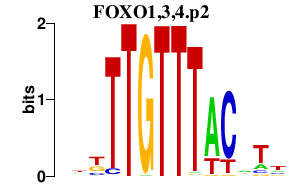
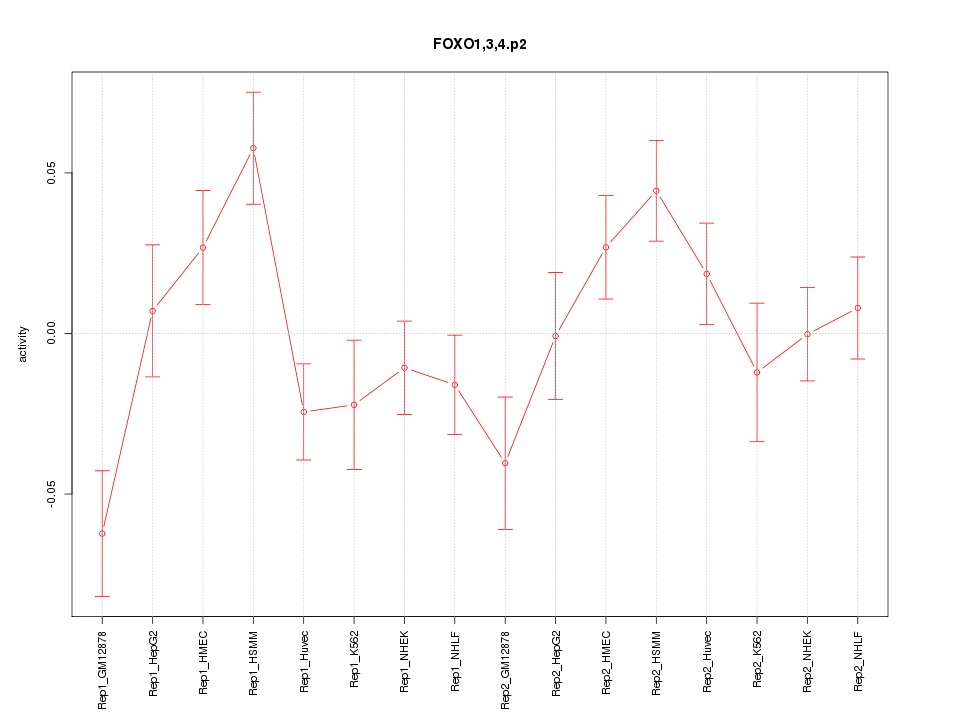
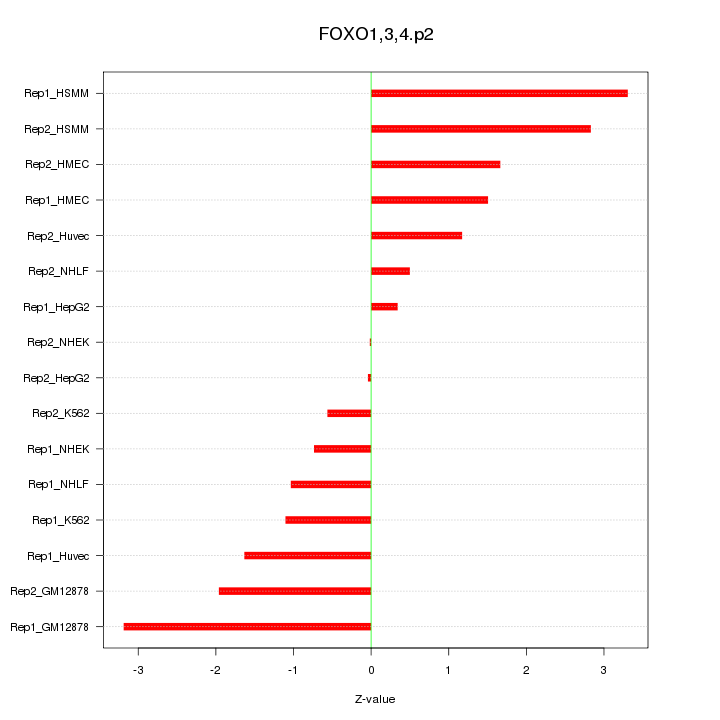
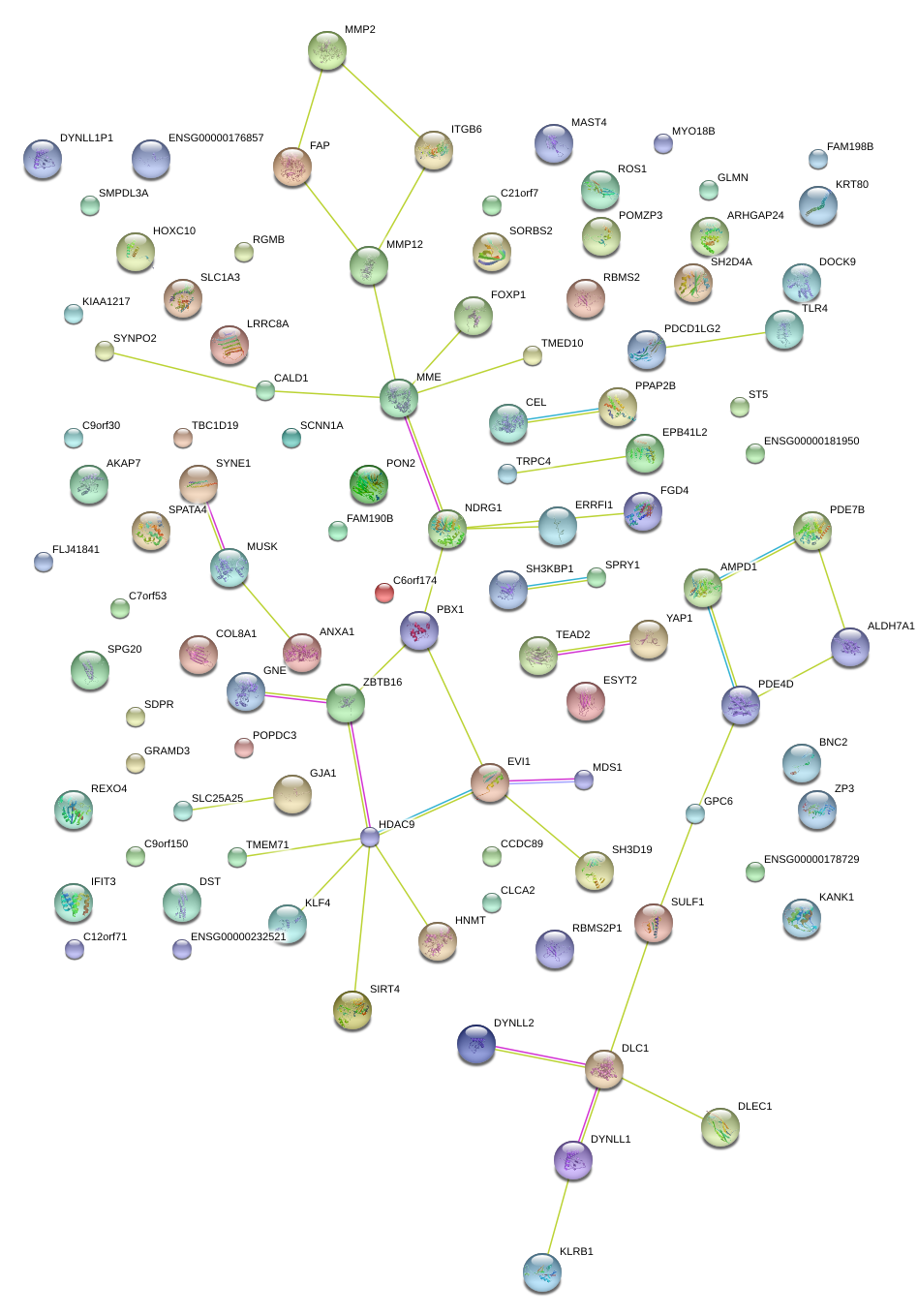
|
chr12_+_32578513
|
3.480
|
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr10_+_86174665
|
3.299
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr6_-_152744143
|
3.249
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr5_+_66160359
|
3.132
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr6_-_48186868
|
2.786
|
|
C6orf138
|
chromosome 6 open reading frame 138
|
|
chr7_+_134114957
|
2.743
|
|
CALD1
|
caldesmon 1
|
|
chr4_-_186934059
|
2.693
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_58607673
|
2.644
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_+_120167264
|
2.571
|
|
SYNPO2
|
synaptopodin 2
|
|
chr5_-_59100194
|
2.513
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr9_+_74956614
|
2.464
|
|
ANXA1
|
annexin A1
|
|
chr9_+_130719034
|
2.421
|
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr4_+_26194643
|
2.321
|
NM_018317
|
TBC1D19
|
TBC1 domain family, member 19
|
|
chrX_-_19598912
|
2.266
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr13_-_98428244
|
2.191
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr9_+_102231616
|
2.187
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr6_-_105734535
|
2.150
|
NM_022361
|
POPDC3
|
popeye domain containing 3
|
|
chr2_-_162808123
|
2.089
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr4_+_120167721
|
1.974
|
|
SYNPO2
|
synaptopodin 2
|
|
chr5_+_36644187
|
1.968
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr6_-_152744022
|
1.966
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr7_+_75892187
|
1.901
|
NM_001110354
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor)
|
|
chr4_-_159311959
|
1.892
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr3_+_100840129
|
1.862
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr9_+_119506428
|
1.825
|
|
TLR4
|
toll-like receptor 4
|
|
chr8_-_13178399
|
1.792
|
|
DLC1
|
deleted in liver cancer 1
|
|
chr9_-_16717887
|
1.721
|
|
BNC2
|
basonuclin 2
|
|
chr2_-_192419896
|
1.701
|
|
SDPR
|
serum deprivation response
|
|
chr9_+_119506446
|
1.681
|
|
TLR4
|
toll-like receptor 4
|
|
chr9_+_119506274
|
1.670
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr11_+_113436438
|
1.670
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr4_-_152368492
|
1.669
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr4_+_86968069
