr19_+_46416975
|
0.795
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr17_+_72827185
|
0.789
|
NM_006640
|
SEPT9
|
septin 9
|
|
chr17_+_77261447
|
0.789
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr12_-_10766965
|
0.782
|
|
CSDA
|
cold shock domain protein A
|
|
chr1_+_152644668
|
0.768
|
|
IL6R
|
interleukin 6 receptor
|
|
chr2_+_219852268
|
0.752
|
NM_001039550
NM_006736
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr19_+_46416971
|
0.749
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr17_-_18102743
|
0.748
|
|
FLII
|
flightless I homolog (Drosophila)
|
|
chr5_+_147562531
|
0.747
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr11_-_102174032
|
0.744
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr17_+_77261477
|
0.737
|
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chrX_-_129072152
|
0.734
|
NM_001421
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr7_+_39739690
|
0.723
|
|
NCRNA00265
|
non-protein coding RNA 265
|
|
chr17_-_7434113
|
0.719
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|