|
chr1_-_153098811
|
6.002
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr7_+_50318801
|
5.567
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chrX_-_48661226
|
5.458
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr16_+_23754997
|
4.922
|
|
PRKCB
|
protein kinase C, beta
|
|
chr6_-_33016561
|
4.912
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr16_+_23754860
|
4.061
|
|
PRKCB
|
protein kinase C, beta
|
|
chr5_-_176868483
|
3.983
|
|
DOK3
|
docking protein 3
|
|
chr10_-_98021144
|
3.968
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr11_+_63829616
|
3.875
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr3_+_99733432
|
3.772
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
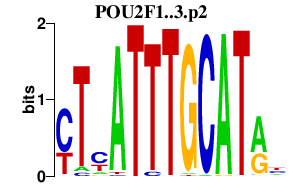
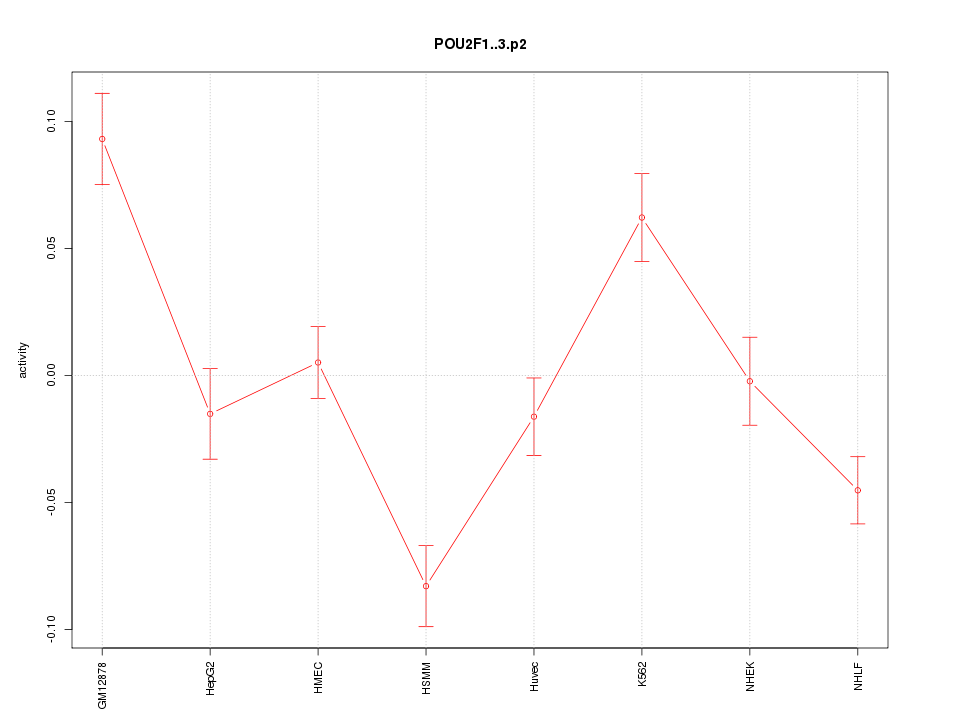
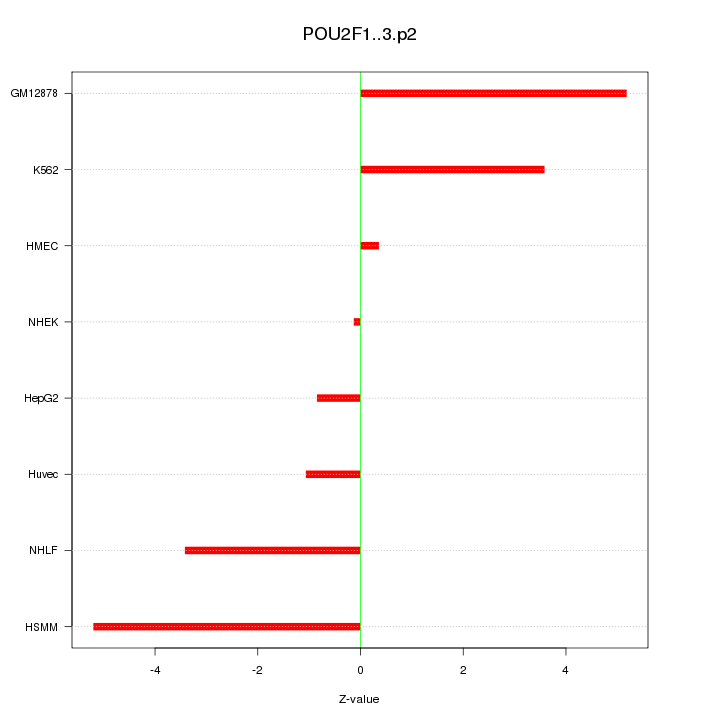
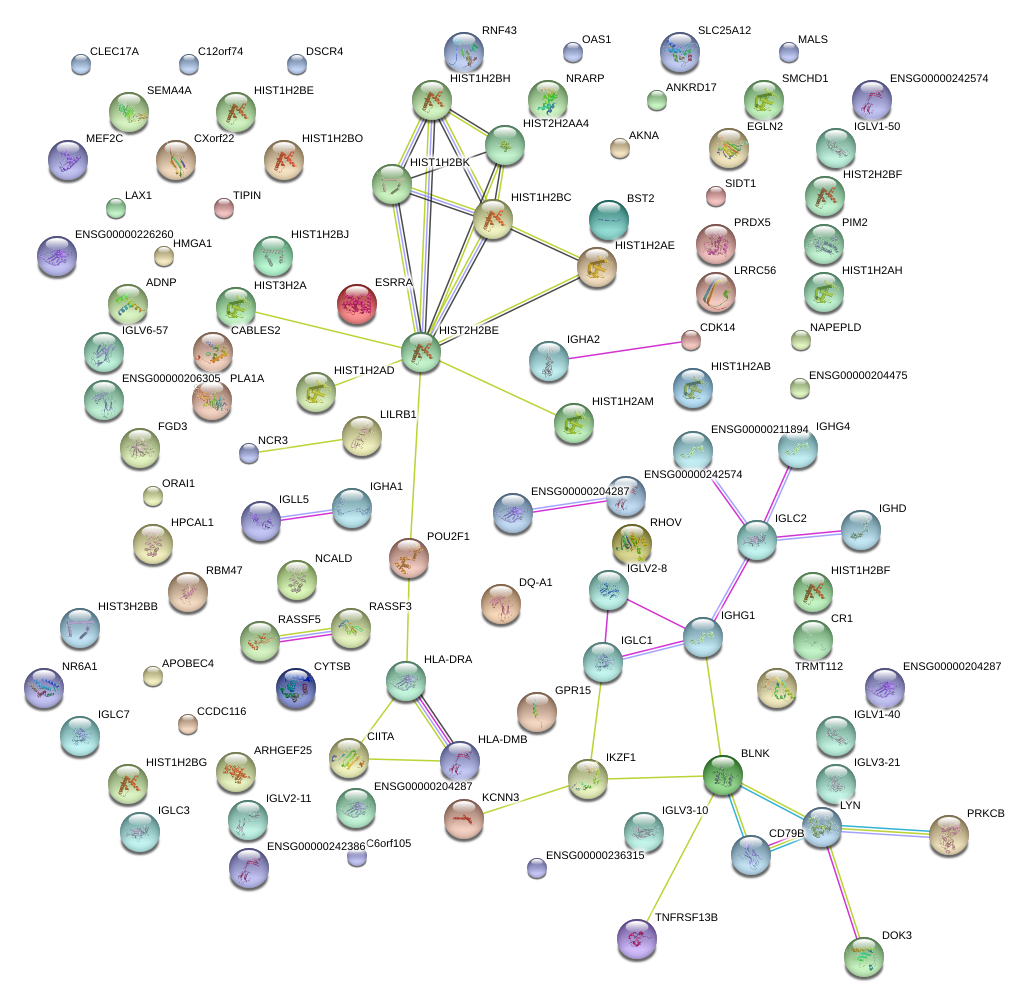
chr1_+_165565006
|
3.743
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_+_165564904
|
3.615
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr16_+_23754822
|
3.586
|
|
PRKCB
|
protein kinase C, beta
|
|
chr4_-_40212670
|
3.585
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr6_+_26325124
|
3.572
|
NM_021052
|
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr1_+_226712430
|
3.463
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr1_+_202000906
|
3.461
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr1_-_226712140
|
3.375
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr16_+_23754772
|
3.254
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr1_+_154386358
|
3.253
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr6_+_26359805
|
3.231
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr5_-_88155360
|
3.227
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr19_-_17377383
|
3.101
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr22_+_21370394
|
2.944
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr19_+_17377502
|
2.927
|
|
LOC100507535
|
hypothetical LOC100507535
|
|
chr1_+_204797079
|
2.902
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr17_-_59363352
|
2.899
|
NM_000626
NM_001039933
NM_021602
|
CD79B
|
CD79b molecule, immunoglobulin-associated beta
|
|
chr12_+_91620749
|
2.887
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr22_+_20715340
|
2.832
|
|
|
|
|
chr8_+_56954946
|
2.812
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr8_+_56954899
|
2.796
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr7_+_26404711
|
2.720
|
|
LOC441204
|
hypothetical locus LOC441204
|
|
chr8_+_56954953
|
2.720
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr17_-_16816113
|
2.692
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr6_-_11887265
|
2.602
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr6_+_27222839
|
2.576
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr6_+_34312906
|
2.555
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_-_27968906
|
2.545
|
NM_003514
|
HIST1H2AM
|
histone cluster 1, H2am
|
|
chr6_-_27222556
|
2.480
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr1_-_181889070
|
2.479
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr11_+_527521
|
2.478
