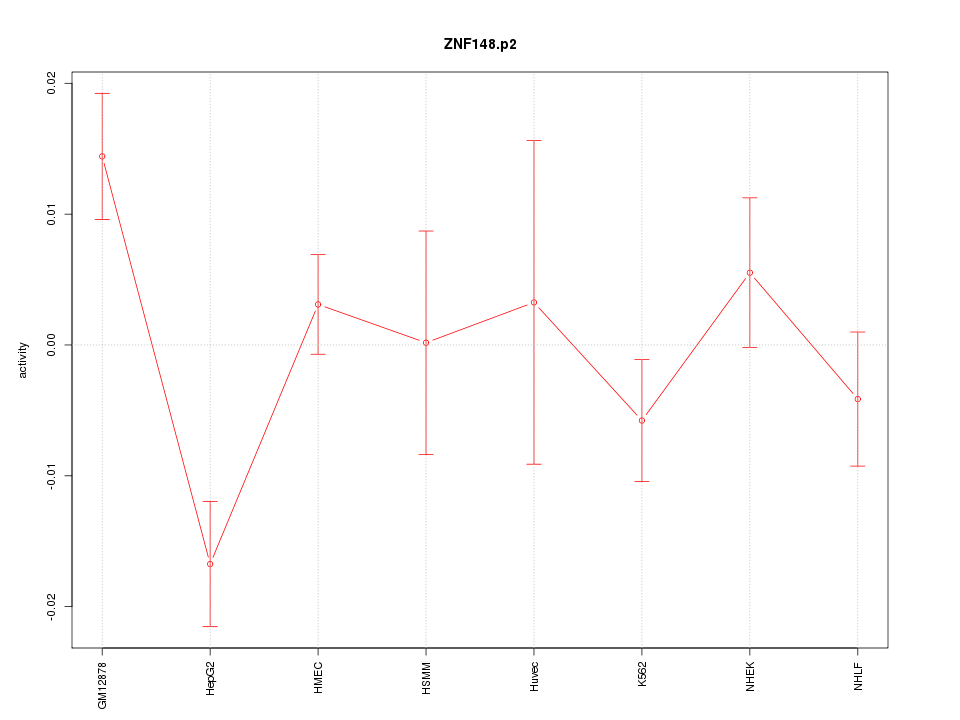
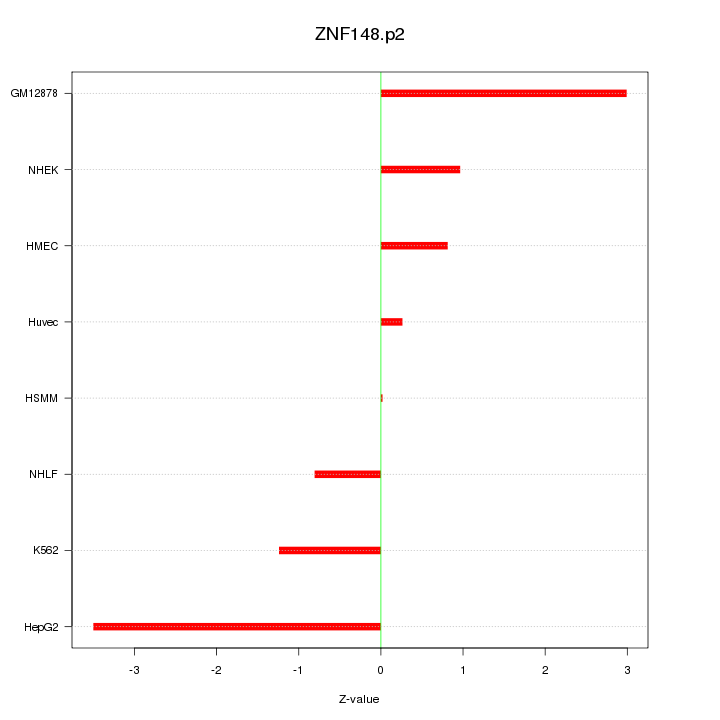
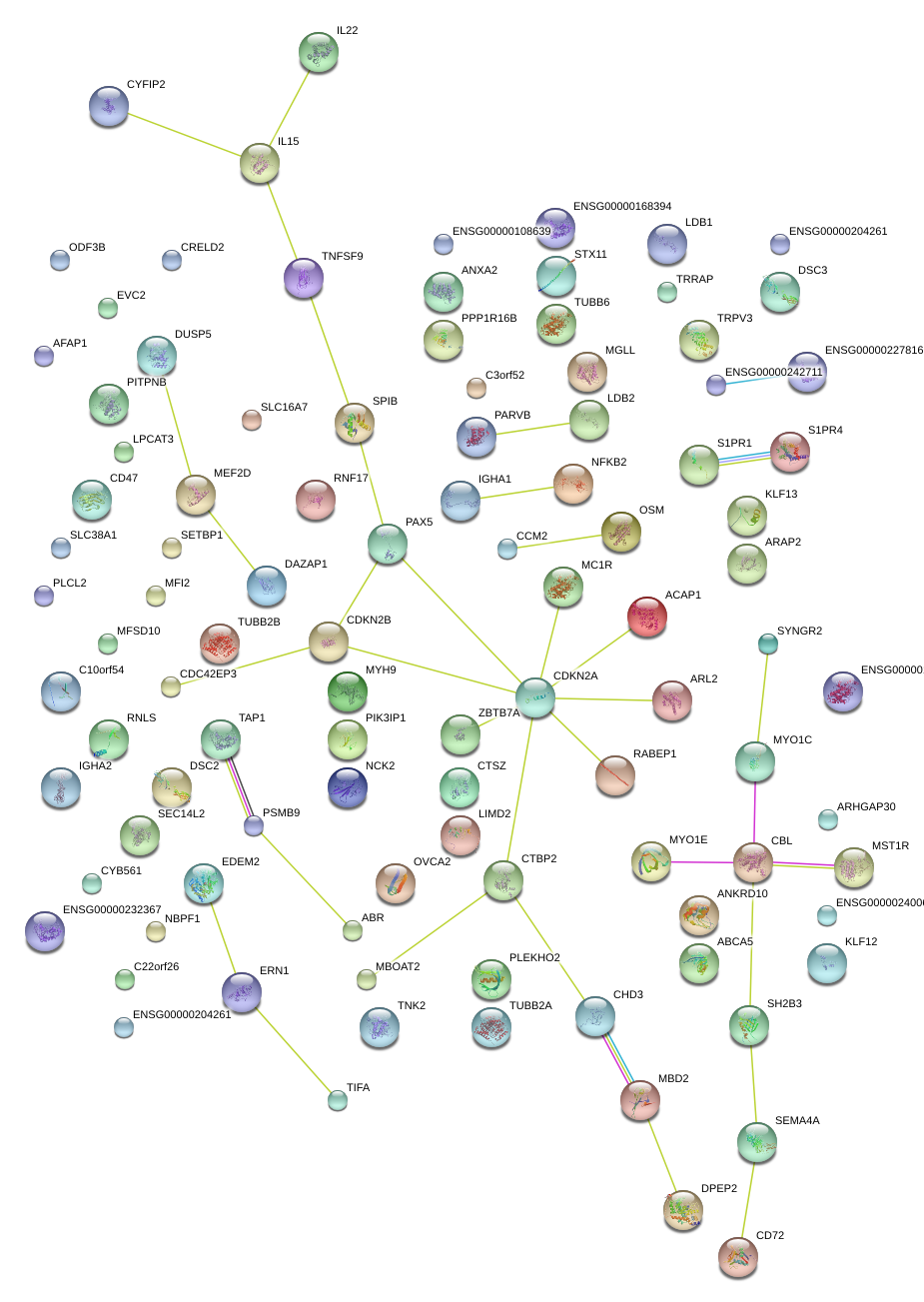
|
chr22_+_42751487
|
1.760
|
NM_013327
|
PARVB
|
parvin, beta
|
|
chr20_-_57015547
|
1.561
|
|
CTSZ
|
cathepsin Z
|
|
chr17_-_3407780
|
1.421
|
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr6_-_32929570
|
1.394
|
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr17_-_3408038
|
1.358
|
NM_145068
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr3_-_49916304
|
1.314
|
NM_002447
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase)
|
|
chr17_-_1029758
|
1.241
|
NM_021962
|
ABR
|
active BCR-related gene
|
|
chr20_+_36867708
|
1.234
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr6_-_32929721
|
1.228
|
NM_000593
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr12_+_110328126
|
1.176
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr3_+_198214173
|
1.128
|
|
LOC100507057
MFI2
|
hypothetical LOC100507057
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5
|
|
chr6_-_32929390
|
1.097
|
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr4_+_142777172
|
1.075
|
NM_000585
NM_172174
|
IL15
|
interleukin 15
|
|
chr6_+_32929902
|
1.072
|
NM_002800
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2)
|
|
chr22_-_49317408
|
1.047
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr4_-_2906361
|
1.044
|
NM_001146069
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr12_+_58276114
|
1.035
|
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr10_-_73203202
|
1.035
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr3_+_16901455
|
1.031
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr15_+_29406335
|
1.010
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr3_-_129023850
|
0.982
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr6_+_144513324
|
0.975
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr4_-_2906256
|
0.968
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr4_-_2906335
|
0.966
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr17_-_59561207
|
0.964
|
NM_001433
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1
|
|
chr22_-_28992787
|
0.959
|
NM_020530
|
OSM
|
oncostatin M
|
|
chr17_+_7180571
|
0.923
|
NM_014716
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1
|
|
chr15_-_58477407
|
0.921
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr18_-_26876622
|
0.915
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr22_-_35113947
|
0.872
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr17_-_1335763
|
0.871
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr5_+_156625629
|
0.866