|
1.669
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr9_+_112470871
|
1.652
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr13_+_92677012
|
1.649
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr8_-_13178327
|
1.648
|
|
|
|
|
chr1_+_162795538
|
1.639
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr8_+_70567634
|
1.632
|
|
SULF1
|
sulfatase 1
|
|
chr16_+_54070302
|
1.606
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr9_+_74956601
|
1.531
|
|
ANXA1
|
annexin A1
|
|
chr5_-_125958503
|
1.517
|
NM_001182
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr1_-_7998265
|
1.509
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr9_+_5500544
|
1.493
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr6_-_117853710
|
1.477
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chr2_-_192420168
|
1.457
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr10_-_29963847
|
1.419
|
|
|
|
|
chr9_+_696744
|
1.413
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr7_+_18515440
|
1.412
|
|
HDAC9
|
histone deacetylase 9
|
|
chr7_-_158304062
|
1.401
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr7_+_134114912
|
1.399
|
|
CALD1
|
caldesmon 1
|
|
chr19_-_54557450
|
1.383
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr7_+_18515488
|
1.358
|
|
HDAC9
|
histone deacetylase 9
|
|
chr6_+_131498540
|
1.355
|
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr5_-_125956996
|
1.318
|
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr6_-_131333173
|
1.272
|
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr5_+_125786975
|
1.263
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr7_-_94902149
|
1.258
|
NM_000305
NM_001018161
|
PON2
|
paraoxonase 2
|
|
chr10_+_91077581
|
1.251
|
NM_001549
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr11_+_101488380
|
1.240
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr3_+_156280497
|
1.227
|
|
MME
|
membrane metallo-endopeptidase
|
|
chr4_+_124540114
|
1.206
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr2_-_160764819
|
1.205
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr3_+_156280539
|
1.205
|
|
MME
|
membrane metallo-endopeptidase
|
|
chr8_-_134378630
|
1.204
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr9_+_74956494
|
1.199
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr12_+_52665196
|
1.191
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr9_-_109291575
|
1.176
