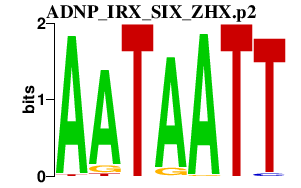
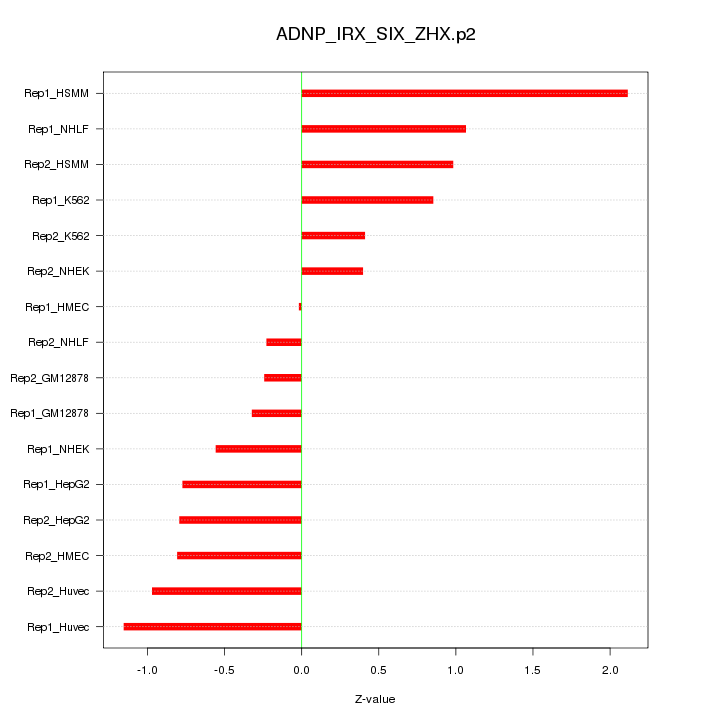
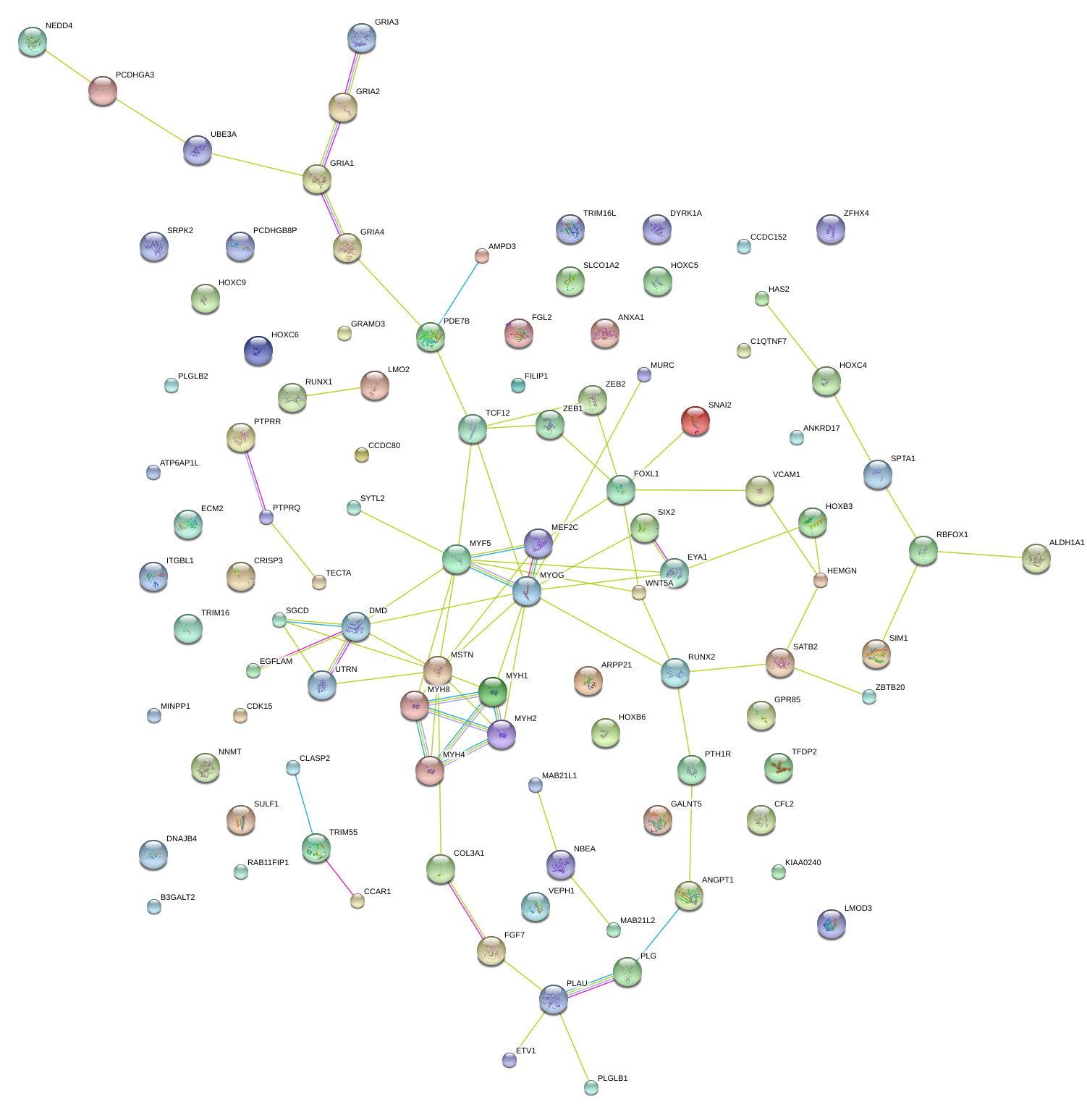
|
chr3_-_55489973
|
2.282
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr3_-_115960807
|
1.558
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr16_+_7322751
|
1.405
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr12_+_52697160
|
1.397
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr11_+_10433241
|
1.276
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr3_-_143230124
|
1.255
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr9_+_102380156
|
1.224
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr5_+_125786975
|
1.169
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr8_+_77756061
|
1.153
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr12_+_52696908
|
1.139
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr2_-_190635566
|
1.132
|
NM_005259
|
MSTN
|
myostatin
|
|
chr15_+_47502666
|
1.115
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr13_+_100902942
|
1.092
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr15_-_53996597
|
1.086
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr8_+_77756077
|
1.081
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr11_-_85107569
|
1.063
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_+_155686333
|
1.029
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr8_-_72431274
|
1.009
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr17_-_10265991
|
1.009
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr1_-_191422346
|
0.979
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr2_-_144994038
|
0.944
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_+_556968
|
0.849
|
|
|
|
|
chr2_+_157822355
|
0.823
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr4_-_74307637
|
0.802
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr16_+_85169615
|
0.801
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr15_+_55298908
|
0.749
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr12_+_52680143
|
0.726
|
NM_006897
|
HOXC9
|
homeobox C9
|
|
chr11_+_113672378
|
0.719
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr1_+_78243181
|
0.714
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr6_-_101018271
|
0.710
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr8_-_49996353
|
0.704
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr11_+_113672407
|
0.701
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr2_+_200031333
|
0.699
|
|
|
|
|
chr2_-_200030412
|
0.696
|
|
SATB2
|
SATB homeobox 2
|
|
chr2_-_200033445
|
0.687
|
|
SATB2
|
SATB homeobox 2
|
|
chr3_-_33675484
|
0.655
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chrX_-_153698930
|
0.648
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr9_-_99740243
|
0.647
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr11_-_33870411
|
0.644
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr13_-_34948758
|
0.638
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr11_-_85108030
|
0.633
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr8_-_37847095
|
0.627
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr5_+_38481392
|
0.611
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr13_+_34948977
|
0.595
|
|
NBEA
|
neurobeachin
|
|
chr3_-_113841200
|
0.587
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_-_76667045
|
0.583
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr3_+_35696119
|
0.579
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr15_-_23201740
|
0.579
|
NM_130838
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr7_-_13992351
|
0.567
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr5_+_42792676
|
0.565
|
NM_001134848
|
CCDC152
|
coiled-coil domain containing 152