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr2_-_9061193
|
0.866
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr1_+_154390939
|
0.863
|
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr19_+_3129735
|
0.859
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chr2_+_105727836
|
0.851
|
NM_001004722
NM_003581
|
NCK2
|
NCK adaptor protein 2
|
|
chr1_+_101474892
|
0.850
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr4_-_113426332
|
0.845
|
NM_052864
|
TIFA
|
TRAF-interacting protein with forkhead-associated domain
|
|
chr6_-_20320607
|
0.843
|
NM_001080480
|
MBOAT1
|
membrane bound O-acyltransferase domain containing 1
|
|
chr22_+_48698337
|
0.842
|
|
CRELD2
|
cysteine-rich with EGF-like domains 2
|
|
chr10_+_104145421
|
0.840
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr4_-_2906341
|
0.836
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr17_-_59131194
|
0.811
|
|
LIMD2
|
LIM domain containing 2
|
|
chr19_+_45807909
|
0.799
|
|
|
|
|
chr4_-_35922288
|
0.793
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr3_-_109292444
|
0.793
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr4_+_2906403
|
0.784
|
|
C4orf10
|
chromosome 4 open reading frame 10
|
|
chr3_+_113287864
|
0.780
|
NM_001171747
NM_024616
|
C3orf52
|
chromosome 3 open reading frame 52
|
|
chr18_-_50004770
|
0.780
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr2_-_37752731
|
0.778
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr22_-_35113876
|
0.776
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr22_-_35113923
|
0.774
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr10_-_90332927
|
0.771
|
NM_001031709
NM_018363
|
RNLS
|
renalase, FAD-dependent amine oxidase
|
|
chr17_+_1892026
|
0.770
|
NM_080822
|
OVCA2
|
ovarian tumor suppressor candidate 2
|
|
chr17_-_64834780
|
0.769
|
NM_172232
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr17_+_73676219
|
0.768
|
NM_004710
|
SYNGR2
|
synaptogyrin 2
|
|
chr10_-_103869923
|
0.759
|
NM_001113407
|
LDB1
|
LIM domain binding 1
|
|
chr22_+_44828338
|
0.754
|
|
|
|
|
chr1_-_154727014
|
0.752
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr9_-_21984414
|
0.751
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr18_+_12298189
|
0.749
|
|
TUBB6
|
tubulin, beta 6
|
|
chr4_-_7991992
|
0.744
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr10_+_112247614
|
0.740
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr18_+_12298242
|
0.740
|
NM_032525
|
TUBB6
|
tubulin, beta 6
|
|
chr22_+_48698282
|
0.738
|
NM_001135101
NM_024324
|
CRELD2
|
cysteine-rich with EGF-like domains 2
|
|
chr22_-_44828636
|
0.737
|
NM_018280
|
C22orf26
|
chromosome 22 open reading frame 26
