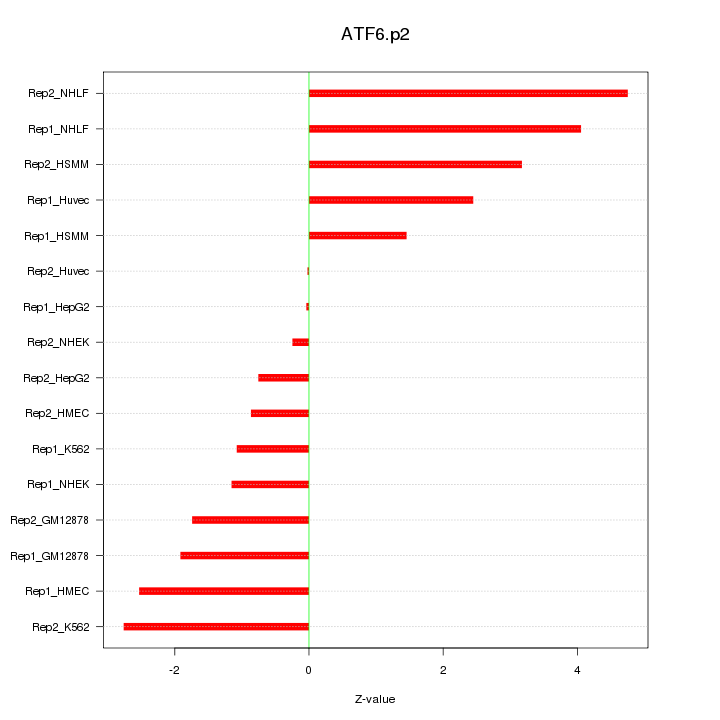
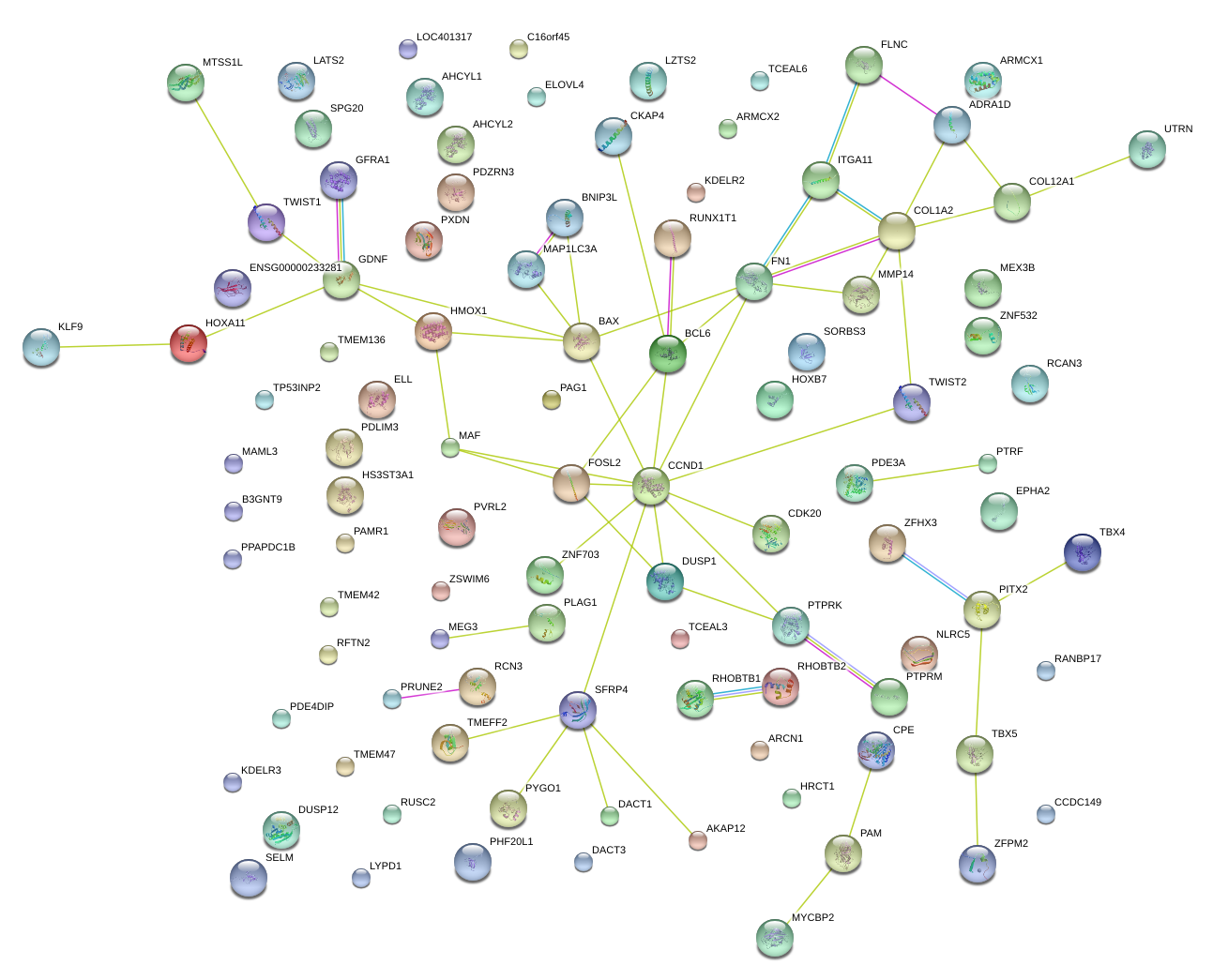
|
chr13_-_20533629
|
3.224
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr19_+_54723044
|
3.139
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr3_-_73756668
|
3.005
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr14_+_100362204
|
2.819
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr7_+_27191534
|
2.703
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr4_-_186693574
|
2.695
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chrX_+_100692071
|
2.619
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr4_-_187884721
|
2.533
|
|
|
|
|
chr13_-_35818777
|
2.439
|
NM_001142295
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr2_+_28469199
|
2.432
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr2_+_28469275
|
2.429
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr4_-_24523605
|
2.324
|
NM_001130726
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr12_-_123568781
|
2.318
|
|
|
|
|
chr2_+_28469172
|
2.295
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr17_-_13445942
|
2.280
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr7_+_93861808
|
2.227
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr14_+_100362236
|
2.216
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr13_-_35818603
|
2.172
|
NM_001142296
NM_015087
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr14_+_22375632
|
2.097
|
NM_004995
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr8_+_106400322
|
2.081
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr14_+_22375656
|
2.002
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr6_+_144649142
|
1.988
|
|
UTRN
|
utrophin
|
|
chr13_-_35818491
|
1.987
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr14_+_22375747
|
1.965
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr3_-_188945921
|
1.963
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr13_-_35818396
|
1.963
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr7_-_27191287
|
1.954
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr14_+_22375753
|
1.951
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr17_+_56884560
|
1.941
|
|
TBX4
|
T-box 4
|
|
chr12_-_113328271
|
1.907
|
NM_181486
|
TBX5
|
T-box 5
|
|
chr9_+_35528890
|
1.905
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr6_+_1336077
|
1.904
|
|
|
|
|
chr7_-_19123787
|
1.892
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr6_-_75972473
|
1.879
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr10_-_62373828
|
1.872
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chrX_-_100801472
|
1.868
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr7_-_19123765
|
1.855
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr8_-_38245694
|
1.780
|
NM_001102559
NM_001102560
NM_032483
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B
|
|
chr7_+_28415495
|
1.758
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr13_-_20533703
|
1.745
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr16_-_65742323
|
1.737
|
NM_033309
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9
|
|
chrX_-_34585287
|
1.732
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr2_-_198248828
|
1.710
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr6_-_128883125
|
1.709
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr15_-_53668341
|
1.678
|
NM_015617
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr10_-_118022965
|
1.672
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr5_+_102229425
|
1.659
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr5_+_170221584
|
1.655
|
NM_022897
|
RANBP17
|
RAN binding protein 17
|
|
chr3_+_44878380
|
1.653
|
NM_144638
|
TMEM42
|
transmembrane protein 42
|
|
chr7_-_6490214
|
1.621
|
NM_001100603
NM_006854
