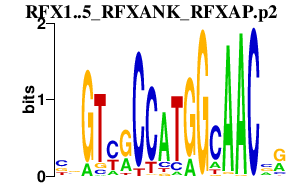
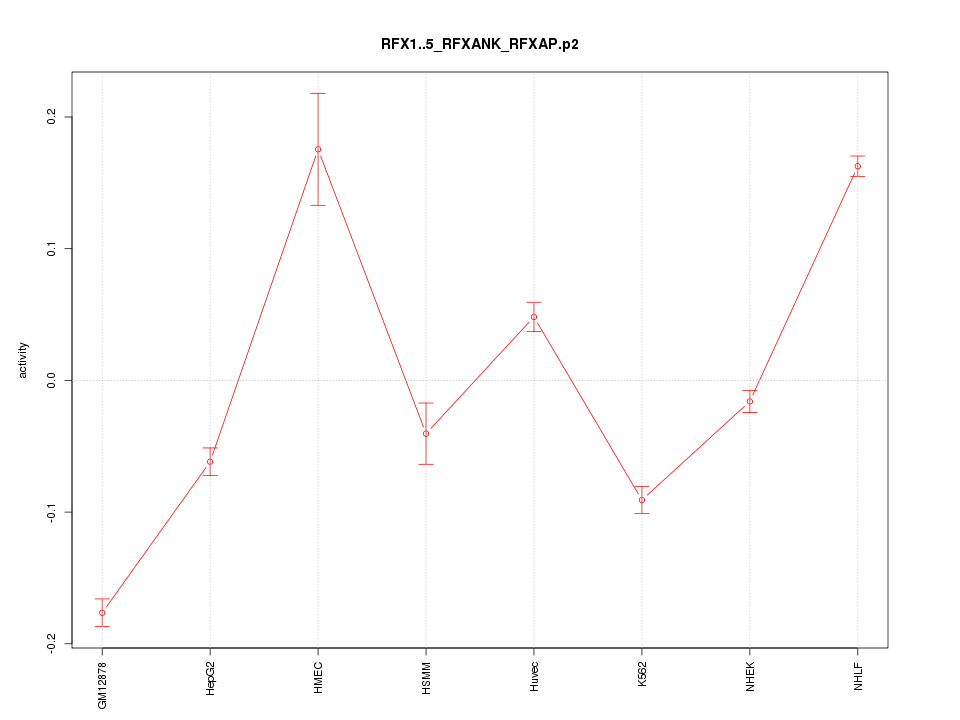
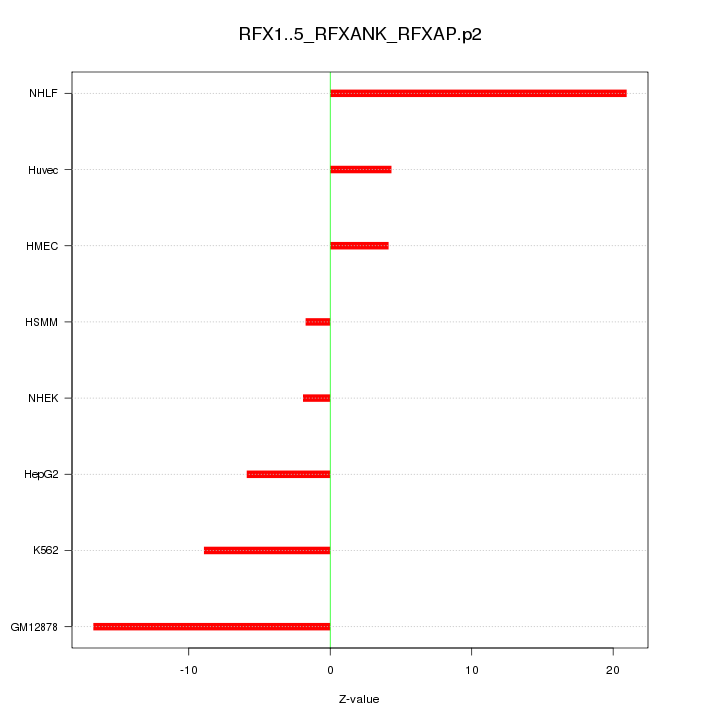
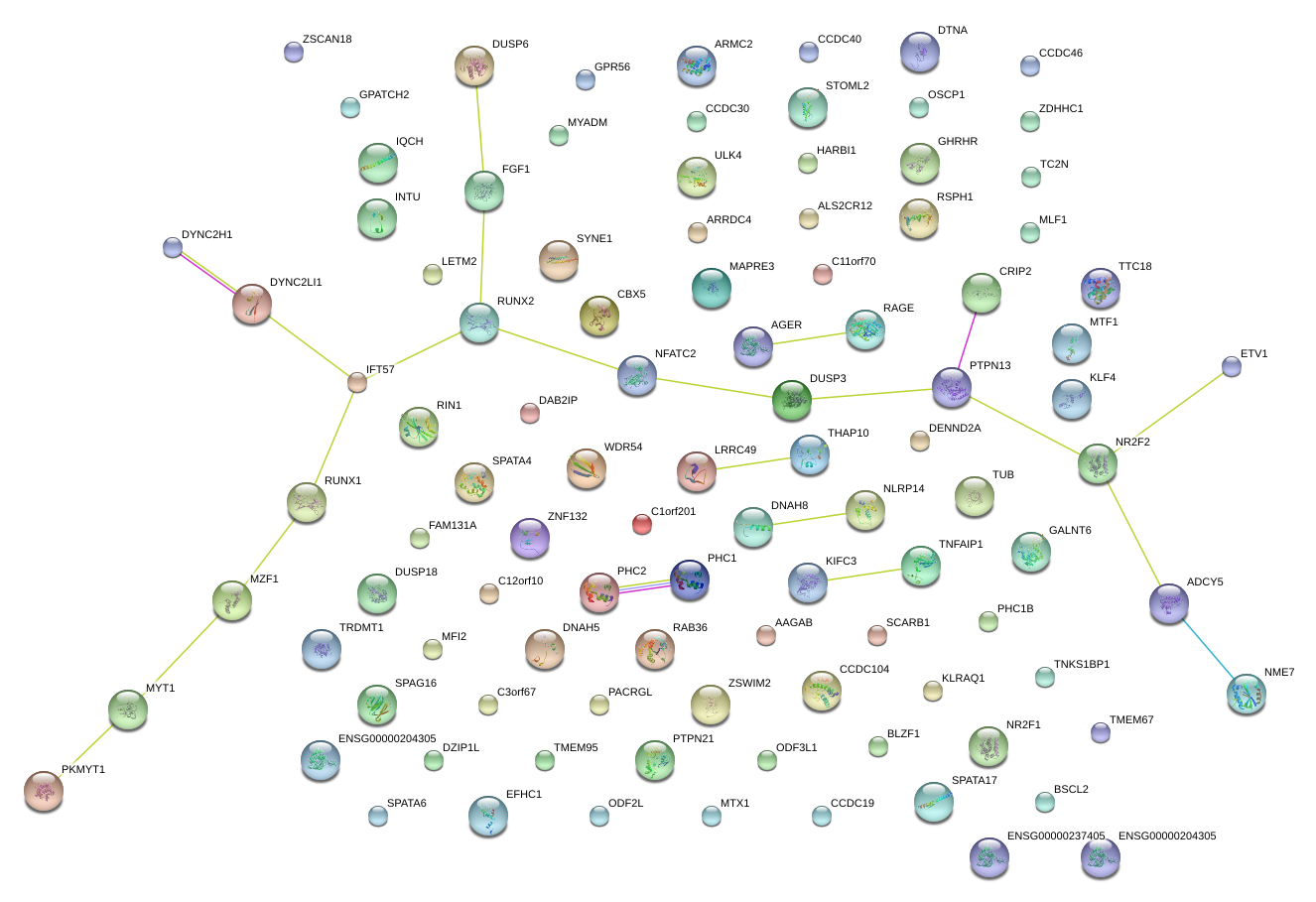
|
chr1_+_42701460
|
17.451
|
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr11_+_102485293
|
16.053
|
NM_001080463
NM_001377
|
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1
|
|
chr22_+_21817512
|
15.990
|
NM_004914
|
RAB36
|
RAB36, member RAS oncogene family
|
|
chr5_-_14064846
|
15.134
|
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr2_+_27046952
|
14.011
|
NM_012326
|
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3
|
|
chr14_+_105012106
|
13.893
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr2_-_187422141
|
13.275
|
NM_182521
|
ZSWIM2
|
zinc finger, SWIM-type containing 2
|
|
chr15_+_68971835
|
12.773
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr2_+_74502364
|
12.607
|
NM_032118
|
WDR54
|
WD repeat domain 54
|
|
chr18_+_30544193
|
11.950
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr15_-_68971665
|
11.838
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr4_+_87734489
|
11.822
|
NM_006264
NM_080683
NM_080684
NM_080685
|
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase)
|
|
chr11_-_65860450
|
11.463
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr4_-_177353707
|
9.440
|
NM_144644
|
SPATA4
|
spermatogenesis associated 4
|
|
chr17_-_61618276
|
9.386
|
NM_145036
|
CCDC46
|
coiled-coil domain containing 46
|
|
chr19_-_63776547
|
9.369
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr3_-_139315011
|
9.223
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr22_+_29393914
|
9.180
|
|
|
|
|
chr14_-_101841222
|
8.896
|
NM_014226
|
RAGE
|
renal tumor antigen
|
|
chr6_+_109276280
|
8.880
|
NM_032131
|
ARMC2
|
armadillo repeat containing 2
|
|
chr4_+_128773529
|
8.663
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr7_-_139987044
|
8.653
|
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr1_-_24612742
|
8.629
|
NM_001199013
NM_001199014
NM_178122
|
C1orf201
|
chromosome 1 open reading frame 201
|
|
chr15_+_73803351
|
8.561
|
NM_175881
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1
|
|
chr2_+_48521411
|
8.556
|
NM_001135629
NM_001193475
NM_152994
|
KLRAQ1
|
KLRAQ motif containing 1
|
|
chr9_-_35093189
|
8.487
|
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr19_+_59062962
|
8.456
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_+_59062937
|
8.382
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr22_-_29393786
|
8.362
|
NM_152511
|
DUSP18
|
dual specificity phosphatase 18
|
|
chr3_-_59010777
|
8.224
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr12_-_52939636
|
8.061
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr6_-_152744022
|
7.947
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr19_-_63776442
|
7.862
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr22_-_29393591