|
|
chr1_-_159306037
|
0.737
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr19_-_4016467
|
0.714
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr4_-_5761194
|
0.713
|
NM_147127
|
EVC2
|
Ellis van Creveld syndrome 2
|
|
chr2_-_37752272
|
0.712
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr11_+_64538161
|
0.709
|
NM_001667
|
ARL2-SNX15
ARL2
|
ARL2-SNX15 read-through transcript
ADP-ribosylation factor-like 2
|
|
chr17_+_5126275
|
0.709
|
NM_001083585
NM_004703
|
RABEP1
|
rabaptin, RAB GTPase binding effector protein 1
|
|
chr4_-_2905649
|
0.706
|
NM_001120
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr1_-_159306273
|
0.700
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr19_+_1358750
|
0.698
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr11_+_118582184
|
0.695
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr3_-_197106828
|
0.687
|
NM_001010938
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr22_-_35113797
|
0.682
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr17_+_7728819
|
0.680
|
NM_001005271
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr17_-_58877276
|
0.680
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr1_-_159306210
|
0.679
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr20_-_33329319
|
0.678
|
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr22_-_26645150
|
0.678
|
|
PITPNB
|
phosphatidylinositol transfer protein, beta
|
|
chr12_-_44949046
|
0.677
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr13_-_73606362
|
0.675
|
|
KLF12
|
Kruppel-like factor 12
|
|
chr19_+_55614006
|
0.662
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr19_+_6482009
|
0.659
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr11_+_118582231
|
0.658
|
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr17_-_59561094
|
0.658
|
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1
|
|
chr14_-_105245959
|
0.657
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr22_-_30016707
|
0.656
|
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr7_+_98314020
|
0.652
|
NM_003496
|
TRRAP
|
transformation/transcription domain-associated protein
|
|
chr18_+_40513889
|
0.651
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr10_-_126839056
|
0.651
|
NM_001329
|
CTBP2
|
C-terminal binding protein 2
|
|
chr9_-_35608300
|
0.648
|
NM_001782
|
CD72
|
CD72 molecule
|
|
chr13_-_110365416
|
0.647
|
NM_017664
|
ANKRD10
|
ankyrin repeat domain 10
|
|
chr15_+_62921134
|
0.645
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr9_-_122731263
|
0.643
|
NM_001190945
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr2_-_24436795
|