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr7_-_6490375
|
1.609
|
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr3_-_8518327
|
1.605
|
|
LOC100288428
|
hypothetical LOC100288428
|
|
chr3_-_188946168
|
1.601
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr9_+_35528628
|
1.600
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr11_+_119701047
|
1.597
|
NM_001198670
NM_001198671
NM_001198672
NM_001198673
NM_001198674
NM_001198675
NM_174926
|
TMEM136
|
transmembrane protein 136
|
|
chr18_+_7557313
|
1.594
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr16_-_78192111
|
1.582
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chrX_+_102749489
|
1.582
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr5_+_102229612
|
1.582
|
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr5_-_37875538
|
1.542
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr12_-_105165805
|
1.530
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr4_-_141293572
|
1.522
|
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr9_-_78710698
|
1.478
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr7_+_128257698
|
1.471
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr16_-_71639670
|
1.463
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr9_+_35896188
|
1.446
|
NM_001039792
|
HRCT1
|
histidine rich carboxyl terminus 1
|
|
chr2_-_216008689
|
1.428
|
|
FN1
|
fibronectin 1
|
|
chr1_+_110328850
|
1.427
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr17_-_37828643
|
1.413
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr22_-_29833484
|
1.372
|
NM_080430
|
SELM
|
selenoprotein M
|
|
chr11_+_69165119
|
1.370
|
|
CCND1
|
cyclin D1
|
|
chr8_+_22479115
|
1.353
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr5_+_60663842
|
1.345
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr11_+_69165018
|
1.334
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr4_+_166519392
|
1.331
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr6_+_151602787
|
1.328
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr11_-_35503720
|
1.324
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr17_-_44043361
|
1.321
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr11_+_69165065
|
1.321
|
|
CCND1
|
cyclin D1
|
|
chr16_+_55580910
|
1.320
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr20_-_4177519
|
1.317
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr15_-_66511488
|
1.311
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr6_-_75972286
|
1.295
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr20_+_32755778
|
1.294
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr12_+_20413445
|
1.282
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr9_-_89779413
|
1.275
|
NM_001039803
NM_001170639
NM_001170640
NM_012119
NM_178432
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr17_-_44043278
|
1.250
|
|
HOXB7
|
homeobox B7
|
|
chr8_+_26296439
|
1.248
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr2_-_192767844
|
1.244
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr2_-_192767494
|
1.239
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr1_+_110328922
|
1.239
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr3_-_131312963
|
1.238
|
|
|
|
|
chr14_+_58174489
|
1.220
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr8_-_93184629
|
1.216
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_-_172130766
|
1.204
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr2_-_216009015
|
1.199
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr16_-_69277355
|
1.199
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr20_+_32610161
|
1.197
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr11_+_117948350
|
1.195
|
|
ARCN1
|
archain 1
|
|
chr2_-_133144094
|
1.192
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr5_+_170221441
