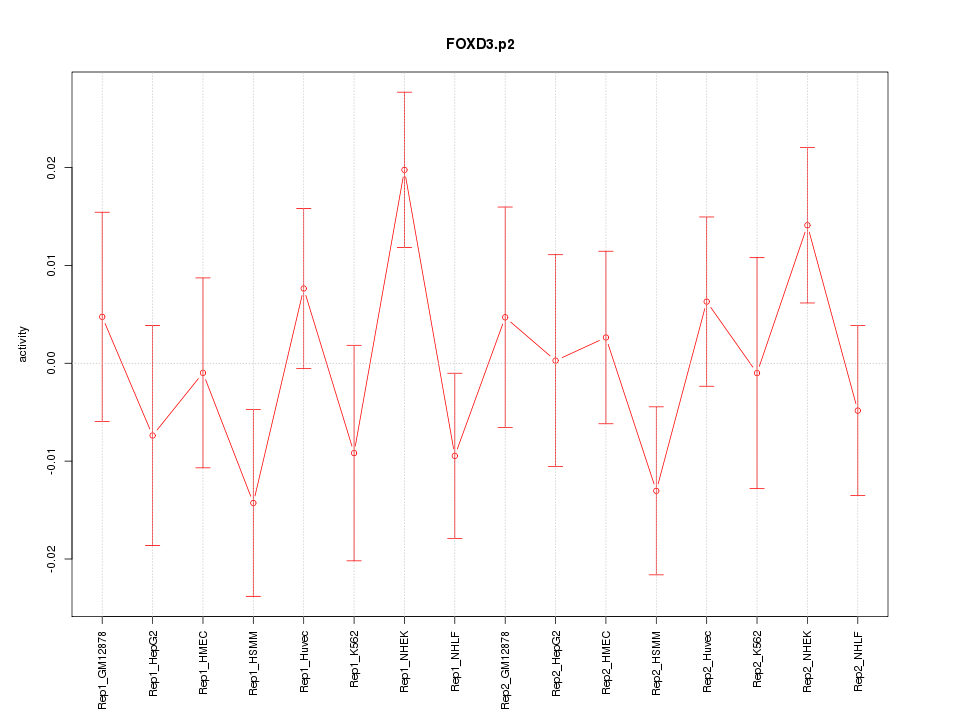
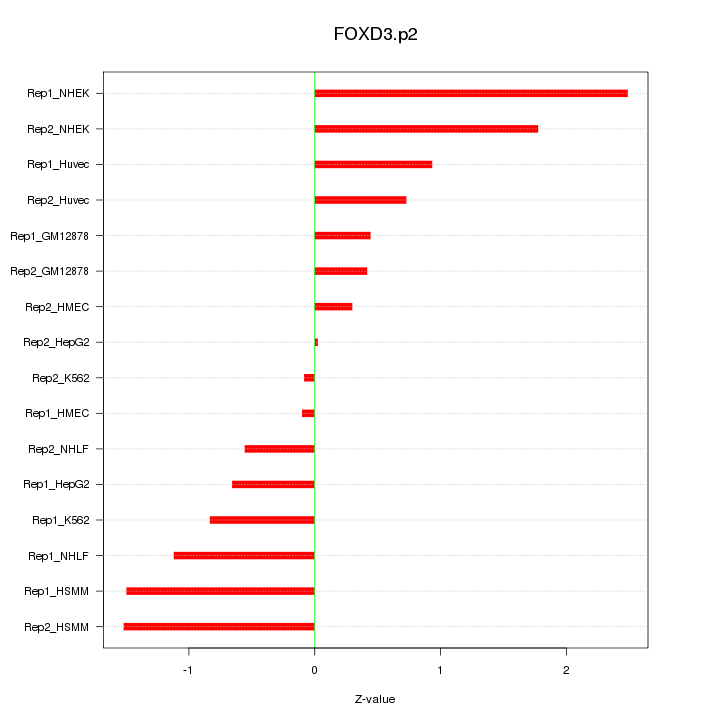
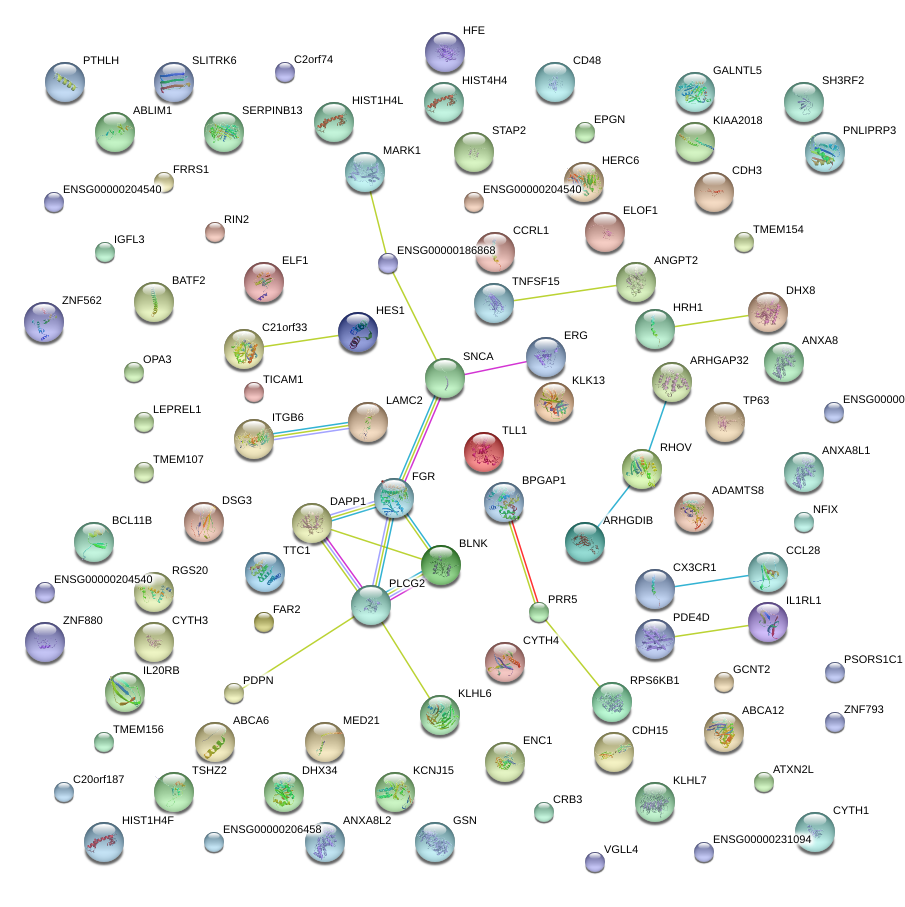
|
chr20_+_19863716
|
2.394
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr3_+_138159396
|
1.948
|
NM_144717
|
IL20RB
|
interleukin 20 receptor beta
|
|
chr13_-_40491491
|
1.794
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr18_+_59405513
|
1.682
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr3_+_11242666
|
1.647
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr13_-_40491405
|
1.497
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr18_+_27281729
|
1.473
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr4_+_167013859
|
1.468
|
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr3_+_195336625
|
1.434
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr1_+_218768170
|
1.380
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr14_-_98807307
|
1.351
|
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr19_-_4782711
|
1.263
|
NM_182919
|
TICAM1
|
toll-like receptor adaptor molecule 1
|
|
chr1_+_218768146
|
1.211
|
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr10_+_118177371
|
1.166
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr4_-_38710376
|
1.121
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr9_-_116608113
|
1.084
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr5_+_159368733
|
1.051
|
NM_003314
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr4_+_75393053
|
1.034
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr5_-_59100194
|
1.030
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_159368721
|
1.002
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr3_+_190990142
|
0.986
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr5_+_159368690
|
0.982
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr21_+_38550739
|
0.970
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr1_-_158948176
|
0.962
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr1_+_181421796
|
0.961
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr16_+_67235651
|
0.950
|
NM_001793
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr3_+_195336631
|
0.933
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr19_-_4289750
|
0.915
|
NM_001013841
NM_017720
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr21_-_38792173
|
0.911
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr16_+_80370400
|
0.864
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr22_+_36008511
|
0.843
|
|
CYTH4
|
cytohesin 4
|
|
chr3_+_133798770
|
0.830
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr1_+_13784553
|
0.809
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr6_+_49626071
|
0.798
|
NM_001145652
|
C6orf141
|
chromosome 6 open reading frame 141
|
|
chr9_+_676640
|
0.791
|
|
|
|
|
chr1_-_100003936
|
0.789
|
NM_001013660
|
FRRS1
|
ferric-chelate reductase 1
|
|
chr22_+_36008465
|
0.787
|
|
CYTH4
|
cytohesin 4
|
|
chr19_+_12995782
|
0.784
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr22_+_36008369
|
0.751
|
NM_013385
|
CYTH4
|
cytohesin 4
|
|
chr1_-_27825178
|
0.749
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr4_-_90978469
|