0.642
|
NM_006277
NM_019595
NM_147152
|
ITSN2
|
intersectin 2
|
|
chr2_+_238265791
|
0.640
|
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr17_+_6858779
|
0.628
|
NM_001142798
NM_001142799
NM_174893
|
C17orf49
|
chromosome 17 open reading frame 49
|
|
chr1_+_101475149
|
0.628
|
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr15_-_32417233
|
0.622
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr19_-_50838824
|
0.620
|
NM_001193269
|
EML2
|
echinoderm microtubule associated protein like 2
|
|
chr12_+_622374
|
0.618
|
|
LOC100049716
|
hypothetical LOC100049716
|
|
chr5_+_108112537
|
0.618
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr1_+_27972269
|
0.616
|
NM_177424
|
STX12
|
syntaxin 12
|
|
chr19_+_627346
|
0.615
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr7_+_128365226
|
0.613
|
NM_001098629
NM_001098630
|
IRF5
|
interferon regulatory factor 5
|
|
chr17_-_1336073
|
0.612
|
|
MYO1C
|
myosin IC
|
|
chr12_-_44948976
|
0.611
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr20_-_4752067
|
0.608
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr7_+_90063543
|
0.604
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr3_+_160964574
|
0.602
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr1_-_159306369
|
0.602
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr6_-_30762957
|
0.601
|
|
KIAA1949
|
KIAA1949
|
|
chr8_+_32525269
|
0.600
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr1_-_159306327
|
0.600
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr9_-_21984379
|
0.599
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr1_-_26105477
|
0.591
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr9_+_138900795
|
0.589
|
|
TRAF2
|
TNF receptor-associated factor 2
|
|
chr10_+_16518947
|
0.589
|
NM_001001484
NM_030664
|
PTER
|
phosphotriesterase related
|
|
chr1_+_158441770
|
0.587
|
|
PEA15
|
phosphoprotein enriched in astrocytes 15
|
|
chr13_-_110365336
|
0.587
|
|
ANKRD10
|
ankyrin repeat domain 10
|
|
chr7_-_102044374
|
0.584
|
|
RASA4
|
RAS p21 protein activator 4
|
|
chr15_+_73281286
|
0.576
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr7_-_3049872
|
0.574
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr9_-_21984329
|
0.572
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr10_+_31648103
|
0.571
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr2_+_84986273
|
0.570
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr2_+_84986510
|
0.568
|
|
TMSB10
|
thymosin beta 10
|
|
chr10_+_88720477
|
0.567
|
NM_133447
|
AGAP11
|
ankyrin repeat and GTPase domain Arf GTPase activating protein 11
|
|
chr11_+_100063061
|
0.560
|
NM_152432