|
7.856
|
|
DUSP18
|
dual specificity phosphatase 18
|
|
chr6_-_152744143
|
7.814
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr3_-_59010797
|
7.763
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr10_-_17283623
|
7.701
|
|
TRDMT1
|
tRNA aspartic acid methyltransferase 1
|
|
chr19_-_63776740
|
7.575
|
NM_003422
NM_198055
|
MZF1
|
myeloid zinc finger 1
|
|
chr1_-_36688556
|
7.547
|
NM_145047
NM_206837
|
OSCP1
|
organic solute carrier partner 1
|
|
chr9_-_35093144
|
7.539
|
NM_013442
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr12_+_8958397
|
7.538
|
NM_004426
|
PHC1
|
polyhomeotic homolog 1 (Drosophila)
|
|
chr19_+_59062982
|
7.518
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr21_-_42789369
|
7.499
|
NM_080860
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas)
|
|
chr9_-_109291866
|
7.394
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr8_+_94836247
|
7.391
|
NM_001142301
NM_153704
|
TMEM67
|
transmembrane protein 67
|
|
chr11_-_56846246
|
7.272
|
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr11_-_62230078
|
7.222
|
NM_032667
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr3_-_41978464
|
7.034
|
NM_017886
|
ULK4
|
unc-51-like kinase 4 (C. elegans)
|
|
chr3_-_109423858
|
6.977
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr12_-_88270375
|
6.973
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr9_-_109291575
|
6.924
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr7_-_13997389
|
6.909
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr3_-_59010740
|
6.839
|
NM_198463
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr1_+_167604056
|
6.825
|
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr16_-_56393871
|
6.794
|
|
KIFC3
|
kinesin family member C3
|
|
chr11_+_101423378
|
6.745
|
NM_001195005
NM_032930
|
C11orf70
|
chromosome 11 open reading frame 70
|
|
chr9_-_35093089
|
6.726
|
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr16_+_56219802
|
6.682
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr1_-_167603623
|
6.651
|
NM_013330
NM_197972
|
NME7
|
non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr21_-_35181094
|
6.635
|
|
|
|
|
chr1_-_38097811
|
6.597
|
NM_005955
|
MTF1
|
metal-regulatory transcription factor 1
|
|
chr1_+_167603817
|
6.589
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr15_+_94674849
|
6.570
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr17_+_7199178
|
6.433
|
NM_198154
|
TMEM95
|
transmembrane protein 95
|
|
chr12_+_51979721
|
6.415
|
NM_021640
|
C12orf10
|
chromosome 12 open reading frame 10
|
|
chr8_+_38363150
|
6.397
|
NM_144652
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr17_-_39211706
|
6.394
|
NM_004090
|
DUSP3
|
dual specificity phosphatase 3
|
|
chr4_+_20311121
|
6.378
|
NM_001130727
NM_145048
|
PACRGL
|
PARK2 co-regulated-like
|
|
chr21_-_35181278
|
6.336