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr3_+_120799411
|
2.441
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr20_-_60415729
|
2.440
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr1_+_204797120
|
2.414
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr6_+_34312979
|
2.350
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr3_+_114733907
|
2.300
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr6_-_26324850
|
2.293
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr6_+_34312667
|
2.221
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_+_27969181
|
2.220
|
NM_003527
|
HIST1H2BO
|
histone cluster 1, H2bo
|
|
chr9_-_139316523
|
2.157
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr6_+_34313007
|
2.132
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr19_+_14554895
|
2.099
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr6_+_34312641
|
2.059
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_-_26141774
|
1.997
|
NM_003513
|
HIST1H2AB
|
histone cluster 1, H2ab
|
|
chr9_-_116200569
|
1.928
|
|
AKNA
|
AT-hook transcription factor
|
|
chr7_+_90176647
|
1.909
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr6_+_26291936
|
1.882
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr17_-_53849888
|
1.881
|
|
RNF43
|
ring finger protein 43
|
|
chr15_-_38953628
|
1.880
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr2_-_172458970
|
1.822
|
NM_003705
|
SLC25A12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12
|
|
chr4_-_74307637
|
1.790
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr9_+_94766379
|
1.788
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr6_+_34312633
|
1.751
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr16_+_10878539
|
1.736
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr21_-_38415273
|
1.728
|
NM_005867
|
DSCR4
|
Down syndrome critical region gene 4
|
|
chr17_-_53849929
|
1.721
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr22_+_20316968
|
1.716
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr12_+_111829191
|
1.703
|
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr19_+_59820413
|
1.694
|
NM_006669
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1
|
|
chr11_+_63842178
|
1.676
|
|
PRDX5
|
peroxiredoxin 5
|
|
chr11_-_63841556
|
1.643
|
NM_016404
|
TRMT112
|
tRNA methyltransferase 11-2 homolog (S. cerevisiae)
|
|
chr9_-_126573349
|
1.641
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr19_+_45996881
|
1.603
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr6_+_34312625
|
1.593
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr12_+_56290229
|
1.578
|
NM_001111270
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chr12_+_111829121
|
1.570
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr17_+_19999784
|
1.565
|
NM_001033554
NM_001033555
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr11_+_63842135
|
1.558
|
NM_012094
NM_181651
NM_181652
|
PRDX5
|
peroxiredoxin 5
|
|
chr15_-_64436045
|
1.557
|
NM_017858
|
TIPIN
|
TIMELESS interacting protein
|
|
chr20_-_48980858
|
1.546
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr14_-_105623358
|
1.545
|
|
IGHA1