|
ARHGAP42
|
Rho GTPase activating protein 42
|
|
chr6_-_30762385
|
0.557
|
|
KIAA1949
|
KIAA1949
|
|
chr10_-_31647925
|
0.557
|
|
LOC220930
|
hypothetical LOC220930
|
|
chr21_-_45173061
|
0.556
|
NM_001127491
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr19_-_48976852
|
0.556
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr4_-_53220073
|
0.555
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr5_+_127447306
|
0.554
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr6_-_137154859
|
0.553
|
|
|
|
|
chr2_+_219453487
|
0.553
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr11_-_118582098
|
0.553
|
|
CCDC153
|
coiled-coil domain containing 153
|
|
chr8_+_86563382
|
0.551
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr15_+_73281249
|
0.550
|
NM_015492
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr11_+_527521
|
0.550
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr15_+_73281288
|
0.550
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr1_+_13899272
|
0.543
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr18_+_12298236
|
0.541
|
|
TUBB6
|
tubulin, beta 6
|
|
chr1_+_231530136
|
0.540
|
NM_032435
|
KIAA1804
|
mixed lineage kinase 4
|
|
chr11_+_66813337
|
0.540
|
NM_207354
|
ANKRD13D
|
ankyrin repeat domain 13 family, member D
|
|
chr1_+_15609009
|
0.539
|
|
EFHD2
|
EF-hand domain family, member D2
|
|
chrX_-_153767625
|
0.538
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr19_+_2047792
|
0.536
|
NM_001031735
NM_001039846
|
IZUMO4
|
IZUMO family member 4
|
|
chr4_-_2905448
|
0.536
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr8_-_135794386
|
0.533
|
NM_001174157
NM_020863
|
ZFAT
|
zinc finger and AT hook domain containing
|
|
chr11_+_2355099
|
0.531
|
|
CD81
|
CD81 molecule
|
|
chr21_+_44560779
|
0.529
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr6_-_86409701
|
0.525
|
NM_001159677
NM_006372
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr5_-_127447197
|
0.524
|
|
FLJ33630
|
hypothetical LOC644873
|
|
chr5_-_127446557
|
0.523
|
|
FLJ33630
|
hypothetical LOC644873
|
|
chr14_+_60858151
|
0.522
|
NM_006255
|
PRKCH
|
protein kinase C, eta
|
|
chr1_+_26479030
|
0.520
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr11_-_33752468
|
0.519
|
NM_012175
NM_033406
|
FBXO3
|
F-box protein 3
|
|
chr10_+_47875229
|
0.519
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr5_+_154115222
|
0.518
|
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr1_+_15608974
|
0.515
|
NM_024329
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr22_+_40800193
|
0.513
|
NM_001002034
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr8_+_90983934