0.731
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr3_-_184756101
|
0.720
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr13_-_85271329
|
0.712
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr2_-_215711313
|
0.707
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr15_-_38953628
|
0.698
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr4_-_153820535
|
0.692
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr19_+_6415259
|
0.691
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr5_-_43432969
|
0.685
|
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr12_+_27066749
|
0.680
|
NM_004264
|
MED21
|
mediator complex subunit 21
|
|
chr4_+_100957003
|
0.679
|
NM_014395
|
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides
|
|
chr20_+_51023159
|
0.674
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr11_-_128399218
|
0.670
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr12_+_27066768
|
0.650
|
|
MED21
|
mediator complex subunit 21
|
|
chr10_-_116276560
|
0.640
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr7_+_151284396
|
0.630
|
NM_145292
|
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5
|
|
chr2_+_61225710
|
0.630
|
NM_001143960
|
C2orf74
|
chromosome 2 open reading frame 74
|
|
chr6_+_29963328
|
0.612
|
|
HLA-H
|
major histocompatibility complex, class I, H (pseudogene)
|
|
chr8_-_6407882
|
0.609
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr2_-_160765008
|
0.606
|
|
ITGB6
|
integrin, beta 6
|
|
chr12_-_28016182
|
0.606
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr6_+_10629550
|
0.593
|
NM_145649
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr10_+_47216925
|
0.591
|
NM_001630
|
ANXA8L2
ANXA8
|
annexin A8-like 2
annexin A8
|
|
chr3_-_191321367
|
0.587
|
|
LEPREL1
|
leprecan-like 1
|
|
chr10_-_46593923
|
0.583
|
NM_001040084
NM_001098845
|
ANXA8L2
LOC653110
ANXA8
ANXA8L1
|
annexin A8-like 2
hypothetical LOC653110
annexin A8
annexin A8-like 1
|
|
chr3_-_12561962
|
0.576
|
NM_001195279
|
LOC100129480
|
hypothetical protein LOC100129480
|
|
chr19_-_51319769
|
0.575
|
NM_207393
|
IGFL3
|
IGF-like family member 3
|
|
chr19_+_57564981
|
0.574
|
NM_001145434
|
ZNF880
|
zinc finger protein 880
|
|
chr19_+_42689881
|
0.572
|
|
ZNF793
|
zinc finger protein 793
|
|
chr12_+_29193399
|
0.572
|
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr22_+_43527101
|
0.563
|
NM_001017526
NM_001198726
NM_181335
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chr10_+_47875229
|
0.548
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr10_-_98021144
|
0.543
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr3_-_11736946
|
0.536
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr12_-_15005761
|
0.533
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr4_+_89519004
|
0.530
|
|
HERC6
|
hect domain and RLD 6
|
|
chr2_+_102320148
|
0.530
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr6_+_31190505
|
0.527
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr6_+_26348539
|
0.527
|
NM_003540
|
HIST1H4F
|
histone cluster 1, H4f
|
|
chr5_+_145296313
|
0.526
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr1_+_61697209
|
0.522
|
