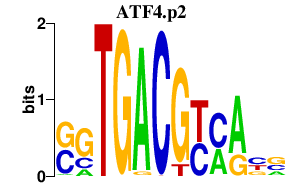
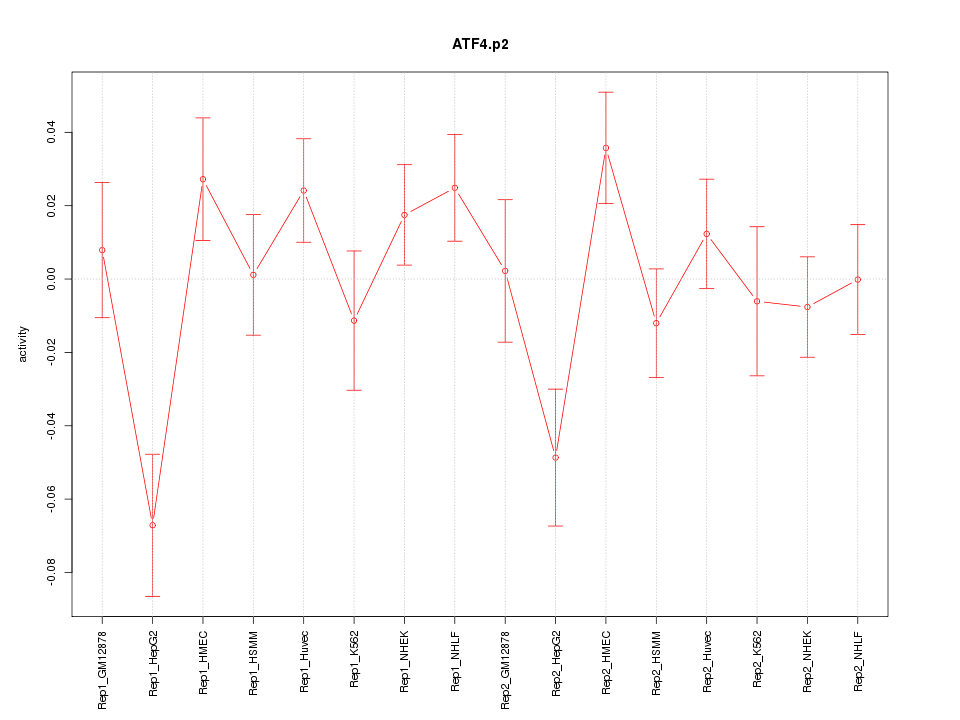
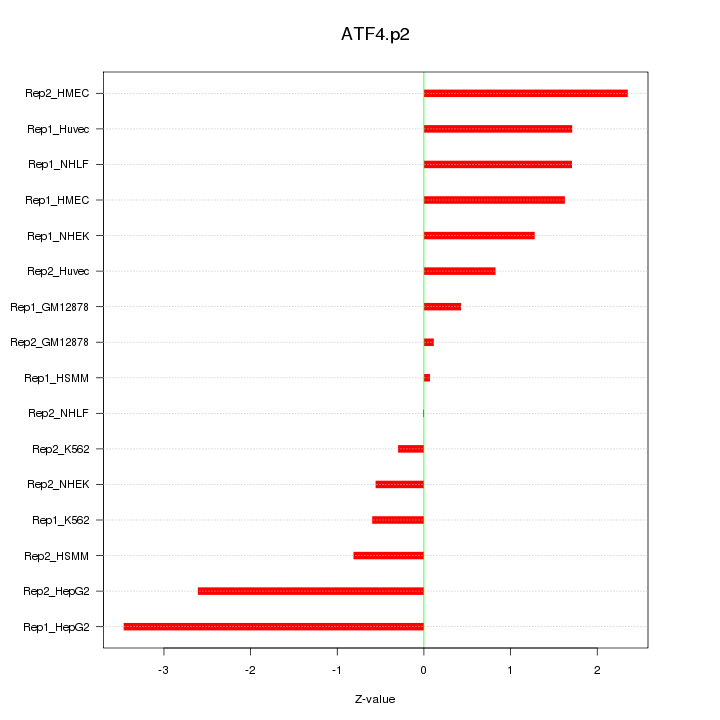
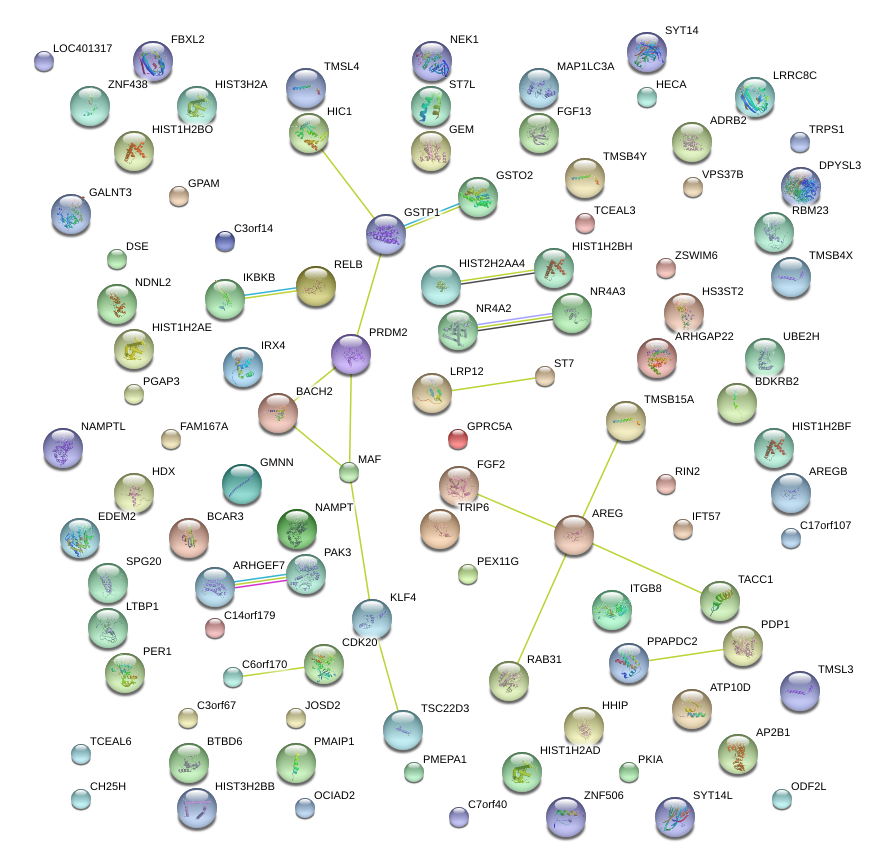
|
chr20_+_32610161
|
6.079
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr7_-_44992729
|
4.511
|
|
C7orf40
|
chromosome 7 open reading frame 40
|
|
chr6_+_139497933
|
3.591
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr12_-_121946635
|
2.846
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr9_-_109291575
|
2.621
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr9_-_109291866
|
2.571
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr20_+_19863716
|
2.430
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr12_-_121946581
|
2.400
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr8_-_95343696
|
2.287
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr8_-_95343716
|
2.267
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chrX_-_83644054
|
2.235
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr7_+_28415495
|
2.115
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr2_-_156897286
|
2.057
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr18_+_55718171
|
2.018
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr14_+_75521806
|
2.008
|
NM_001102564
NM_052873
|
C14orf179
|
chromosome 14 open reading frame 179
|
|
chr4_+_75529716
|
1.941
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr5_+_60663842
|
1.924
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr4_+_75699652
|
1.855
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr4_-_48603501
|
1.828
|
NM_001014446
NM_152398
|
OCIAD2
|
OCIA domain containing 2
|
|
chr7_+_20336849
|
1.767
|
|
ITGB8
|
integrin, beta 8
|
|
chr1_-_86634511
|
1.675
|
NM_001007022
NM_001184765
NM_001184766
NM_020729
|
ODF2L
|
outer dense fiber of sperm tails 2-like
|
|
chr11_+_67107852
|
1.653
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr20_-_33329319
|
1.642
|
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chrX_+_12903145
|
1.618
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr8_+_128875902
|
1.604
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr12_-_121946561
|
1.603
|
|
|
|
|
chr1_+_13899272
|
1.568
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr10_-_90957028
|
1.542
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr20_-_55718351
|
1.537
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr10_-_49483002
|
1.501
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr8_+_38763878
|
1.469
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr12_+_12935619
|
1.422
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr10_+_106018620
|
1.354
|
NM_001191013
NM_183239
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr19_-_7459867
|
1.341
|
NM_080662
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr5_-_1935764
|
1.327
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr8_+_38763911
|
1.326
|
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chrX_+_102749489
|
1.306
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr4_+_145786622
|
1.299
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr10_-_113933457
