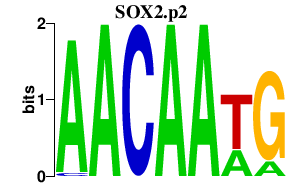
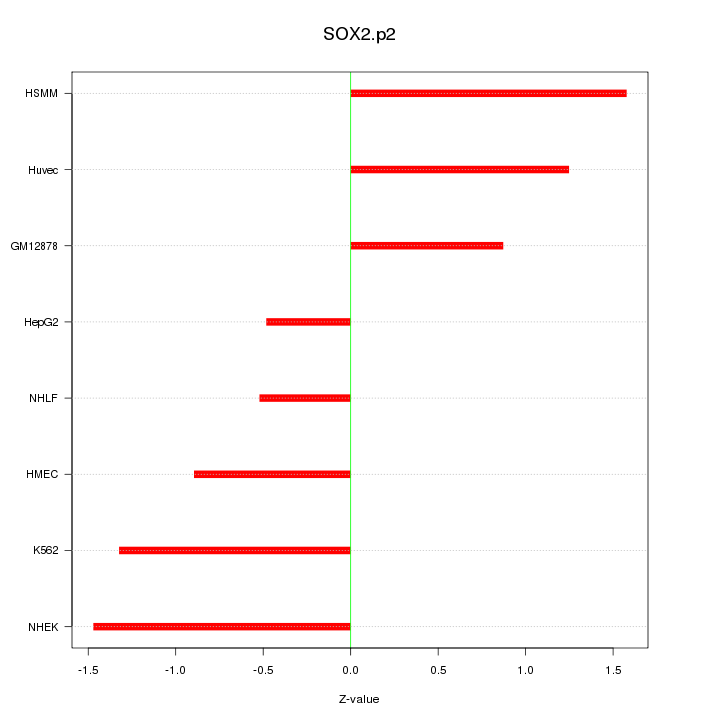
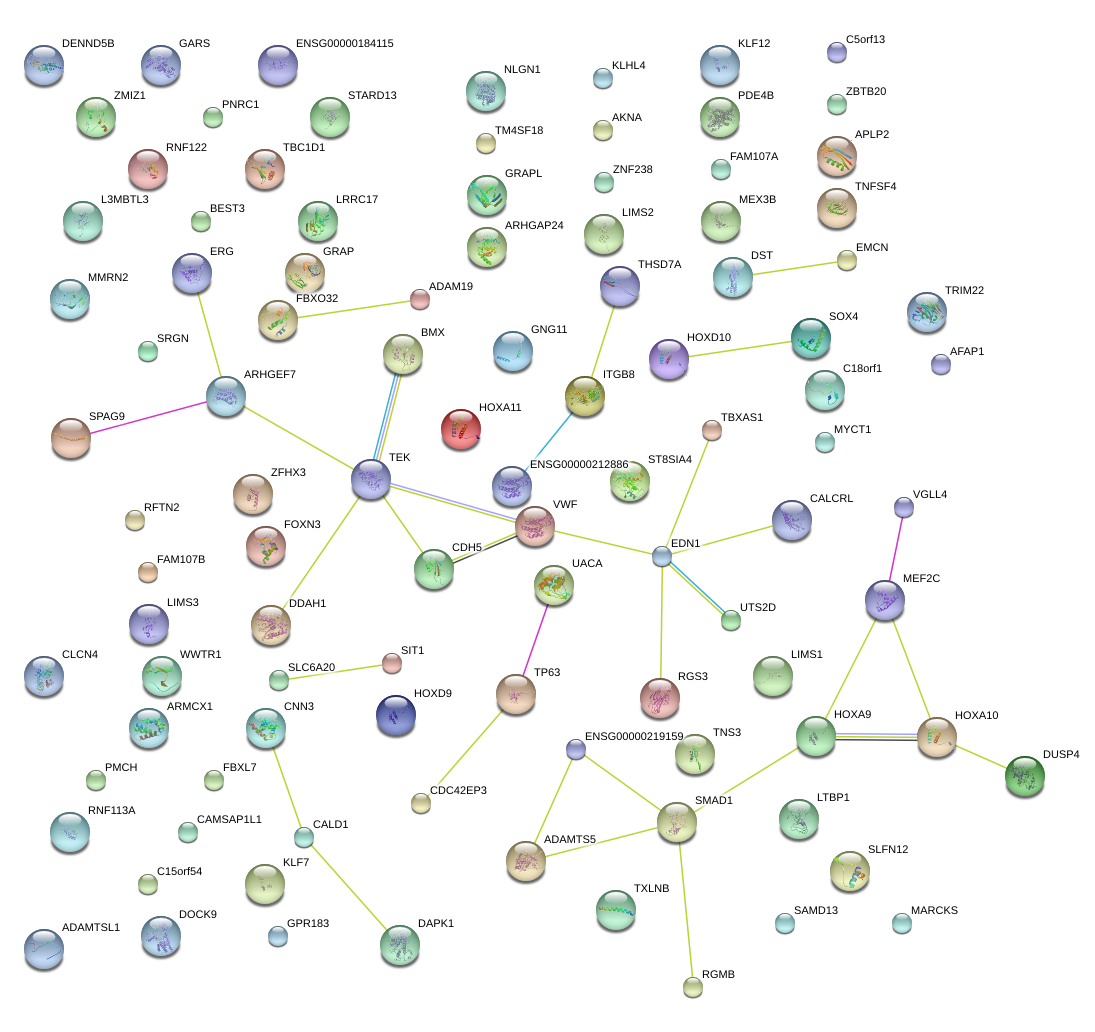
|
chr6_-_139654807
|
2.559
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr3_-_150533948
|
2.270
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr7_-_27186400
|
2.076
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr4_-_101658094
|
1.618
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr6_+_130381408
|
1.405
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr1_-_171441234
|
1.382
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr4_+_146623405
|
1.349
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr4_+_86918882
|
1.312
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_+_66570363
|
1.279
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr2_-_37752272
|
1.278
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr1_-_197173159
|
1.270
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr12_-_6103945
|
1.214
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr7_-_27180398
|
1.212
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr2_-_188021117
|
1.206
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr13_-_98428244
|
1.205
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr7_+_102340579
|
1.130
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr6_+_12398514
|
1.107
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr7_+_27190688
|
1.089
|
|
|
|
|
chrX_+_15428820
|
1.071
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr21_-_38792173
|
1.069
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr1_-_95165089
|
1.069
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr17_-_18890963
|
1.066
|
NM_006613
|
GRAP
|
GRB2-related adaptor protein
|
|
chr6_+_114285209
|
1.050
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr9_+_18464138
|
1.050
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr9_+_18464090
|
1.047
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr12_-_101115677
|
1.042
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr7_-_27190882
|
1.028
|
|
HOXA11
|
homeobox A11
|
|
chr6_+_153060722
|
1.023
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr13_-_73606362
|
1.021
|
|
KLF12
|
Kruppel-like factor 12
|
|
chr16_-_71639670
|
1.000
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chrX_+_86659370
|
0.992
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr1_-_95165037
|
0.977
|
|
CNN3
|
calponin 3, acidic
|
|
chr5_+_15553736
|
0.976
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr9_-_35640852
|
0.973
|
NM_014450
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1
|
|
chr5_+_15553288
|
0.956
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr11_+_5667494
|
0.934
|
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr5_-_100266769
|
0.925
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr5_-_111119846
|
0.924
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_-_27171673
|
0.908
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr3_-_116272911
|
0.908
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_+_15553539
|
0.887
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr8_-_124622434
|
0.887
|
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr10_+_22765996
|
0.885
|
|
LOC100289455
|
hypothetical LOC100289455
|
|
chr10_+_70517863
|
0.867
|
|
SRGN
|
serglycin
|
|
chr12_-_68369322
|
0.843
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr9_-_116190038
|
0.842
|
|
AKNA
|
AT-hook transcription factor
|
|
chr6_-_56824371
|
0.837
|
|
DST
|
dystonin
|
|
chr10_+_70517823
|
0.835
|
NM_002727
|
SRGN
|
serglycin
|
|
chr13_-_98757634
|
0.833
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr15_-_80125403
|
0.831
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr15_-_80125514