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_13782838
|
0.520
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr4_+_89518898
|
0.508
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr14_-_60817946
|
0.506
|
|
|
|
|
chr14_-_60818263
|
0.506
|
NM_001017970
|
TMEM30B
|
transmembrane protein 30B
|
|
chr21_+_41719847
|
0.503
|
NM_002462
NM_001178046
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse)
|
|
chr18_+_59705918
|
0.497
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr17_-_53847937
|
0.489
|
|
RNF43
|
ring finger protein 43
|
|
chr14_-_98807574
|
0.484
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr11_-_107969554
|
0.482
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr3_-_191321601
|
0.472
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr1_-_159275334
|
0.469
|
|
TSTD1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1
|
|
chr3_-_194118321
|
0.463
|
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr6_-_10520264
|
0.458
|
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr6_+_121798443
|
0.457
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr5_-_94442941
|
0.448
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr19_+_42689680
|
0.441
|
NM_001013659
|
ZNF793
|
zinc finger protein 793
|
|
chr20_-_23014828
|
0.441
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr19_+_48772791
|
0.440
|
NM_001193621
|
LOC390940
|
hypothetical protein LOC390940
|
|
chr7_+_113842470
|
0.439
|
|
FOXP2
|
forkhead box P2
|
|
chr15_-_32398220
|
0.434
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_+_34993234
|
0.433
|
NM_005268
|
GJB5
|
gap junction protein, beta 5, 31.1kDa
|
|
chr12_+_46799279
|
0.431
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chr2_-_20390601
|
0.429
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr15_-_61460967
|
0.427
|
|
CA12
|
carbonic anhydrase XII
|
|
chr1_+_179719338
|
0.426
|
NM_000721
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr6_+_148705817
|
0.425
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr19_+_3087026
|
0.423
|
NM_002068
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class)
|
|
chr7_-_87180499
|
0.413
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr2_-_160764819
|
0.404
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr12_+_91620749
|
0.399
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr19_+_59820413
|
0.395
|
NM_006669
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1
|
|
chr15_-_61461127
|
0.395
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr4_-_87989291
|
0.395
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr19_-_14853166
|
0.393
|
NM_030901
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17
|
|
chrX_+_123062143
|
0.391
|
|
STAG2
|
stromal antigen 2
|
|
chr9_-_25668230
|
0.390
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr10_+_71231693
|
0.389
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr17_-_26665194
|
0.389
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr19_-_61876057
|
0.388
|
NM_001005850
|
ZNF835
|
zinc finger protein 835
|
|
chr5_+_68746694
|
0.386
|
NM_001038603
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr20_-_45848043
