|
chr9_-_109290107
|
3.688
|
|
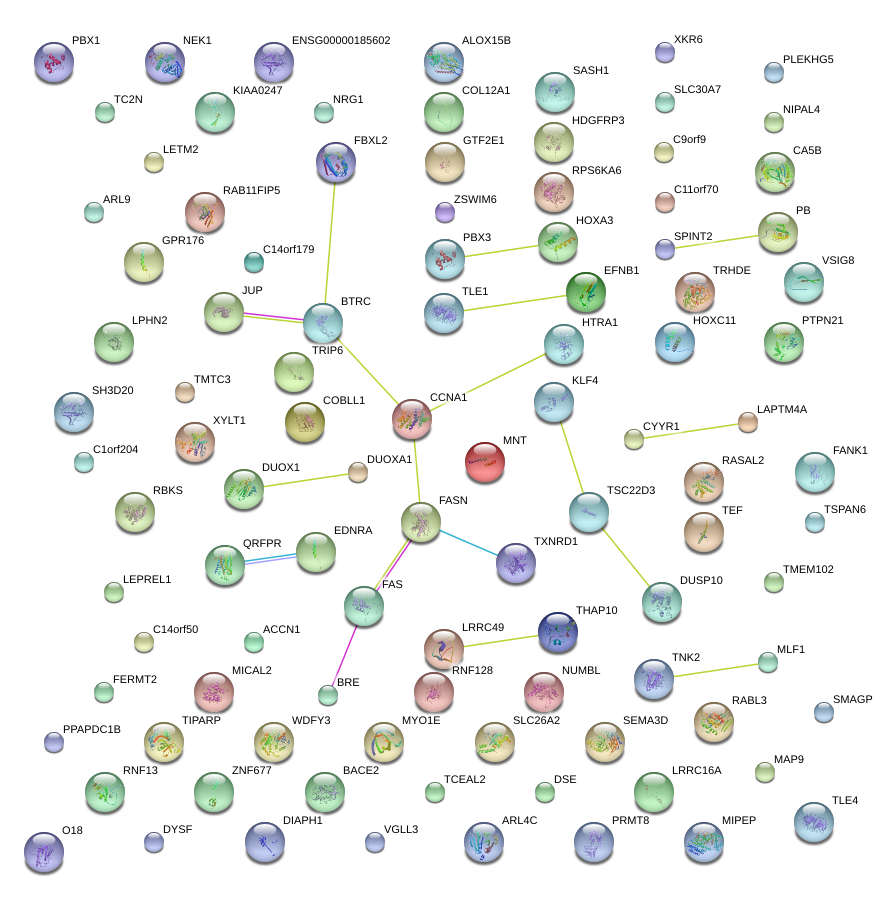
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr10_+_124211353
|
2.652
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chrX_-_106846253
|
2.267
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chrX_-_106846684
|
2.202
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr3_-_191321367
|
2.111
|
|
LEPREL1
|
leprecan-like 1
|
|
chr7_-_27120015
|
1.989
|
|
HOXA3
|
homeobox A3
|
|
chr3_+_33293915
|
1.948
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr15_+_68971835
|
1.883
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr14_+_75521806
|
1.864
|
NM_001102564
NM_052873
|
C14orf179
|
chromosome 14 open reading frame 179
|
|
chrX_+_67965517
|
1.704
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr12_+_56406296
|
1.521
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr3_-_87122936
|
1.433
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr3_-_87122958
|
1.412
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr15_-_68971665
|
1.400
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr19_-_58449752
|
1.378
|
|
ZNF677
|
zinc finger protein 677
|
|
chr8_-_11096257
|
1.361
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr2_-_27966715
|
1.354
|
NM_022128
|
RBKS
|
ribokinase
|
|
chr12_+_3470685
|
1.352
|
NM_019854
|
PRMT8
|
protein arginine methyltransferase 8
|
|
chr4_+_57066131
|
1.285
|
NM_206919
|
ARL9
|
ADP-ribosylation factor-like 9
|
|
chr6_+_148705646
|
1.269
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr1_+_32992131
|
1.235
|
|
|
|
|
chr11_+_12088639
|
1.230
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr3_-_191321601
|
1.222
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr6_+_116798791
|
1.210
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr3_+_159771646
|
1.186
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr13_+_35903966
|
1.159
|
NM_001111046
NM_001111047
|
CCNA1
|
cyclin A1
|
|
chr8_-_38245694
|
1.141
|
NM_001102559
NM_001102560
NM_032483
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B
|
|
chrX_-_83329562
|
1.137
|
NM_014496
|
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6
|
|
chr2_+_27966985
|
1.135
|
NM_199193
NM_199194
NM_004899
NM_199191
NM_199192
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr8_+_32525269
|
1.097
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr4_+_148621567
|
1.091
|
|
EDNRA
|
endothelin receptor type A
|
|
chr2_+_27967116
|
1.088
|
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr2_+_27967102
|
1.083
|
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr9_+_127550298
|
1.080
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr15_-_57452361
|
1.054
|
NM_004998
|
MYO1E
|
myosin IE
|
|
chr10_+_103103797
|
1.042
|
NM_003939
NM_033637
|
BTRC
|
beta-transducin repeat containing
|
|
chr15_-_57452345
|
1.034
|
|
MYO1E
|
myosin IE
|
|
chr11_+_101423378
|
1.030
|
NM_001195005
NM_032930
|
C11orf70
|
chromosome 11 open reading frame 70
|
|
chr15_-_57452357
|
1.028
|
|
MYO1E
|
myosin IE
|
|
chr6_-_75972286
|
1.019
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr15_-_37999717
|
1.018
|
|
GPR176
|
G protein-coupled receptor 176
|
|
chr1_+_82038669
|
1.016
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr13_+_20770248
|
1.007
|
|
LOC650794
|
hypothetical LOC650794
|
|
chr15_+_43209422
|
0.999
|
NM_017434
NM_175940
|
DUOX1
|
dual oxidase 1
|
|
chr4_-_156517394
|
0.977
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr17_+_7883076
|
0.976
|
NM_001039130
NM_001039131
NM_001141
|
ALOX15B
|
arachidonate 15-lipoxygenase, type B
|
|
chr9_+_81377289
|
0.934
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr14_-_91372365
|
0.923
|
NM_001128595
NM_152332
|
TC2N
|
tandem C2 domains, nuclear
|
|
chr21_-_26867138
|
0.921
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chrX_+_15666312
|
0.919
|
NM_007220
|
CA5B
|
carbonic anhydrase VB, mitochondrial
|
|
chr7_-_84654106
|
0.911
|
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr1_-_219982081