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_-_33669190
|
6.220
|
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr20_-_49612745
|
6.203
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr16_-_56393881
|
6.139
|
NM_001130100
NM_005550
|
KIFC3
|
kinesin family member C3
|
|
chr1_-_86634511
|
6.113
|
NM_001007022
NM_001184765
NM_001184766
NM_020729
|
ODF2L
|
outer dense fiber of sperm tails 2-like
|
|
chr9_+_123501186
|
6.088
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr1_-_215870935
|
6.038
|
NM_018040
|
GPATCH2
|
G patch domain containing 2
|
|
chr17_+_75625025
|
6.029
|
NM_017950
|
CCDC40
|
coiled-coil domain containing 40
|
|
chr15_+_65334169
|
5.997
|
NM_001031715
NM_022784
|
IQCH
|
IQ motif containing H
|
|
chr6_+_52392907
|
5.992
|
NM_018100
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr12_-_88270369
|
5.987
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr1_+_215871288
|
5.981
|
NM_138796
|
SPATA17
|
spermatogenesis associated 17
|
|
chr15_-_65334022
|
5.972
|
NM_024666
|
AAGAB
|
alpha- and gamma-adaptin binding protein
|
|
chr3_+_159771646
|
5.970
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr2_+_43854681
|
5.958
|
NM_001193464
NM_015522
NM_016008
|
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1
|
|
chr11_+_7997366
|
5.952
|
|
TUB
|
tubby homolog (mouse)
|
|
chr17_+_23686674
|
5.918
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr1_+_153445113
|
5.898
|
NM_002455
NM_198883
|
MTX1
|
metaxin 1
|
|
chr16_-_66007650
|
5.877
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr1_-_158136521
|
5.874
|
NM_012337
|
CCDC19
|
coiled-coil domain containing 19
|
|
chr5_-_142045770
|
5.840
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr12_-_52939584
|
5.818
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr1_-_48710263
|
5.806
|
|
SPATA6
|
spermatogenesis associated 6
|
|
chr14_-_88087575
|
5.753
|
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr1_-_48710378
|
5.714
|
NM_019073
|
SPATA6
|
spermatogenesis associated 6
|
|
chr20_-_49612574
|
5.701
|
NM_001136021
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr2_+_213857347
|
5.664
|
NM_001025436
NM_024532
|
SPAG16
|
sperm associated antigen 16
|
|
chr3_+_185538998
|
5.659
|
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr19_-_63301418
|
5.651
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr2_+_55600220
|
5.609
|
NM_080667
|
CCDC104
|
coiled-coil domain containing 104
|
|
chr19_-_63301402
|
5.602
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr12_-_88270128
|
5.578
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr2_-_201930311
|
5.545
|
NM_001127391
NM_139163
|
ALS2CR12
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 12
|
|
chr19_-_63643400
|
5.532
|
NM_003433
|
ZNF132
|
zinc finger protein 132
|
|
chr11_+_6998275
|
5.492
|
NM_176822
|
NLRP14
|
NLR family, pyrin domain containing 14