|
0.820
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr5_-_88155360
|
0.815
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr17_-_59818033
|
0.814
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr7_+_93388951
|
0.798
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr11_+_129496792
|
0.795
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr13_-_32658215
|
0.795
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr1_-_85816520
|
0.782
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_+_108637698
|
0.772
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_-_198248828
|
0.771
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr9_+_89302574
|
0.771
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr18_+_13601664
|
0.764
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr15_-_68781662
|
0.755
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr13_+_110653407
|
0.754
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr3_+_148593911
|
0.754
|
|
|
|
|
chr3_+_190831909
|
0.750
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr1_-_85703198
|
0.747
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_+_108637916
|
0.738
|
|
LIMS1
LIMS3
|
LIM and senescent cell antigen-like domains 1
LIM and senescent cell antigen-like domains 3
|
|
chr17_-_59817456
|
0.734
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr3_+_182900081
|
0.729
|
|
SOX2OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chr17_-_59817722
|
0.727
|
NM_000442
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr13_-_32678142
|
0.724
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_-_192531018
|
0.721
|
NM_198152
|
UTS2D
|
urotensin 2 domain containing
|
|
chr2_-_56266408
|
0.709
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr17_-_59817654
|
0.705
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr13_-_73605914
|
0.705
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr9_+_115303527
|
0.705
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr6_-_56816403
|
0.701
|
|
DST
|
dystonin
|
|
chr10_+_80672842
|
0.701
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr9_+_89302239
|
0.697
|
|
DAPK1
|
death-associated protein kinase 1
|
|
chr17_-_59817575
|
0.697
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr17_+_18971506
|
0.688
|
NM_001129778
|
GRAPL
|
GRB2-related adaptor protein-like
|
|
chr1_+_84540921
|
0.672
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr2_-_110665188
|
0.669
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr17_-_30783631
|
0.667
|
NM_018042
|
SLFN12
|
schlafen family member 12
|
|
chr12_-_31635089
|
0.660
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr2_+_110013297
|
0.659
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr4_+_37655385
|
0.659
|
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1
|
|
chr5_+_98132898
|
0.652
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr2_-_207739815
|
0.644
|
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr15_+_37330176
|
0.642
|
NM_207445
|
C15orf54
|
chromosome 15 open reading frame 54
|
|
chr7_+_20337249
|
0.642
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr1_+_242281199
|
0.640
|
|
ZNF238
|
zinc finger protein 238
|
|
chr6_+_21701942
|
0.640
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr2_-_37752731
|
0.639
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr8_-_29262240
|
0.639
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr7_-_47588266
|
0.638
|
|
TNS3
|
tensin 3
|
|
chr6_-_56824672
|
0.637
|
|
DST
|
dystonin
|
|
chr5_+_15553768
|
0.606
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr7_+_134114912
|
0.603
|
|
CALD1
|
caldesmon 1
|
|
chr9_+_27099138
|
0.598
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr17_-_46479127