|
0.909
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr4_-_86106567
|
0.906
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr21_+_41461962
|
0.902
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chrX_+_105856531
|
0.891
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr17_-_40866062
|
0.877
|
NM_174919
|
SH3D20
|
SH3 domain containing 20
|
|
chr17_-_2250967
|
0.868
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr15_-_43209348
|
0.863
|
NM_144565
|
DUOXA1
|
dual oxidase maturation factor 1
|
|
chr4_-_156517423
|
0.859
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr17_-_40865987
|
0.846
|
|
SH3D20
|
SH3 domain containing 20
|
|
chr5_-_140978664
|
0.836
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr17_-_60208083
|
0.836
|
|
LOC146880
|
hypothetical LOC146880
|
|
chr2_-_73193581
|
0.826
|
NM_015470
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr1_-_219982049
|
0.825
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr9_+_81377419
|
0.821
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chrX_+_101267315
|
0.820
|
NM_080390
|
TCEAL2
|
transcription elongation factor A (SII)-like 2
|
|
chr1_-_158091760
|
0.820
|
NM_001134233
|
C1orf204
|
chromosome 1 open reading frame 204
|
|
chr17_-_37196463
|
0.816
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr12_+_70952729
|
0.808
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr2_-_73193419
|
0.807
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr1_-_219982016
|
0.803
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr17_+_7279485
|
0.793
|
NM_178518
|
TMEM102
|
transmembrane protein 102
|
|
chr15_-_37999519
|
0.782
|
|
|
|
|
chr1_+_101134276
|
0.757
|
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr2_-_73193566
|
0.756
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr2_+_71534260
|
0.755
|
NM_001130976
NM_001130977
NM_001130978
NM_001130979
NM_001130980
NM_001130981
NM_003494
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr5_+_156819782
|
0.754
|
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr19_+_43447262
|
0.752
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr5_+_60663842
|
0.746
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr1_+_101134259
|
0.743
|
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr9_+_81376666
|
0.743
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr15_-_38000379
|
0.742
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr2_+_27967159
|
0.739
|
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr1_+_176329895
|
0.736
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr17_-_28644118
|
0.732
|
NM_183377
|
ACCN1
|
amiloride-sensitive cation channel 1, neuronal
|
|
chr14_+_69148066
|
0.725
|
|
KIAA0247
|
KIAA0247
|
|
chr1_+_101134237
|
0.724
|
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr1_+_101134177
|
0.721
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr12_+_52653176
|
0.720
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr1_+_176329925
|
0.719
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr3_+_121944247
|
0.716
|
NM_005513
|
GTF2E1
|
general transcription factor IIE, polypeptide 1, alpha 56kDa
|
|
chr12_+_87060176
|
0.708
|
NM_181783
|
TMTC3
|
transmembrane and tetratricopeptide repeat containing 3
|
|
chr12_-_49950236
|
0.704
|
NM_001033873
NM_001031628
|
SMAGP
|
small cell adhesion glycoprotein
|
|
chr16_-_17472238
|
0.701
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr12_-_70952572
|
0.701
|
|
LOC283392
|
hypothetical LOC283392
|
|
chr14_+_64086366
|
0.701
|
NM_172365
|
C14orf50
|
chromosome 14 open reading frame 50
|
|
chr1_+_176329860
|
0.685
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr13_-_23361430
|
0.683
|
NM_005932
|
MIPEP
|
mitochondrial intermediate peptidase
|
|
chr7_+_100302885
|
0.683
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr2_-_165406098
|
0.680
|
|
COBLL1
|
COBL-like 1
|
|
chr4_-_122521562
|
0.677
|
NM_198179
|
QRFPR
|