|
|
chr10_-_74788508
|
5.412
|
NM_145170
|
TTC18
|
tetratricopeptide repeat domain 18
|
|
chr12_-_123568781
|
5.392
|
|
|
|
|
chr15_+_96304911
|
5.378
|
NM_183376
|
ARRDC4
|
arrestin domain containing 4
|
|
chr18_+_76006795
|
5.378
|
|
LOC100130522
|
hypothetical LOC100130522
|
|
chr1_+_201862845
|
5.367
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr2_-_202216451
|
5.362
|
NM_001044385
|
ALS2CR4
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4
|
|
chrX_+_101910713
|
5.344
|
|
LOC100287765
|
hypothetical LOC100287765
|
|
chr19_-_63301386
|
5.312
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr20_-_13567529
|
5.301
|
NM_017714
|
TASP1
|
taspase, threonine aspartase, 1
|
|
chr11_-_6998041
|
5.279
|
NM_013249
|
ZNF214
|
zinc finger protein 214
|
|
chr20_+_43425501
|
5.258
|
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr2_+_38956606
|
5.210
|
NM_001145450
|
MORN2
|
MORN repeat containing 2
|
|
chr5_+_92944680
|
5.198
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr22_-_36679479
|
5.097
|
NM_032561
|
C22orf23
|
chromosome 22 open reading frame 23
|
|
chr2_-_38956496
|
5.093
|
NM_198963
|
DHX57
|
DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57
|
|
chr1_+_15725894
|
5.074
|
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr17_-_23686566
|
5.041
|
NM_174887
|
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas)
|
|
chr6_+_117044334
|
4.979
|
NM_001010892
NM_001161664
|
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas)
|
|
chr9_-_35093048
|
4.978
|
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr16_-_74147608
|
4.972
|
NM_001077416
NM_001077418
NM_001077419
|
CHST5
TMEM231
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5
transmembrane protein 231
|
|
chr11_-_82582328
|
4.955
|
|
DKFZp547G183
|
hypothetical LOC55525
|
|
chr1_+_37795101
|
4.906
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr7_+_150356435
|
4.900
|
NM_007188
|
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8
|
|
chr16_+_4485859
|
4.810
|
NM_001127205
|
HMOX2
|
heme oxygenase (decycling) 2
|
|
chr11_+_85243791
|
4.761
|
NM_173556
|
CCDC83
|
coiled-coil domain containing 83
|
|
chr5_-_142046175
|
4.746
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr14_+_59628381
|
4.740
|
NM_022495
|
C14orf135
|
chromosome 14 open reading frame 135
|
|
chr15_+_96304942
|
4.724
|
|
ARRDC4
|
arrestin domain containing 4
|
|
chr11_+_93394025
|
4.721
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr17_+_260408
|
4.713
|
NM_001013672
|
C17orf97
|
chromosome 17 open reading frame 97
|
|
chr1_+_15725935
|
4.699
|
NM_015291
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr2_+_228444570
|
4.697
|
NM_178821
|
WDR69
|
WD repeat domain 69
|
|
chr11_+_82582399
|
4.661
|
|
ANKRD42
|
ankyrin repeat domain 42
|
|
chr4_+_123873306
|
4.650
|
NM_001178007
NM_152618
|
BBS12
|
Bardet-Biedl syndrome 12