|
1.295
|
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr12_+_12935759
|
1.294
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr8_-_11361594
|
1.289
|
NM_053279
|
FAM167A
|
family with sequence similarity 167, member A
|
|
chr16_+_22732982
|
1.287
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr7_-_129379947
|
1.276
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast)
|
|
chr7_+_100302885
|
1.271
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr8_+_79591074
|
1.261
|
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chrX_-_106846684
|
1.260
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chrX_+_110226030
|
1.229
|
NM_002578
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr4_+_123967389
|
1.223
|
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr3_-_109423858
|
1.202
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr14_+_95740916
|
1.145
|
|
BDKRB2
|
bradykinin receptor B2
|
|
chr15_-_27349300
|
1.138
|
NM_138704
|
NDNL2
|
necdin-like 2
|
|
chrX_+_103103848
|
1.123
|
NM_194324
|
TMSB15B
|
thymosin beta 15B
|
|
chr4_+_123967265
|
1.118
|
NM_002006
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr1_-_93919789
|
1.108
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr19_-_55706156
|
1.090
|
NM_138334
|
JOSD2
|
Josephin domain containing 2
|
|
chr9_-_89779180
|
1.087
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr7_+_39739690
|
1.087
|
|
NCRNA00265
|
non-protein coding RNA 265
|
|
chr8_+_42248008
|
1.082
|
NM_001190721
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr20_+_55718619
|
1.081
|
|
|
|
|
chr6_-_91063181
|
1.078
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr20_+_55718718
|
1.074
|
|
|
|
|
chr5_-_146869811
|
1.072
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr17_+_30938391
|
1.057
|
NM_001030006
NM_001282
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr8_+_94998258
|
1.037
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr7_+_20337249
|
1.030
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chrX_-_106846924
|
1.029
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr3_-_59010797
|
1.027
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr4_+_47181987
|
1.019
|
NM_020453
|
ATP10D
|
ATPase, class V, type 10D
|
|
chr8_+_42247978
|
1.018
|
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr16_-_78192111
|
1.014
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr10_-_31360778
|
1.005
|
NM_001143766
NM_001143767
NM_001143768
NM_001143770
NM_001143771
NM_182755
|
ZNF438
|
zinc finger protein 438
|
|
chr14_+_104785898
|
1.003
|
NM_033271
|
BTBD6
|
BTB (POZ) domain containing 6
|
|
chr2_-_166358951
|
0.999
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr17_+_4743645
|
0.996
|
NM_001145536
|
C17orf107
|
chromosome 17 open reading frame 107
|
|
chr1_+_89871281
|
0.994
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr1_+_226712430
|
0.990
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr3_-_59010777
|
0.979
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr13_-_35818491
|
0.967
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr17_+_30938433