pyroglutamylated RFamide peptide receptor
|
|
chr10_+_127575097
|
0.674
|
NM_145235
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1
|
|
chr19_-_45888259
|
0.674
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr1_-_6473216
|
0.673
|
NM_001042664
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr15_-_81667773
|
0.671
|
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr3_-_197106828
|
0.670
|
NM_001010938
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr5_-_140978747
|
0.665
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr10_+_90740364
|
0.662
|
|
FAS
|
Fas (TNF receptor superfamily, member 6)
|
|
chr14_-_88087575
|
0.656
|
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr3_+_157876051
|
0.652
|
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr3_-_121944052
|
0.649
|
NM_173825
|
RABL3
|
RAB, member of RAS oncogene family-like 3
|
|
chr22_+_40107878
|
0.649
|
NM_003216
|
TEF
|
thyrotrophic embryonic factor
|
|
chr2_-_235070431
|
0.643
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr12_+_103133631
|
0.636
|
NM_001093771
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr14_-_52487022
|
0.632
|
|
FERMT2
|
fermitin family member 2
|
|
chr4_-_156517544
|
0.630
|
NM_001039580
|
MAP9
|
microtubule-associated protein 9
|
|
chrX_-_99778340
|
0.628
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr3_+_151013691
|
0.627
|
|
RNF13
|
ring finger protein 13
|
|
chr3_+_151013672
|
0.625
|
|
RNF13
|
ring finger protein 13
|
|
chr4_-_170770111
|
0.625
|
NM_012224
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr2_-_20114761
|
0.624
|
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr6_+_25387630
|
0.620
|
NM_001173977
NM_017640
|
LRRC16A
|
leucine rich repeat containing 16A
|
|
chr5_+_149320480
|
0.614
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr8_+_38363150
|
0.606
|
NM_144652
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr2_-_20114881
|
0.603
|
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr9_+_134743532
|
0.603
|
|
C9orf9
|
chromosome 9 open reading frame 9
|
|
chr6_+_25387273
|
0.600
|
|
LRRC16A
|
leucine rich repeat containing 16A
|
|
chr3_-_109423858
|
0.600
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr9_+_6747919
|
0.596
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr6_+_30002084
|
0.595
|
|
|
|
|
chr2_-_202216451
|
0.594
|
NM_001044385
|
ALS2CR4
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4
|
|
chr18_-_50004770
|
0.590
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr22_+_27798887
|
0.589
|
NM_001039570
NM_032045
|
KREMEN1
|
kringle containing transmembrane protein 1
|
|
chr14_+_69148062
|
0.588
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr9_-_34448477
|
0.586
|
NM_001184940
NM_001184941
NM_001184942
NM_001184943
NM_001184944
NM_001184945
NM_147202
|
C9orf25
|
chromosome 9 open reading frame 25
|
|
chr2_-_20115269
|
0.584
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr11_+_7997366
|
0.579
|
|
TUB
|
tubby homolog (mouse)
|
|
chr7_-_82629735
|
0.575
|
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chrX_-_15263596
|
0.574
|
NM_002641
NM_020473
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr11_+_65311104
|
0.569
|
NM_004561
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chrX_-_106846924
|
0.564
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr5_-_140978751
|
0.560
|
NM_001079812
NM_005219
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chrX_-_15263555
|
0.560
|
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr10_-_30065760
|
0.552
|
|
SVIL
|
supervillin
|
|
chr5_+_174084137
|
0.551
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr12_+_32151351
|
0.551
|
NM_001003398
NM_001714
|
BICD1
|
bicaudal D homolog 1 (Drosophila)
|
|
chrX_-_15263586
|
0.548
|
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr3_-_121944040
|
0.548
|
|
|
|
|
chr12_-_87059877
|
0.543
|
NM_025114
|
CEP290
|
centrosomal protein 290kDa
|
|
chr1_-_158091296
|
0.541
|
|
C1orf204
|
chromosome 1 open reading frame 204