|
0.384
|
NM_018837
|
SULF2
|
sulfatase 2
|
|
chr5_+_108111421
|
0.383
|
NM_005246
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr6_+_153060722
|
0.383
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr17_-_26665213
|
0.380
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr1_-_159275357
|
0.379
|
NM_001113205
NM_001113206
NM_001113207
|
TSTD1
F11R
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1
F11 receptor
|
|
chr3_+_123385219
|
0.379
|
NM_000388
|
CASR
|
calcium-sensing receptor
|
|
chr1_-_25164061
|
0.377
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr9_+_27099138
|
0.375
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr8_+_69026895
|
0.374
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr5_-_58607673
|
0.374
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_+_149895390
|
0.373
|
NM_018326
|
GIMAP4
|
GTPase, IMAP family member 4
|
|
chr17_-_26665144
|
0.373
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr12_+_53178834
|
0.372
|
NM_001184976
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr8_+_54926920
|
0.370
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr1_-_168310232
|
0.366
|
NM_014970
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr16_+_67328805
|
0.360
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr12_-_83954185
|
0.360
|
NM_001100917
|
TSPAN19
|
tetraspanin 19
|
|
chr4_-_64957595
|
0.358
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr8_+_27687821
|
0.358
|
NM_001017420
|
ESCO2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae)
|
|
chr9_-_98815676
|
0.356
|
|
HIATL2
|
hippocampus abundant transcript-like 2
|
|
chrX_+_99726445
|
0.350
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr21_+_38550533
|
0.348
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr9_-_205892
|
0.348
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr2_-_158008763
|
0.347
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr6_-_32892705
|
0.346
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr17_-_26665220
|
0.341
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr10_+_71231649
|
0.341
|
NM_001130103
NM_080798
NM_080800
NM_080801
NM_080802
NM_080805
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr7_+_113842284
|
0.337
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr14_+_20319049
|
0.337
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr3_+_123526700
|
0.336
|
NM_005213
|
CSTA
|
cystatin A (stefin A)
|
|
chr9_+_96176638
|
0.336
|
NM_032558
|
HIATL1
|
hippocampus abundant transcript-like 1
|
|
chr3_-_57505107
|
0.334
|
NM_178504
NM_198564
|
DNAH12
|
dynein, axonemal, heavy chain 12
|
|
chr6_+_126282078
|
0.333
|
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr8_-_68821131
|
0.333
|
NM_020361
|
CPA6
|
carboxypeptidase A6
|
|
chr2_+_237143118
|
0.333
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr6_+_114285209
|
0.332
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr1_+_107485071
|
0.332
|
|
NTNG1
|
netrin G1
|
|
chr14_+_101345878
|
0.332
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr2_-_133145539
|
0.331
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr16_-_3567354
|
0.330