|
|
chr8_+_67201684
|
0.564
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr2_-_200030944
|
0.549
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr6_-_76259976
|
0.548
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr5_-_88235624
|
0.548
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_+_70541631
|
0.547
|
|
SULF1
|
sulfatase 1
|
|
chr7_-_112514990
|
0.527
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr5_+_152850276
|
0.526
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr12_+_79362256
|
0.523
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr11_+_113671744
|
0.522
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr14_-_34252695
|
0.516
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr9_-_74757735
|
0.515
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr21_-_35343456
|
0.514
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr9_-_94338072
|
0.513
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr7_-_104696678
|
0.512
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr2_-_87102453
|
0.509
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr12_+_79634820
|
0.509
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr5_+_81636921
|
0.509
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr21_-_35343428
|
0.508
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr21_+_37713076
|
0.498
|
NM_130436
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr4_+_14985272
|
0.493
|
NM_001135171
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr2_+_189547377
|
0.488
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr21_-_35343500
|
0.487
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr11_+_120478584
|
0.482
|
NM_005422
|
TECTA
|
tectorin alpha
|
|
chr17_-_44037332
|
0.480
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr21_-_35343448
|
0.477
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr12_-_21379098
|
0.476
|
NM_021094
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr7_-_112515068
|
0.471
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr1_+_557269
|
0.469
|
|
|
|
|
chr6_+_136214495
|
0.468
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr10_+_89257569
|
0.465
|
NM_001178118
|
MINPP1
|
multiple inositol-polyphosphate phosphatase 1
|
|
chr8_-_122722674
|
0.464
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr6_+_42896771
|
0.460
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chrX_-_33139349
|
0.459
|
NM_004006
|
DMD
|
dystrophin
|
|
chr17_-_44006808
|
0.456
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr21_-_35343331
|
0.456
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr17_-_15495534
|
0.453
|
|
TRIM16
|
tripartite motif containing 16
|
|
chr8_-_72436519
|
0.452
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr21_-_35174742
|
0.452
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr13_+_34948885
|
0.449
|
|
NBEA
|
neurobeachin
|
|
chr8_+_70541412
|
0.444
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr7_-_13996166
|
0.440
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr3_-_113841448
|
0.440
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr2_-_45090025
|
0.434
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr22_-_34564611
|
0.433
|
|
RPL41P5
|
ribosomal protein L41 pseudogene 5
|
|
chr5_+_140772691
|
0.431
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr3_+_35697432
|
0.430
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr3_-_69254226
|
0.427
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr17_-_10362583
|
0.427
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr1_-_201321999
|
0.427
|
NM_002479
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr17_+_59875623
|
0.426
|
NM_001085423
|
C17orf60
|