|
|
chr14_+_23908141
|
4.646
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr7_-_13997574
|
4.631
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr6_-_46728188
|
4.562
|
NM_016593
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1
|
|
chr5_-_16795336
|
4.542
|
|
MYO10
|
myosin X
|
|
chr7_+_76589869
|
4.519
|
NM_020879
|
CCDC146
|
coiled-coil domain containing 146
|
|
chr13_-_43259032
|
4.518
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr8_-_133756943
|
4.481
|
NM_012472
|
LRRC6
|
leucine rich repeat containing 6
|
|
chr6_-_121697305
|
4.429
|
NM_152730
|
C6orf170
|
chromosome 6 open reading frame 170
|
|
chr11_+_131285747
|
4.414
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr19_-_55220495
|
4.366
|
NM_001025778
NM_016440
|
VRK3
|
vaccinia related kinase 3
|
|
chr19_+_55221023
|
4.356
|
NM_001006656
NM_015428
|
ZNF473
|
zinc finger protein 473
|
|
chr3_-_132228273
|
4.354
|
NM_014065
|
ASTE1
|
asteroid homolog 1 (Drosophila)
|
|
chr21_-_45532176
|
4.345
|
NM_015227
NM_133635
|
POFUT2
|
protein O-fucosyltransferase 2
|
|
chr3_+_115258348
|
4.339
|
|
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1
|
|
chr2_-_110319828
|
4.309
|
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr6_-_161614962
|
4.301
|
NM_020133
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
|
|
chr4_-_153920326
|
4.280
|
NM_145720
|
TIGD4
|
tigger transposable element derived 4
|
|
chr9_-_129537372
|
4.255
|
NM_001085347
NM_001134430
NM_001134431
NM_130459
|
TOR2A
|
torsin family 2, member A
|
|
chr22_-_43272567
|
4.245
|
NM_032287
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr7_+_30777512
|
4.214
|
NM_032222
|
FAM188B
|
family with sequence similarity 188, member B
|
|
chr3_+_132228383
|
4.193
|
NM_001146003
NM_024800
NM_145910
|
NEK11
|
NIMA (never in mitosis gene a)- related kinase 11
|
|
chr13_-_30634031
|
4.185
|
NM_006644
|
HSPH1
|
heat shock 105kDa/110kDa protein 1
|
|
chr3_+_48482213
|
4.175
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr20_+_43425153
|
4.162
|
NM_001099791
NM_033542
|
SYS1-DBNDD2
SYS1
|
SYS1-DBNDD2 read-through transcript
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr10_-_25344871
|
4.153
|
NM_145010
|
ENKUR
|
enkurin, TRPC channel interacting protein
|
|
chr2_-_110319866
|
4.095
|
NM_000272
NM_001128178
NM_001128179
NM_207181
|
NPHP1
|
nephronophthisis 1 (juvenile)
|
|
chr1_-_20707196
|
4.094
|
NM_024544
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1
|
|
chr4_+_153920563
|
4.081
|
|
ARFIP1
|
ADP-ribosylation factor interacting protein 1
|
|
chr19_-_6344005
|
4.078
|
NM_002096
|
GTF2F1
|
general transcription factor IIF, polypeptide 1, 74kDa
|
|
chr4_+_114433551
|
4.077
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_-_16281287
|
4.074
|
NM_001047434
NM_206831
|
DPH3
|
DPH3, KTI11 homolog (S. cerevisiae)
|
|
chr3_+_115258270
|
4.051
|
NM_024638