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr22_+_20880159
|
1.537
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr6_-_31668476
|
1.530
|
NM_001145466
NM_001145467
NM_147130
|
NCR3
|
natural cytotoxicity triggering receptor 3
|
|
chr22_+_21495273
|
1.522
|
|
|
|
|
chr12_+_120548831
|
1.521
|
NM_032790
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1
|
|
chr7_-_102576570
|
1.516
|
NM_001122838
NM_198990
|
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D
|
|
chr1_+_205736095
|
1.511
|
NM_000573
NM_000651
|
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group)
|
|
chr6_-_27208507
|
1.478
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr6_+_32515596
|
1.474
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr14_-_106064887
|
1.469
|
|
LOC100126583
IGHV3-48
IGHA1
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 3-48
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr4_-_186970366
|
1.468
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr12_-_31370387
|
1.454
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr10_+_124897627
|
1.439
|
NM_005519
|
HMX2
|
H6 family homeobox 2
|
|
chr8_+_86287161
|
1.436
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr6_-_26232109
|
1.420
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr6_+_43247168
|
1.410
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr12_-_31370351
|
1.375
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr6_+_34312554
|
1.373
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr2_+_96790298
|
1.365
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr6_+_26381122
|
1.356
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr22_+_20880112
|
1.355
|
|
|
|
|
chr6_+_43247179
|
1.348
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr6_+_27208763
|
1.335
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr6_+_26266311
|
1.304
|
NM_021063
NM_138720
|
HIST1H2BD
|
histone cluster 1, H2bd
|
|
chr6_+_26232351
|
1.291
|
NM_003512
|
HIST1H2AC
|
histone cluster 1, H2ac
|
|
chr22_+_21340757
|
1.290
|
|
|
|
|
chr6_+_27890772
|
1.275
|
NM_003521
|
HIST1H2BM
|
histone cluster 1, H2bm
|
|
chr22_+_21116283
|
1.246
|
|
|
|
|
chr17_-_4792542
|
1.245
|
NM_005022
|
PFN1
|
profilin 1
|
|
chr17_-_70639382
|
1.243
|
NM_014595
|
NT5C
|
5', 3'-nucleotidase, cytosolic
|
|
chr3_+_188130967
|
1.239
|
NM_173216
NM_173217
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr13_-_45640679
|
1.215
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr19_+_45996940
|
1.215
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr12_+_60940404
|
1.209
|
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr6_+_27941012
|
1.204
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr6_-_27890496
|
1.192
|
NM_021066
|
HIST1H2AJ
|
histone cluster 1, H2aj
|
|
chr10_-_64246129
|
1.190
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr12_-_31370305
|
1.177
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr19_-_18253413
|
1.159
|
NM_005354
|
JUND
|
jun D proto-oncogene
|
|
chr19_-_60611136
|
1.156
|
NM_014501
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chrX_+_12903145
|
1.145
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr14_-_105763152
|