|
0.598
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr5_-_156935283
|
0.597
|
|
|
|
|
chr14_-_88953059
|
0.595
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr7_-_11838260
|
0.594
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr8_-_33544184
|
0.592
|
NM_024787
|
RNF122
|
ring finger protein 122
|
|
chr3_-_115826481
|
0.588
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_+_134114957
|
0.573
|
|
CALD1
|
caldesmon 1
|
|
chr4_+_146622154
|
0.571
|
NM_005900
|
SMAD1
|
SMAD family member 1
|
|
chrX_+_10084984
|
0.569
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr2_-_128138435
|
0.568
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr3_-_150858274
|
0.563
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr3_-_11660395
|
0.562
|
NM_001128219
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr21_-_27261276
|
0.562
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr5_-_156935317
|
0.561
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr5_+_95093267
|
0.561
|
|
|
|
|
chr7_-_47588160
|
0.557
|
|
TNS3
|
tensin 3
|
|
chr10_-_14686243
|
0.557
|
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr4_-_7991992
|
0.548
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr7_+_139175420
|
0.547
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr16_+_64958025
|
0.544
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr3_-_58538110
|
0.543
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr3_+_174598937
|
0.543
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr3_-_58538530
|
0.540
|
NM_001076778
NM_007177
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr1_+_198975227
|
0.538
|
NM_203459
|
CAMSAP1L1
|
calmodulin regulated spectrin-associated protein 1-like 1
|
|
chr16_+_64958060
|
0.536
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr2_-_207738858
|
0.531
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr6_+_89847019
|
0.531
|
NM_006813
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr2_+_33213111
|
0.531
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr10_-_88707299
|
0.530
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chrX_+_100692071
|
0.529
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr4_-_140696740
|
0.526
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr4_+_86918870
|
0.521
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr9_+_4480193
|
0.520
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr8_-_6407882
|
0.518
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr16_+_64958086
|
0.517
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr5_-_71838875
|
0.517
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr11_+_113436438
|
0.514
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr1_+_242283129
|
0.514
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr1_+_43539239
|
0.513
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr19_+_13767207
|
0.511
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr8_-_108579270
|
0.509
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr5_-_111120929
|
0.502
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr6_+_89847183
|
0.502
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr3_+_25444837
|
0.501
|
NM_000965
|
RARB
|
retinoic acid receptor, beta
|
|
chr15_-_81667773
|
0.501
|
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr6_+_44295219
|
0.501
|
NM_001078174
NM_004955
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr15_-_81667169
|
0.498
|
NM_016073
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr1_-_26105954
|
0.498
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr1_-_224993498
|
0.495
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|
|
chr10_+_104525877
|
0.493
