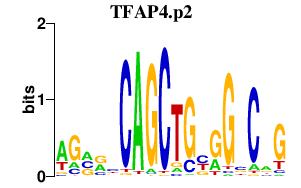
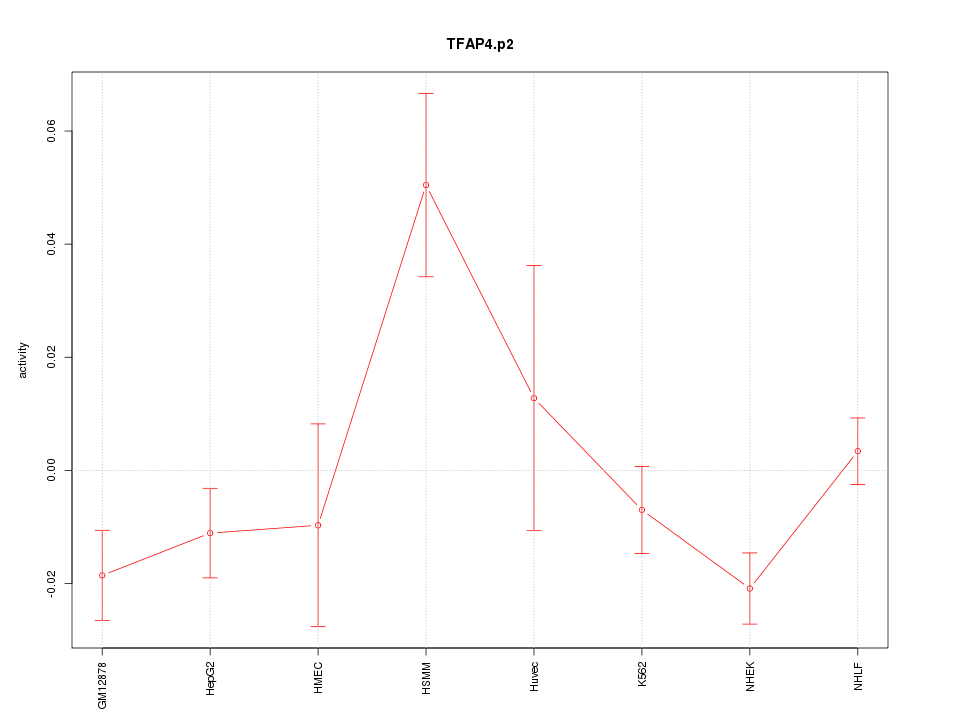
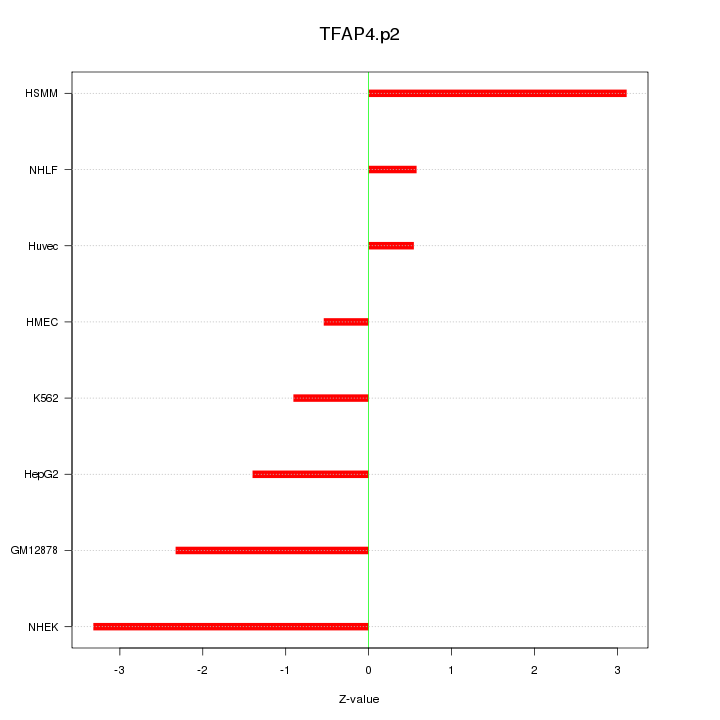
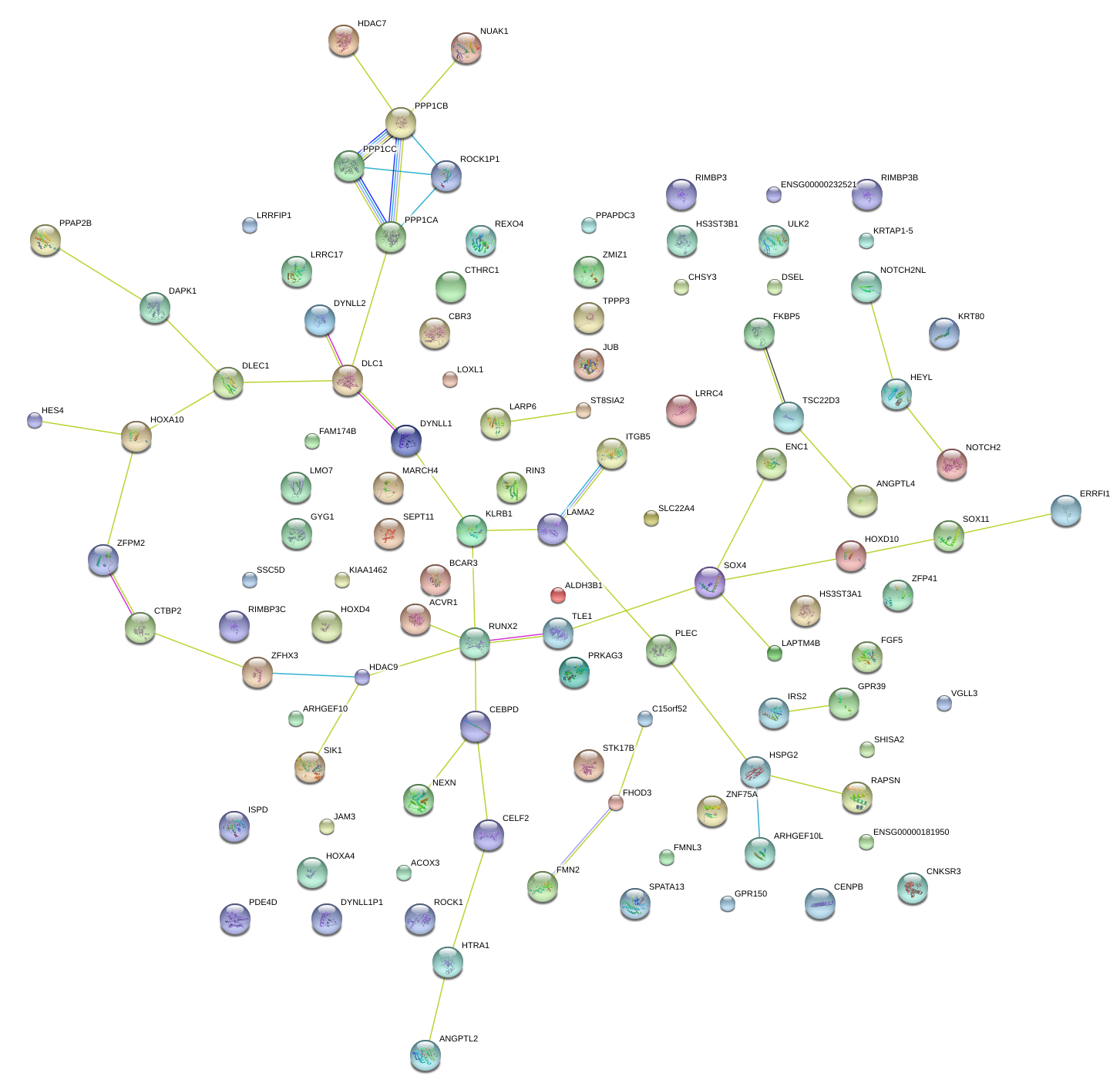
|
chr3_-_87122936
|
4.482
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr1_+_78126787
|
3.393
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr3_-_87122958
|
3.304
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr11_-_47427263
|
2.946
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr12_-_50871953
|
2.883
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr9_+_133154896
|
2.807
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr1_-_93919789
|
2.730
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr6_-_154873344
|
2.231
|
|
CNKSR3
|
CNKSR family member 3
|
|
chr4_+_78089931
|
2.193
|
|
SEPT11
|
septin 11
|
|
chr8_-_145090892
|
2.105
|
NM_201381
|
PLEC
|
plectin
|
|
chr6_+_45498354
|
2.103
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr8_+_104452877
|
2.101
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr12_-_46499834
|
2.082
|
NM_001098416
NM_015401
|
HDAC7
|
histone deacetylase 7
|
|
chr6_-_154873480
|
2.069
|
|
CNKSR3
|
CNKSR family member 3
|
|
chr22_+_20068039
|
2.007
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr10_-_126839056
|
2.000
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr6_+_129245978
|
1.947
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr16_+_3295433
|
1.910
|
NM_153028
|
ZNF75A
|
zinc finger protein 75a
|
|
chr5_+_129268352
|
1.870
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr16_-_71650034
|
1.821
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr3_-_126088530
|
1.787
|
|
ITGB5
|
integrin, beta 5
|
|
chr11_+_67534364
|
1.787
|
NM_000694
NM_001030010
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr1_-_925332
|
1.769
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr11_+_67534371
|
1.741
|
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr5_-_58370693
|
1.710
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_27180398
|
1.706
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr12_+_62523648
|
1.684
|
|
|
|
|
chr13_-_25523164
|
1.682
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr15_+_72006015
|
1.623
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr15_+_90738108
|
1.605
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr4_+_78089968
|
1.602
|
|
SEPT11
|
septin 11
|
|
chr4_+_78089879
|
1.576
|
NM_018243
|
SEPT11
|
septin 11
|
|
chr21_+_36429079
|
1.544
|
NM_001236
|
CBR3
|
carbonyl reductase 3
|
|
chr10_-_30388439
|
1.534
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr1_+_17817397
|
1.506
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr2_-_216944911
|
1.505
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr8_+_98857190
|
1.495
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr16_-_3295375
|
1.486
|
|
|
|
|
chrX_-_106846253
|
1.480
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr22_-_20235372
|
1.473
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr6_-_154873396
|
1.440
|
NM_173515
|
CNKSR3
|
CNKSR family member 3
|
|