chromosome 17 open reading frame 60
|
|
chr5_+_140698537
|
0.424
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr8_-_108579270
|
0.424
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr10_+_70172257
|
0.415
|
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|
|
chr10_+_75343352
|
0.413
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr22_+_21006814
|
0.413
|
|
|
|
|
chr3_-_158704056
|
0.412
|
NM_001167915
NM_001167916
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr1_+_100957883
|
0.409
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr5_-_134291882
|
0.407
|
|
|
|
|
chr6_-_49820030
|
0.405
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr3_+_46894239
|
0.403
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr7_-_112515035
|
0.398
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr1_-_156923111
|
0.397
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr2_+_202379442
|
0.395
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr11_+_65027721
|
0.394
|
|
|
|
|
chr11_-_71424614
|
0.390
|
|
|
|
|
chr9_+_74956614
|
0.385
|
|
ANXA1
|
annexin A1
|
|
chr3_-_113841967
|
0.384
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr6_-_28429950
|
0.383
|
NM_145909
|
ZNF323
|
zinc finger protein 323
|
|
chr11_+_124539768
|
0.381
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr1_+_202033273
|
0.378
|
NM_001174108
|
ZBED6
|
zinc finger, BED-type containing 6
|
|
chr1_-_53972464
|
0.375
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr9_+_117955890
|
0.374
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr10_+_49279647
|
0.373
|
NM_002750
NM_139046
NM_139047
NM_139049
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr5_-_75044001
|
0.372
|
NM_152408
|
POC5
|
POC5 centriolar protein homolog (Chlamydomonas)
|
|
chr5_+_125786717
|
0.369
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr3_-_166278969
|
0.366
|
NM_001041
|
SI
|
sucrase-isomaltase (alpha-glucosidase)
|
|
chr15_+_46285789
|
0.364
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr3_-_71262426
|
0.363
|
|
FOXP1
|
forkhead box P1
|
|
chr5_+_71438796
|
0.356
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr3_-_113841941
|
0.353
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr4_-_153493133
|
0.347
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr11_+_123600553
|
0.344
|
NM_001007249
|
OR8G2
|
olfactory receptor, family 8, subfamily G, member 2
|
|
chr12_-_52281050
|
0.344
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr11_+_117982422
|
0.341
|
NM_015157
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr17_-_44026321
|
0.341
|
|
HOXB5
|
homeobox B5
|
|
chr11_+_65023556
|
0.340
|
|
|
|
|
chr17_-_10393664
|
0.339
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr10_+_68355797
|
0.339
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chrX_-_77801480
|
0.339
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr2_-_99284070
|
0.335
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr6_-_89984214
|
0.334
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr8_+_32905027
|
0.334
|
|
|
|
|
chr1_+_160868822
|
0.333
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr4_-_152366970
|
0.328
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr1_+_160868932
|
0.325
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr12_-_68379331
|
0.322
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr2_+_220087135
|
0.318
|
NM_018674
NM_182847
|
ACCN4
|
amiloride-sensitive cation channel 4, pituitary
|
|
chr3_-_71262405
|
0.317
|
|
FOXP1
|
forkhead box P1
|
|
chr1_-_115039614
|
0.316
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr9_+_129900637
|
0.315
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr10_+_44726635
|
0.314