|
0.967
|
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr7_-_105712565
|
0.966
|
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr3_+_33293915
|
0.965
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr13_-_35818396
|
0.963
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr5_+_148186348
|
0.962
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr1_-_226712140
|
0.961
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr8_+_42247954
|
0.961
|
NM_001190722
NM_001556
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr8_+_42247991
|
0.959
|
NM_001190720
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr19_+_47593137
|
0.950
|
|
|
|
|
chr14_+_104785936
|
0.948
|
|
BTBD6
|
BTB (POZ) domain containing 6
|
|
chr6_+_116798791
|
0.947
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr3_-_59010740
|
0.945
|
NM_198463
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr18_+_9698299
|
0.943
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr3_+_62279707
|
0.937
|
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr17_-_8000362
|
0.931
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr6_-_91063301
|
0.930
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr4_-_170770111
|
0.920
|
NM_012224
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr14_-_22458134
|
0.915
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr19_-_19793512
|
0.911
|
NM_001099269
NM_001145404
|
ZNF506
|
zinc finger protein 506
|
|
chr14_+_95740768
|
0.910
|
NM_000623
|
BDKRB2
|
bradykinin receptor B2
|
|
chr6_-_121697305
|
0.903
|
NM_152730
|
C6orf170
|
chromosome 6 open reading frame 170
|
|
chr2_+_33025872
|
0.892
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr16_+_22733307
|
0.868
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr8_-_116750277
|
0.853
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr1_+_208178198
|
0.852
|
|
SYT14
|
synaptotagmin XIV
|
|
chr19_+_50196526
|
0.839
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr7_+_116447499
|
0.833
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr17_+_1906262
|
0.814
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr5_-_16561936
|
0.812
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr1_+_91739241
|
0.809
|
NM_001134420
|
CDC7
|
cell division cycle 7 homolog (S. cerevisiae)
|
|
chr11_-_76862197
|
0.804
|
NM_001128620
NM_002576
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr5_+_174084137
|
0.803
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr10_+_17312654
|
0.799
|
|
VIM
|
vimentin
|
|
chr12_+_12935222
|
0.786
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr6_+_56927882
|
0.784
|
|
BEND6
|
BEN domain containing 6
|
|
chr6_-_38715901
|
0.783
|
NM_001099272
NM_052893
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr17_+_58058479
|
0.780
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr1_+_110328922
|
0.779
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr18_+_22060383
|
0.777
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr20_+_19903778
|
0.777
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr5_-_133589670
|
0.773
|