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr7_-_25234582
|
0.330
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr8_+_123863224
|
0.329
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr21_-_42219849
|
0.329
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr22_+_44828338
|
0.328
|
|
|
|
|
chr12_+_21175394
|
0.327
|
NM_006446
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1
|
|
chr12_+_111375382
|
0.327
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr11_+_20005704
|
0.320
|
|
NAV2
|
neuron navigator 2
|
|
chr13_+_20081940
|
0.320
|
|
|
|
|
chr12_-_120591938
|
0.320
|
NM_173855
|
MORN3
|
MORN repeat containing 3
|
|
chr9_+_92629525
|
0.319
|
NM_001174167
|
SYK
|
spleen tyrosine kinase
|
|
chr16_+_67328737
|
0.317
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr3_+_123779122
|
0.316
|
NM_001113523
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr6_-_26358811
|
0.314
|
NM_021018
|
HIST1H3F
|
histone cluster 1, H3f
|
|
chr1_-_150044365
|
0.311
|
NM_001004432
|
LINGO4
|
leucine rich repeat and Ig domain containing 4
|
|
chr11_-_64327197
|
0.311
|
NM_004579
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2
|
|
chr20_-_14266200
|
0.310
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr5_+_55182963
|
0.310
|
NM_139017
|
IL31RA
|
interleukin 31 receptor A
|
|
chr7_-_20223467
|
0.310
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr11_-_26550243
|
0.310
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr5_+_147562531
|
0.309
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr9_+_27514311
|
0.309
|
NM_020124
|
IFNK
|
interferon, kappa
|
|
chrX_-_138552517
|
0.309
|
NM_001171877
NM_001171878
NM_001171879
NM_005369
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr19_-_19793512
|
0.309
|
NM_001099269
NM_001145404
|
ZNF506
|
zinc finger protein 506
|
|
chr2_-_157890400
|
0.308
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr1_-_24112345
|
0.308
|
NM_001841
|
CNR2
|
cannabinoid receptor 2 (macrophage)
|
|
chr6_-_44341296
|
0.308
|
NM_004556
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr8_-_42742729
|
0.307
|
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr12_+_47584159
|
0.307
|
NM_033124
|
CCDC65
|
coiled-coil domain containing 65
|
|
chr18_+_59726183
|
0.304
|
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr1_+_84382575
|
0.303
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr10_-_11614279
|
0.302
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr10_+_8137356
|
0.297
|
|
GATA3
|
GATA binding protein 3
|
|
chr1_+_32512298
|
0.297
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr22_+_23753940
|
0.295
|
NM_001145206
|
KIAA1671
|
KIAA1671
|
|
chr6_-_136889302
|
0.294
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr2_+_64534774
|
0.294
|
NM_014181
|
HSPC159
|
galectin-related protein
|
|
chr3_-_9265592
|
0.293
|
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr11_+_112763722
|
0.292
|
NM_178510
|
ANKK1
|
ankyrin repeat and kinase domain containing 1
|
|
chr1_+_190811479
|
0.292
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr2_-_218739891
|
0.290
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|