|
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1
|
|
chr12_+_46863632
|
4.039
|
NM_001013635
|
C12orf68
|
chromosome 12 open reading frame 68
|
|
chr7_+_138469029
|
4.035
|
NM_001144920
NM_001144923
NM_024926
|
TTC26
|
tetratricopeptide repeat domain 26
|
|
chr19_-_7459867
|
3.987
|
NM_080662
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr1_+_74436507
|
3.983
|
NM_001112808
NM_003838
|
FPGT-TNNI3K
FPGT
|
FPGT-TNNI3K fusion protein
fucose-1-phosphate guanylyltransferase
|
|
chr8_-_94781836
|
3.940
|
|
LOC642924
|
hypothetical LOC642924
|
|
chr14_+_99329198
|
3.912
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr8_+_94781910
|
3.909
|
NM_145269
|
FAM92A1
|
family with sequence similarity 92, member A1
|
|
chr22_-_16887151
|
3.905
|
NM_001122731
NM_001136004
NM_015241
|
MICAL3
|
microtubule associated monoxygenase, calponin and LIM domain containing 3
|
|
chr16_+_28893525
|
3.893
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr17_-_19206475
|
3.892
|
NM_015681
|
B9D1
|
B9 protein domain 1
|
|
chr1_+_201862552
|
3.880
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr20_+_42593849
|
3.858
|
NM_007066
NM_181804
NM_181805
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr9_+_115396020
|
3.847
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr6_+_125346208
|
3.841
|
NM_152553
|
RNF217
|
ring finger protein 217
|
|
chr1_-_20707103
|
3.831
|
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1
|
|
chr3_-_115258053
|
3.821
|
NM_020817
|
KIAA1407
|
KIAA1407
|
|
chr1_+_67163165
|
3.819
|
NM_001077700
NM_001077702
NM_001077703
NM_001146110
NM_001146111
NM_001146112
NM_001146113
NM_020948
|
MIER1
|
mesoderm induction early response 1 homolog (Xenopus laevis)
|
|
chr1_-_67163001
|
3.814
|
NM_024763
NM_207014
|
WDR78
|
WD repeat domain 78
|
|
chr15_-_80342045
|
3.808
|
|
EFTUD1
|
elongation factor Tu GTP binding domain containing 1
|
|
chr11_-_114880275
|
3.802
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chrX_+_135079461
|
3.797
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_+_170457763
|
3.770
|
NM_032448
|
FAM120B
|
family with sequence similarity 120B
|
|
chr6_+_46728590
|
3.753
|
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr5_+_72148173
|
3.720
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr11_-_65869113
|
3.717
|
NM_001024957
NM_015399
|
BRMS1
|
breast cancer metastasis suppressor 1
|
|
chr18_-_45630154
|
3.710
|
|
MYO5B
|
myosin VB
|
|
chr13_-_35818396
|
3.698
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr17_+_52026058
|
3.691
|
NM_005450
|
NOG
|
noggin
|
|
chr6_+_170457774
|
3.686
|
|
FAM120B
|
family with sequence similarity 120B
|
|
chr22_-_29082880
|
3.680
|
NM_001005409
NM_005877
|
SF3A1
|
splicing factor 3a, subunit 1, 120kDa
|
|
chr14_+_74539364
|
3.668
|
NM_014239
|
EIF2B2
|
eukaryotic translation initiation factor 2B, subunit 2 beta, 39kDa
|