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr5_-_147266257
|
0.770
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr8_-_145097031
|
0.769
|
NM_201380
|
PLEC
|
plectin
|
|
chr3_+_129927716
|
0.756
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr7_+_5598979
|
0.750
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr5_-_146869613
|
0.743
|
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr1_+_40193368
|
0.742
|
NM_001136493
NM_032793
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr1_+_109090868
|
0.741
|
|
STXBP3
|
syntaxin binding protein 3
|
|
chr4_+_170778265
|
0.740
|
NM_001829
NM_173872
|
CLCN3
|
chloride channel 3
|
|
chr1_+_19843261
|
0.734
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr7_+_5598964
|
0.734
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr1_+_154297588
|
0.726
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr22_-_37062114
|
0.724
|
|
CSNK1E
|
casein kinase 1, epsilon
|
|
chr5_+_108112537
|
0.724
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr14_-_60817621
|
0.722
|
|
|
|
|
chr12_+_70519753
|
0.718
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr19_+_55671600
|
0.716
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr15_-_27349233
|
0.711
|
|
NDNL2
|
necdin-like 2
|
|
chr6_+_1556543
|
0.702
|
|
FOXC1
|
forkhead box C1
|
|
chr1_+_40193389
|
0.696
|
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr1_-_208024491
|
0.695
|
NM_152485
|
C1orf74
|
chromosome 1 open reading frame 74
|
|
chr7_-_105712646
|
0.686
|
NM_005746
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr1_+_40193424
|
0.684
|
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr1_+_44171578
|
0.684
|
NM_001136215
NM_057090
NM_057091
|
ARTN
|
artemin
|
|
chr7_+_100111712
|
0.680
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr14_-_87529222
|
0.679
|
|
GALC
|
galactosylceramidase
|
|
chr19_+_18529652
|
0.677
|
|
C19orf50
|
chromosome 19 open reading frame 50
|
|
chr14_-_22458220
|
0.675
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr2_+_192251105
|
0.672
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr7_+_151353707
|
0.667
|
NM_022087
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11)
|
|
chr1_-_65204414
|
0.667
|
NM_002227
|
JAK1
|
Janus kinase 1
|
|
chr1_+_109090817
|
0.666
|
|
STXBP3
|
syntaxin binding protein 3
|
|
chr19_-_60610792
|
0.665
|
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr7_+_151353759
|
0.659
|
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11)
|
|
chr6_+_86216457
|
0.656
|
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr1_+_110328850
|
0.654
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chrX_-_50573759
|
0.651
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr8_+_26296439
|
0.648
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr6_-_38715613
|
0.647
|
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr18_+_11841388
|
0.641
|
NM_020412
|
CHMP1B
|
chromatin modifying protein 1B
|
|
chr1_+_152644471
|
0.640
|
|
IL6R
|
interleukin 6 receptor
|
|
chr14_+_100362204
|
0.640
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr3_+_157877146
|
0.639