chr11_+_133444175
|
1.428
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr16_-_65984836
|
1.427
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr11_-_66945190
|
1.411
|
|
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme
|
|
chr15_-_38420414
|
1.393
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr18_-_63334902
|
1.388
|
NM_032160
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr15_-_68933481
|
1.382
|
NM_018357
NM_197958
|
LARP6
|
La ribonucleoprotein domain family, member 6
|
|
chr1_-_39877928
|
1.368
|
NM_014571
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr2_+_56265260
|
1.346
|
|
|
|
|
chr10_+_124210980
|
1.320
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr3_+_150191884
|
1.303
|
NM_001184720
NM_001184721
NM_004130
|
GYG1
|
glycogenin 1
|
|
chr1_-_39877883
|
1.301
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr2_-_196744546
|
1.294
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr1_+_143920424
|
1.291
|
NM_203458
|
NOTCH2NL
|
notch 2 N-terminal like
|
|
chr2_+_5750229
|
1.286
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr9_-_128924782
|
1.277
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr18_-_16945763
|
1.265
|
NM_005406
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1
|
|
chr10_+_11100111
|
1.259
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_+_11099860
|
1.258
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr20_-_3713509
|
1.247
|
|
CENPB
|
centromere protein B, 80kDa
|
|
chr13_-_109236897
|
1.244
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr1_-_22136263
|
1.238
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr17_-_19711580
|
1.235
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr1_-_56817568
|
1.234
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr10_+_80498782
|
1.230
|
NM_020338
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr10_-_126839000
|
1.225
|
|
CTBP2
|
C-terminal binding protein 2
|
|
chr1_-_39871226
|
1.215
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr9_-_83493415
|
1.208
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr1_+_181259317
|
1.191
|
|
|
|
|
chr4_+_81406765
|
1.191
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr9_-_128924819
|
1.191
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr7_-_127458231
|
1.183
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr10_+_11100075
|
1.179
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr8_+_98856984
|
1.177
|
NM_018407
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr6_-_35804337
|
1.172
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr9_-_128924706
|
1.172
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr2_+_238200922
|
1.171
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr11_+_133444029
|
1.162
|
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr11_-_1975265
|
1.153
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr17_-_36436979
|
1.148
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr2_-_196744507
|
1.143
|
|
STK17B
|
serine/threonine kinase 17b
|
|
chr19_+_60691671
|
1.136
|
NM_001144950