|
0.507
|
NM_001126111
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2
|
|
chr7_-_75826060
|
0.505
|
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr15_+_88874098
|
0.503
|
NM_001042574
NM_022769
|
CRTC3
|
CREB regulated transcription coactivator 3
|
|
chr12_-_119826073
|
0.502
|
|
SPPL3
|
signal peptide peptidase 3
|
|
chr1_-_65204791
|
0.500
|
|
JAK1
|
Janus kinase 1
|
|
chr3_-_172660545
|
0.499
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr20_+_32610161
|
0.498
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr1_-_181826633
|
0.494
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr12_-_56157898
|
0.493
|
NM_001080156
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr22_+_40800213
|
0.493
|
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr22_+_48971067
|
0.493
|
|
TRABD
|
TraB domain containing
|
|
chr7_-_102044419
|
0.492
|
NM_001079877
NM_006989
|
RASA4
|
RAS p21 protein activator 4
|
|
chr16_-_4406608
|
0.491
|
|
CORO7
|
coronin 7
|
|
chr11_-_64402501
|
0.489
|
|
EHD1
|
EH-domain containing 1
|
|
chr11_+_63864263
|
0.487
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr19_+_2047930
|
0.485
|
|
IZUMO4
|
IZUMO family member 4
|
|
chr5_+_10617431
|
0.485
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr1_+_30964199
|
0.480
|
|
LOC100129196
|
hypothetical LOC100129196
|
|
chr2_-_166358951
|
0.478
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr7_+_100302885
|
0.478
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr19_-_2047280
|
0.477
|
|
MOBKL2A
|
MOB1, Mps One Binder kinase activator-like 2A (yeast)
|
|
chr14_-_50367513
|
0.477
|
NM_016350
NM_020921
NM_182944
NM_182946
|
NIN
|
ninein (GSK3B interacting protein)
|
|
chr3_+_52254782
|
0.474
|
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M
|
|
chr7_-_143523582
|
0.474
|
NM_001003702
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35
|
|
chr2_-_166359016
|
0.473
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr9_+_133259330
|
0.470
|
|
PRRC2B
|
proline-rich coiled-coil 2B
|
|
chr1_+_198975227
|
0.470
|
NM_203459
|
CAMSAP1L1
|
calmodulin regulated spectrin-associated protein 1-like 1
|
|
chr12_-_44948800
|
0.470
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr11_-_72111027
|
0.468
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chrX_+_129133234
|
0.468
|
NM_004794
|
RAB33A
|
RAB33A, member RAS oncogene family
|
|
chr10_-_103864651
|
0.467
|
NM_003893
|
LDB1
|
LIM domain binding 1
|
|
chr17_+_7151368
|
0.467
|
NM_001970
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr3_-_198767202
|
0.466
|
NM_004051
NM_203314
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|