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr1_-_177378801
|
0.313
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr5_-_111340421
|
0.310
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_15145001
|
0.309
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr1_-_153480980
|
0.309
|
NM_001005741
NM_001005742
NM_001171811
|
GBA
|
glucosidase, beta, acid
|
|
chr17_-_46479127
|
0.305
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr9_+_129900590
|
0.301
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr6_-_46401174
|
0.300
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr1_+_154120170
|
0.299
|
|
SYT11
|
synaptotagmin XI
|
|
chr15_-_70350609
|
0.299
|
NM_020214
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6
|
|
chr22_+_40286957
|
0.297
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chrX_-_19727775
|
0.296
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr12_-_69434330
|
0.294
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr7_-_135083906
|
0.293
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr2_-_144991556
|
0.292
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr5_+_35892747
|
0.292
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chrX_-_135690699
|
0.291
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr15_-_74091817
|
0.290
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr10_+_70745615
|
0.289
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr5_-_178977441
|
0.289
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr4_+_169250281
|
0.289
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr1_-_16217871
|
0.288
|
NM_014424
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr7_+_129057152
|
0.285
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr1_-_103269356
|
0.284
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr11_-_57173823
|
0.284
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr6_+_158990982
|
0.283
|
NM_001009991
|
SYTL3
|
synaptotagmin-like 3
|
|
chr7_-_13995811
|
0.282
|
|
ETV1
|
ets variant 1
|
|
chr12_+_9871343
|
0.282
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr21_+_29439768
|
0.280
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr7_+_106897737
|
0.280
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr5_+_140742585
|
0.279
|
NM_018920
NM_032087
|
PCDHGA7
|
protocadherin gamma subfamily A, 7
|
|
chr3_+_110338250
|
0.278
|
|
FLJ22763
|
hypothetical LOC401081
|
|
chrX_+_77889861
|
0.278
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr7_+_129634935
|
0.277
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr1_+_84402962
|
0.275
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr17_-_64462976
|
0.274
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr11_-_56951628
|
0.274
|
NM_017611
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr10_+_91051685
|
0.274
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chr3_-_197726587
|
0.274
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr4_+_146820805
|
0.272
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr12_+_43972732
|
0.271
|
NM_001142678
|
ANO6
|
anoctamin 6
|
|
chr2_+_151922350
|
0.271
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr17_-_51155073
|
0.269
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr6_+_158653679
|
0.269
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr2_+_189547358
|
0.268
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr21_-_35343065
|
0.268
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr14_-_80934523
|
0.266
|
NM_033104
|
STON2
|
stonin 2
|
|
chr6_+_144227265
|
0.266
|
NM_001013623
|
C6orf94
|
chromosome 6 open reading frame 94
|
|
chr15_-_70838703
|
0.265
|
|
|
|
|
chr1_-_93030538
|
0.265
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|