NM_001195267
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr2_-_219404755
|
1.133
|
NM_017431
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit
|
|
chr7_-_27136876
|
1.129
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr13_+_75108450
|
1.127
|
|
LMO7
|
LIM domain 7
|
|
chr9_+_89302574
|
1.123
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr2_-_158440587
|
1.117
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr18_+_32131628
|
1.115
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr2_+_132890616
|
1.114
|
NM_001508
|
GPR39
|
G protein-coupled receptor 39
|
|
chr5_-_73972055
|
1.098
|
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr5_+_131657964
|
1.089
|
NM_003059
|
SLC22A4
|
solute carrier family 22 (organic cation/ergothioneine transporter), member 4
|
|
chr9_+_89302239
|
1.087
|
|
DAPK1
|
death-associated protein kinase 1
|
|
chr7_-_16427471
|
1.086
|
NM_001101417
NM_001101426
|
ISPD
|
isoprenoid synthase domain containing
|
|
chr1_+_238321794
|
1.077
|
NM_020066
|
FMN2
|
formin 2
|
|
chr8_-_48813236
|
1.073
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr15_-_90999606
|
1.069
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr15_+_72005839
|
1.065
|
NM_005576
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr17_+_14145051
|
1.065
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr11_-_1975144
|
1.058
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr4_-_8481085
|
1.055
|
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr13_+_23451838
|
1.054
|
|
|
|
|
chr1_-_56817717
|
1.053
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr8_+_1759502
|
1.043
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr14_-_22521544
|
1.042
|
NM_032876
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr13_+_23451919
|
1.034
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr8_+_106400322
|
1.013
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr12_-_105057940
|
1.006
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr14_+_92049396
|
1.000
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr8_+_144420939
|
0.996
|
NM_138465
|
GLI4
|
GLI family zinc finger 4
|
|
chr15_+_72005875
|
0.991
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr15_+_72005864
|
0.980
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr21_-_43671355
|
0.979
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr5_+_94981735
|
0.972
|
NM_199243
|
GPR150
|
G protein-coupled receptor 150
|
|
chr1_-_8008979
|
0.971
|
NM_018948
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr7_+_102340579
|
0.970
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr3_-_126088819
|
0.970
|
NM_002213
|
ITGB5
|
integrin, beta 5
|
|
chr8_-_13178399
|
0.968
|
|
DLC1
|
deleted in liver cancer 1
|
|
chr21_+_41461597
|
0.962
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr6_+_45498262
|
0.960
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr11_-_71202538
|
0.954
|
|
LOC285407
|
asparagine-linked glycosylation 1 homolog pseudogene
|
|
chr11_+_65536037
|
0.953
|
NM_001323
|
CST6
|
cystatin E/M
|
|
chr19_-_2378874