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr1_+_66570683
|
0.489
|
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr3_-_115825742
|
0.488
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_+_76418545
|
0.487
|
|
LOC728723
|
hypothetical LOC728723
|
|
chr3_+_25444753
|
0.486
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr4_+_3264509
|
0.486
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr1_-_32574185
|
0.485
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr12_-_41164052
|
0.482
|
NM_001144882
NM_001144883
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr1_-_223907283
|
0.481
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr5_-_111120817
|
0.474
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_+_12904207
|
0.474
|
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chrX_-_128616594
|
0.474
|
NM_017413
|
APLN
|
apelin
|
|
chr6_+_116939500
|
0.473
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr12_-_41164010
|
0.472
|
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chrX_+_28515479
|
0.470
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr11_+_111313067
|
0.470
|
NM_001037954
|
DIXDC1
|
DIX domain containing 1
|
|
chr19_-_14121728
|
0.467
|
|
LPHN1
|
latrophilin 1
|
|
chr15_+_22751162
|
0.465
|
NM_003097
NM_022804
NM_005678
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chr15_-_80123935
|
0.464
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr12_-_10142779
|
0.464
|
NM_016511
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chr13_-_20533703
|
0.461
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr12_-_47869134
|
0.459
|
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr9_+_115382990
|
0.458
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr11_+_111313227
|
0.456
|
|
DIXDC1
|
DIX domain containing 1
|
|
chr14_+_76298693
|
0.453
|
|
VASH1
|
vasohibin 1
|
|
chr10_-_14686413
|
0.453
|
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr9_+_36026900
|
0.452
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr13_-_22905822
|
0.450
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr8_+_67201684
|
0.449
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr15_-_93671314
|
0.448
|
|
LOC400456
|
hypothetical LOC400456
|
|
chr3_+_29297806
|
0.447
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr11_+_128068982
|
0.445
|
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr5_-_111120764
|
0.441
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr17_-_26672757
|
0.440
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr5_-_111782637
|
0.440
|
NM_022140
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A
|
|
chr7_-_47588652
|
0.439
|
|
TNS3
|
tensin 3
|
|
chr5_-_95184221
|
0.438
|
NM_001118890
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr18_-_21186107
|
0.438
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr5_+_72148173
|
0.437
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr4_-_54119102
|
0.436
|
|
LNX1
|
ligand of numb-protein X 1
|
|
chrX_-_50573759
|
0.436
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr8_-_18710575
|
0.434
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr9_-_4289574
|
0.433
|
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr4_-_54119159
|
0.429
|
NM_032622
|
LNX1
|
ligand of numb-protein X 1
|
|
chr12_-_47869067
|
0.428
|
NM_006009
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr1_-_79244948
|
0.426
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr3_-_194118627
|
0.425
|
NM_178496
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr7_-_27171648
|
0.423
|
|
HOXA9
|
homeobox A9
|
|
chr12_+_48443588
|
0.422
|
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr1_-_153107084
|
0.421
|
|
|
|
|
chr3_-_194118321
|
0.420
|
|
MB21D2
|
Mab-21 domain containing 2
|