|
1.187
|
|
RANBP17
|
RAN binding protein 17
|
|
chr19_+_54149843
|
1.180
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr1_-_16355013
|
1.178
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr15_-_53667800
|
1.169
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr6_-_80713589
|
1.160
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr19_-_18493886
|
1.156
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr8_-_82186832
|
1.153
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr9_-_72218098
|
1.151
|
|
KLF9
|
Kruppel-like factor 9
|
|
chr11_+_117948301
|
1.150
|
NM_001142281
NM_001655
|
ARCN1
|
archain 1
|
|
chr22_+_34107087
|
1.148
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr2_-_1727292
|
1.148
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr10_+_102749597
|
1.146
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr8_+_37672427
|
1.128
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr1_+_110328830
|
1.127
|
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr22_+_34107051
|
1.123
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr3_-_131313010
|
1.117
|
|
|
|
|
chr4_-_111763655
|
1.112
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr22_+_37193961
|
1.108
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr19_+_50041400
|
1.099
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr7_-_37922902
|
1.099
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr16_+_15435825
|
1.082
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr19_+_54149941
|
1.079
|
|
BAX
|
BCL2-associated X protein
|
|
chr8_+_133856738
|
1.077
|
NM_016018
NM_032205
NM_198513
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr3_+_127118130
|
1.063
|
|
LOC100125556
|
family with sequence similarity 86, member A pseudogene
|
|
chr1_-_143750988
|
1.056
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr7_+_93862137
|
1.055
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr18_+_54682261
|
1.046
|
|
ZNF532
|
zinc finger protein 532
|
|
chr15_-_80125514
|
1.046
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr3_+_173240126
|
1.038
|
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr15_+_76952356
|
1.033
|
|
MORF4L1
|
mortality factor 4 like 1
|
|
chr11_+_119701214
|
1.031
|
|
TMEM136
|
transmembrane protein 136
|
|
chr2_-_144994038
|
1.021
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr15_-_38000379
|
1.020
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr9_+_136673464
|
1.020
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr19_-_18493847
|
1.016
|
|
ELL
|
elongation factor RNA polymerase II
|
|
chr4_-_111763356
|
1.011
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr12_+_58276114
|
1.001
|
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr9_-_35062643
|
1.000
|
NM_007126
|
VCP
|
valosin containing protein
|
|
chr19_-_45888259
|
0.991
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr15_-_80125403
|
0.987
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr5_+_80292313
|
0.976
|
NM_006909
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr2_+_10969513
|
0.976
|
NM_002236
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1
|
|
chr2_-_1727247
|
0.962
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr10_-_128067062
|
0.958
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr10_-_14090459
|
0.957
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr22_-_35502059
|
0.956
|
NM_001177701
NM_006860
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas)
|
|
chr11_+_101291134
|
0.953
|
|
KIAA1377
|
KIAA1377
|
|
chr1_-_206484258
|
0.953
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr2_-_236741352
|
0.949
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr17_-_72045218
|
0.945
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr5_-_99898788