|
0.946
|
NM_012458
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr5_-_77980097
|
0.945
|
NM_005779
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr4_-_10068055
|
0.942
|
NM_053042
|
ZNF518B
|
zinc finger protein 518B
|
|
chr10_+_102749597
|
0.942
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr10_+_124211353
|
0.933
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr7_+_55400634
|
0.933
|
NM_018697
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial)
|
|
chr1_-_225572419
|
0.932
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr9_-_25668230
|
0.932
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr1_-_199704773
|
0.931
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr10_-_126839546
|
0.929
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr11_-_1974992
|
0.926
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr2_-_161058550
|
0.924
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr13_-_76358370
|
0.923
|
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr3_+_144321286
|
0.922
|
NM_004267
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2
|
|
chr16_-_73842884
|
0.922
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr2_-_11523666
|
0.917
|
|
E2F6
|
E2F transcription factor 6
|
|
chr15_-_53822468
|
0.914
|
NM_173814
|
PRTG
|
protogenin
|
|
chr12_-_6535481
|
0.907
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr3_+_124268545
|
0.906
|
NM_006810
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr14_-_24588916
|
0.905
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr1_-_226202210
|
0.905
|
NM_003395
|
WNT9A
|
wingless-type MMTV integration site family, member 9A
|
|
chr9_-_35679902
|
0.899
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr16_-_30983909
|
0.888
|
NM_001172669
NM_001172670
|
ZNF668
|
zinc finger protein 668
|
|
chr1_+_26220852
|
0.887
|
NM_004455
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr13_+_112704028
|
0.884
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr10_-_118021532
|
0.881
|
NM_145793
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr6_-_132314004
|
0.879
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr6_-_170735593
|
0.877
|
NM_002598
NM_144781
|
PDCD2
|
programmed cell death 2
|
|
chr10_+_4858321
|
0.869
|
NM_001040177
|
AKR1E2
|
aldo-keto reductase family 1, member E2
|
|
chr4_+_39734902
|
0.869
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr2_-_110665188
|
0.868
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr1_-_23376860
|
0.860
|
|
LUZP1
|
leucine zipper protein 1
|
|
chr2_-_11523727
|
0.854
|
NM_198256
|
E2F6
|
E2F transcription factor 6
|
|
chr9_-_35679799
|
0.851
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr15_+_72005847
|
0.851
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr2_-_219791902
|
0.851
|
NM_005689
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr1_-_151848039
|
0.845
|
|
S100A16
|
S100 calcium binding protein A16
|
|
chr13_-_109757442
|
0.844
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr13_+_112704051
|
0.842
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr8_+_9450800