1.125
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG3
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chrX_-_138742050
|
1.121
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr10_-_64246080
|
1.114
|
|
EGR2
|
early growth response 2
|
|
chr13_-_98757634
|
1.111
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr21_-_10120690
|
1.089
|
NM_001187
NM_182484
NM_181704
NM_182481
NM_182482
|
BAGE
BAGE5
BAGE4
BAGE3
BAGE2
|
B melanoma antigen
B melanoma antigen family, member 5
B melanoma antigen family, member 4
B melanoma antigen family, member 3
B melanoma antigen family, member 2
|
|
chr19_-_18253436
|
1.085
|
|
JUND
|
jun D proto-oncogene
|
|
chr3_-_48569213
|
1.080
|
NM_004567
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr12_-_8656705
|
1.055
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr12_+_6923441
|
1.049
|
NM_138425
|
C12orf57
|
chromosome 12 open reading frame 57
|
|
chr22_+_21407066
|
1.032
|
|
|
|
|
chr12_-_120725210
|
1.031
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr10_+_97505398
|
1.013
|
NM_001164178
NM_001164181
NM_001164179
NM_001164182
NM_001164183
NM_001776
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1
|
|
chr19_-_16111695
|
1.010
|
|
|
|
|
chr11_+_74377587
|
1.006
|
NM_006656
|
NEU3
|
sialidase 3 (membrane sialidase)
|
|
chr19_+_17239148
|
0.972
|
NM_001033549
NM_014173
|
BABAM1
|
BRISC and BRCA1 A complex member 1
|
|
chr14_-_106241445
|
0.950
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chrX_+_153767803
|
0.937
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr16_+_28850760
|
0.931
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr2_-_2313894
|
0.930
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr11_-_62125826
|
0.928
|
NM_004739
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr18_+_74841262
|
0.916
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr1_-_68735314
|
0.914
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr22_+_21384862
|
0.912
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr13_+_50925436
|
0.910
|
|
|
|
|
chr11_-_558419
|
0.906
|
|
|
|
|
chr1_-_68735267
|
0.903
|
|
DEPDC1
|
DEP domain containing 1
|
|
chr14_-_106330781
|
0.899
|
|
IGHV5-78
|
immunoglobulin heavy variable 5-78 pseudogene
|
|
chr6_+_27883877
|
0.880
|
NM_003509
|
HIST1H2AI
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H3f
|
|
chr19_-_45288621
|
0.878
|
NM_001010880
NM_001142577
NM_001142578
NM_001142579
|
ZNF780A
|
zinc finger protein 780A
|
|
chr4_-_91979113
|
0.874
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr19_-_45253947
|
0.874
|
NM_001005851
|
ZNF780B
|
zinc finger protein 780B
|
|
chr19_-_8279157
|
0.871
|
NM_001165895
NM_016579
|
CD320
|
CD320 molecule
|
|
chr17_-_7773477
|
0.870
|
NM_004732
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3
|
|
chr14_-_105862080
|
0.848
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr15_+_47249700
|
0.842
|
NM_002044
|
GALK2
|
galactokinase 2
|
|
chr6_-_133161318
|
0.825
|
NM_052831
|
C6orf192
|
chromosome 6 open reading frame 192
|
|
chr12_-_120270035
|
0.825
|
|
ANAPC5
|
anaphase promoting complex subunit 5
|
|
chr19_-_16111594
|
0.808
|
|
|
|
|
chr13_-_50925134