|
0.931
|
|
|
|
|
chr8_+_133856793
|
0.928
|
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr10_+_121400881
|
0.928
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr3_+_156280772
|
0.922
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_+_173240107
|
0.920
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr11_+_69165159
|
0.912
|
|
CCND1
|
cyclin D1
|
|
chr1_+_66772393
|
0.911
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr20_+_43895802
|
0.910
|
NM_001042632
NM_001042633
NM_033421
NM_152897
|
SNX21
|
sorting nexin family member 21
|
|
chr2_+_190916571
|
0.904
|
|
INPP1
|
inositol polyphosphate-1-phosphatase
|
|
chr9_-_35062596
|
0.899
|
|
VCP
|
valosin containing protein
|
|
chr1_-_59020875
|
0.898
|
|
JUN
|
jun proto-oncogene
|
|
chr3_-_75566781
|
0.896
|
|
FAM86DP
|
family with sequence similarity 86, member D, pseudogene
|
|
chr9_+_101623957
|
0.894
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr3_+_49211936
|
0.893
|
NM_001135197
|
CCDC36
|
coiled-coil domain containing 36
|
|
chr10_+_112247614
|
0.889
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr1_-_191422346
|
0.885
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr7_-_27136876
|
0.882
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr13_-_27967215
|
0.881
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr9_-_109291130
|
0.877
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_+_110328941
|
0.875
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr17_-_44042924
|
0.871
|
|
HOXB7
|
homeobox B7
|
|
chr7_-_5428043
|
0.866
|
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr19_+_50041387
|
0.862
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr10_-_131652364
|
0.855
|
|
EBF3
|
early B-cell factor 3
|
|
chr1_+_152459918
|
0.852
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chrX_+_133952633
|
0.851
|
NM_001163438
|
LOC644538
|
hypothetical protein LOC644538
|
|
chr7_+_12692938
|
0.846
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr7_-_37923040
|
0.845
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr18_+_9903954
|
0.836
|
NM_003574
NM_194434
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr5_+_80292213
|
0.836
|
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr14_+_23905984
|
0.835
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr4_-_114901789
|
0.824
|
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr1_-_202595666
|
0.823
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr10_+_121400926
|
0.822
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr12_-_47604930
|
0.810
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr9_-_35062578
|
0.809
|
|
VCP
|
valosin containing protein
|
|
chr9_-_35062545
|
0.803
|
|
VCP
|
valosin containing protein
|
|
chr12_+_70434927
|
0.801
|
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr10_+_121400741
|
0.801
|
NM_004281
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr19_+_54149963
|
0.800
|
|
BAX
|
BCL2-associated X protein
|
|
chr10_-_120830286
|
0.798
|
|
EIF3A
|
eukaryotic translation initiation factor 3, subunit A
|
|
chr19_-_52709975
|
0.798
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr7_-_45095017
|
0.793
|
NM_001146334
|
NACAD
|
NAC alpha domain containing
|
|
chr10_-_128067052
|
0.790
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr17_+_77529070
|
0.788
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr10_-_128067001
|
0.778
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr18_+_9903992
|
0.776
|
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr18_+_9904000
|
0.773
|
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr9_-_89779180
|
0.769
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr2_-_1727324
|
0.761
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|