|
0.842
|
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr7_+_154720416
|
0.840
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr13_-_20533629
|
0.839
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr1_-_8008934
|
0.834
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr19_+_23091616
|
0.832
|
|
ZNF730
|
zinc finger protein 730
|
|
chr8_-_13178327
|
0.830
|
|
|
|
|
chr11_-_1975583
|
0.828
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr11_-_86061003
|
0.826
|
NM_006680
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr10_+_103338030
|
0.824
|
NM_015448
|
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse)
|
|
chr8_-_145085616
|
0.823
|
NM_201384
|
PLEC
|
plectin
|
|
chr16_+_15435825
|
0.823
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr11_-_1974829
|
0.823
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr18_+_7557313
|
0.820
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr1_+_10193267
|
0.816
|
NM_015074
NM_183416
|
KIF1B
|
kinesin family member 1B
|
|
chr1_-_925215
|
0.814
|
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr1_+_176329578
|
0.813
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr6_-_166960690
|
0.811
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr4_-_1655515
|
0.810
|
NM_001013622
NM_001174070
|
FAM53A
|
family with sequence similarity 53, member A
|
|
chr10_+_23768203
|
0.810
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr11_+_76455574
|
0.805
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chr11_-_86060812
|
0.804
|
NM_001014811
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr13_+_112704077
|
0.803
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr9_+_91409746
|
0.802
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr1_-_36589744
|
0.802
|
|
|
|
|
chr12_-_54409107
|
0.796
|
NM_001040034
NM_001780
|
CD63
|
CD63 molecule
|
|
chr16_-_63713382
|
0.792
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr12_+_88626770
|
0.791
|
|
LOC338758
|
hypothetical LOC338758
|
|
chr8_-_41774296
|
0.790
|
NM_000037
NM_020475
NM_020476
NM_020477
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr1_-_94475504
|
0.788
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr10_-_103337968
|
0.787
|
NM_001174084
NM_001174085
NM_013274
|
POLL
|
polymerase (DNA directed), lambda
|
|
chr4_+_2783743
|
0.786
|
NM_001145855
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr11_-_118693021
|
0.784
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr1_+_51206819
|
0.782
|
NM_001262
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr22_+_21094094
|
0.781
|
|
|
|
|
chr3_+_151171755
|
0.780
|
|
LOC646903
|
hypothetical LOC646903
|
|
chr9_+_36126667
|
0.780
|
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr11_-_832507
|
0.775
|
NM_021128
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa
|
|
chr11_+_832821
|
0.775
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr4_-_125853166
|
0.771
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr17_-_1478859
|
0.765
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|