|
|
chr6_+_43151922
|
0.539
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chrX_-_13307425
|
0.539
|
|
|
|
|
chrX_+_17665483
|
0.534
|
NM_001037535
NM_001037536
NM_001037540
NM_006746
|
SCML1
|
sex comb on midleg-like 1 (Drosophila)
|
|
chr5_-_140978744
|
0.534
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr2_-_36384
|
0.533
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr11_-_107969554
|
0.533
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr10_+_124210980
|
0.530
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chrX_-_134013644
|
0.530
|
NM_001078172
NM_001134321
|
FAM127B
|
family with sequence similarity 127, member B
|
|
chr11_+_65357972
|
0.529
|
NM_152760
|
SNX32
|
sorting nexin 32
|
|
chr13_+_44461657
|
0.528
|
NM_018559
|
KIAA1704
|
KIAA1704
|
|
chr2_-_227371698
|
0.521
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr13_-_44461554
|
0.507
|
NM_012345
|
NUFIP1
|
nuclear fragile X mental retardation protein interacting protein 1
|
|
chr19_-_54188376
|
0.504
|
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr12_+_8741746
|
0.503
|
NM_020734
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chr9_-_21984414
|
0.503
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr18_-_63334902
|
0.495
|
NM_032160
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr5_+_149320630
|
0.491
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chrX_+_16714452
|
0.491
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr20_-_49073023
|
0.491
|
NM_002237
|
KCNG1
|
potassium voltage-gated channel, subfamily G, member 1
|
|
chr17_-_40263124
|
0.489
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr10_-_127574957
|
0.487
|
|
DHX32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32
|
|
chr2_-_156897286
|
0.486
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr1_+_218768423
|
0.483
|
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chrX_-_51256166
|
0.483
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr2_+_36436316
|
0.482
|
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr19_-_54188332
|
0.482
|
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr11_+_122031549
|
0.479
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr16_+_49333513
|
0.479
|
NM_001042412
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr15_+_47235243
|
0.478
|
NM_001001556
|
GALK2
|
galactokinase 2
|
|
chr3_-_135575925
|
0.478
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr15_+_71763830
|
0.477
|
|
CD276
|
CD276 molecule
|
|
chr19_-_58449919
|
0.475
|
NM_182609
|
ZNF677
|
zinc finger protein 677
|
|
chr11_-_65094319
|
0.475
|
|
LOC254100
|
hypothetical protein LOC254100
|
|
chr5_+_99899133
|
0.472
|
|
FAM174A
|
family with sequence similarity 174, member A
|
|
chr5_+_122875691
|
0.472
|
NM_001031812
NM_001044722
NM_001044723
NM_004384
|
CSNK1G3
|
casein kinase 1, gamma 3
|
|
chr17_-_10042579
|
0.472
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr13_-_77390907
|
0.468
|
NM_001122659
NM_003991
|
EDNRB
|
endothelin receptor type B
|
|
chr2_-_68547893
|
0.468
|
NM_001024680
|
FBXO48
|
F-box protein 48
|
|
chr19_-_54188378
|
0.466
|
NM_001161587
NM_002103
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr3_-_49370682
|
0.462
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr4_+_148621694
|
0.460
|
|
EDNRA
|
endothelin receptor type A
|
|
chr17_+_45488330
|
0.460
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr19_+_61711008
|
0.455
|
NM_020813
|
ZNF471
|
zinc finger protein 471
|
|
chr12_-_70952410
|
0.455
|
|
LOC283392
|
hypothetical LOC283392
|
|
chr6_+_7486836
|
0.454
|
|
DSP
|
desmoplakin
|
|
chr11_+_66580861
|
0.454
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr11_+_65311142
|
0.453
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr5_+_149320539
|
0.451
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr1_-_203557379
|
0.449
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr6_-_18230829
|
0.448
|
NM_198586
|
NHLRC1
|
NHL repeat containing 1
|