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr14_+_100362236
|
0.638
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr19_-_57082907
|
0.637
|
NM_001135590
NM_032679
|
ZNF577
|
zinc finger protein 577
|
|
chr4_+_159812683
|
0.637
|
NM_004453
|
ETFDH
|
electron-transferring-flavoprotein dehydrogenase
|
|
chr3_+_158026769
|
0.634
|
NM_001004316
NM_001193283
|
LEKR1
|
leucine, glutamate and lysine rich 1
|
|
chr1_-_23367934
|
0.632
|
NM_001142546
NM_033631
|
LUZP1
|
leucine zipper protein 1
|
|
chr9_+_101623957
|
0.631
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr7_-_150847698
|
0.629
|
|
RHEB
|
Ras homolog enriched in brain
|
|
chr3_+_129927668
|
0.628
|
NM_004637
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr14_-_54563554
|
0.627
|
NM_001008396
NM_007086
|
WDHD1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr7_-_150847884
|
0.624
|
NM_005614
|
RHEB
|
Ras homolog enriched in brain
|
|
chr1_-_65204791
|
0.622
|
|
JAK1
|
Janus kinase 1
|
|
chr19_+_55671584
|
0.622
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr17_+_30938470
|
0.622
|
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr12_-_91063441
|
0.621
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr4_+_170778303
|
0.621
|
|
CLCN3
|
chloride channel 3
|
|
chr19_-_60610971
|
0.621
|
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr14_-_87529212
|
0.618
|
|
GALC
|
galactosylceramidase
|
|
chr6_+_86216020
|
0.615
|
NM_002526
|
NT5E
|
5'-nucleotidase, ecto (CD73)
|
|
chr9_+_70978970
|
0.615
|
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr19_+_55671594
|
0.614
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr3_+_185538998
|
0.613
|
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr7_+_151353775
|
0.612
|
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11)
|
|
chr13_-_27967215
|
0.611
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr11_+_101723125
|
0.607
|
|
BIRC2
|
baculoviral IAP repeat containing 2
|
|
chr15_-_50259367
|
0.605
|
NM_006578
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr1_-_143643324
|
0.604
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr8_-_98359347
|
0.603
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr6_-_38715567
|
0.599
|
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr8_-_98359248
|
0.598
|
|
TSPYL5
|
TSPY-like 5
|
|
chr1_+_110328955
|
0.597
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr19_+_55671468
|
0.597
|
NM_175063
NM_206538
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr1_+_110328941
|
0.596
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr14_-_54563520
|
0.594
|
|
WDHD1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr17_+_2443652
|
0.592
|
NM_000430
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr19_-_55671273
|
0.590
|
|
FAM71E1
|
family with sequence similarity 71, member E1
|
|
chr1_-_23367918
|
0.585
|
|
LUZP1
|
leucine zipper protein 1
|
|
chr11_+_101291134
|
0.583
|
|
KIAA1377
|
KIAA1377
|
|
chr13_-_73605914
|
0.582
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr11_+_101723157
|
0.582
|
NM_001166
|
BIRC2
|
baculoviral IAP repeat containing 2
|