|
0.807
|
|
INTS6
|
integrator complex subunit 6
|
|
chr19_-_48815572
|
0.803
|
|
ZNF428
|
zinc finger protein 428
|
|
chr5_+_68702617
|
0.802
|
NM_133340
NM_133341
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chr1_-_148050534
|
0.801
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr6_-_10523424
|
0.783
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr11_-_62125673
|
0.776
|
|
MTA2
|
metastasis associated 1 family, member 2
|
|
chr22_-_34886727
|
0.772
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chr16_-_30014993
|
0.759
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr11_-_3620060
|
0.752
|
NM_053017
NM_001079536
|
ART5
|
ADP-ribosyltransferase 5
|
|
chr3_-_113334657
|
0.751
|
NM_001190259
NM_001190260
NM_152785
|
GCET2
|
germinal center expressed transcript 2
|
|
chr6_-_25834733
|
0.747
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr3_-_122862391
|
0.739
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chrX_+_69591639
|
0.724
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr22_+_20899155
|
0.722
|
|
|
|
|
chr16_+_30015243
|
0.721
|
|
|
|
|
chr2_-_113310796
|
0.718
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr17_-_38529638
|
0.717
|
NM_007298
|
BRCA1
|
breast cancer 1, early onset
|
|
chr5_+_68703036
|
0.715
|
NM_133339
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chr13_-_35327798
|
0.713
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chrX_-_154341451
|
0.711
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chrX_+_80343958
|
0.705
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr8_-_38324809
|
0.703
|
|
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1
|
|
chr12_-_48902692
|
0.699
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr8_-_33490094
|
0.699
|
NM_025115
NM_001102401
|
C8orf41
|
chromosome 8 open reading frame 41
|
|
chrX_+_80344253
|
0.698
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chrX_-_80344096
|
0.691
|
NM_030763
|
HMGN5
|
high-mobility group nucleosome binding domain 5
|
|
chr6_+_25834983
|
0.690
|
NM_170610
|
HIST1H2BA
|
histone cluster 1, H2ba
|
|
chr2_+_173927783
|
0.689
|
NM_031942
NM_145810
|
CDCA7
|
cell division cycle associated 7
|
|
chr14_-_105524207
|
0.689
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr6_-_10523203
|
0.683
|
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr14_+_22422270
|
0.679
|
NM_173527
|
REM2
|
RAS (RAD and GEM)-like GTP binding 2
|
|
chrX_+_80344215
|
0.677
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chrX_+_123062143
|
0.676
|
|
STAG2
|
stromal antigen 2
|
|
chr19_+_60803412
|
0.675
|
NM_153219
|
ZNF524
|
zinc finger protein 524
|
|
chr17_+_77910411
|
0.668
|
NM_207459
|
TEX19
|
testis expressed 19
|
|
chr1_+_165075304
|
0.663
|
|
POGK
|
pogo transposable element with KRAB domain
|
|
chr5_-_141011118
|
0.661
|
|
FCHSD1
|
FCH and double SH3 domains 1
|
|
chr14_-_67353046
|
0.657
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr6_-_35764588
|
0.656
|
|
FKBP5
|
FK506 binding protein 5
|
|
chr2_-_175170674
|
0.656
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr15_+_89248423
|
0.650
|
NM_006122
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|