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr11_-_85074848
|
1.176
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chr4_-_177353707
|
1.156
|
NM_144644
|
SPATA4
|
spermatogenesis associated 4
|
|
chr22_+_24468110
|
1.150
|
NM_032608
|
MYO18B
|
myosin XVIIIB
|
|
chr5_+_98132898
|
1.149
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr22_+_24468168
|
1.134
|
|
MYO18B
|
myosin XVIIIB
|
|
chr4_+_86918870
|
1.129
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr21_+_29439768
|
1.117
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr1_+_86662356
|
1.115
|
NM_006536
|
CLCA2
|
chloride channel accessory 2
|
|
chr7_+_111908143
|
1.102
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr1_-_115039614
|
1.101
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr6_+_136214495
|
1.094
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr12_+_55201854
|
1.079
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr2_+_138438277
|
1.078
|
NM_001024074
NM_001024075
NM_006895
|
HNMT
|
histamine N-methyltransferase
|
|
chr8_-_133841988
|
1.078
|
NM_001145153
NM_144649
|
TMEM71
|
transmembrane protein 71
|
|
chr3_-_71262426
|
1.073
|
|
FOXP1
|
forkhead box P1
|
|
chr3_+_156280604
|
1.065
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr6_-_56615544
|
1.065
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr6_+_123152044
|
1.060
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr3_-_170347019
|
1.059
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_-_71262405
|
1.056
|
|
FOXP1
|
forkhead box P1
|
|
chr14_-_74669864
|
1.055
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr11_-_8695974
|
1.055
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr6_-_127822227
|
1.050
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr3_-_170346760
|
1.036
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr6_+_121798443
|
1.032
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr1_+_162795328
|
1.032
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr12_-_50871953
|
1.026
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr13_-_37341859
|
1.011
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr4_+_86918882
|
0.997
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr10_+_24795465
|
0.992
|
|
KIAA1217
|
KIAA1217
|
|
chr10_+_24795446
|
0.987
|
|
KIAA1217
|
KIAA1217
|
|
chr8_+_70567581
|
0.982
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr12_+_119224506
|
0.982
|
NM_012240
|
SIRT4
|
sirtuin 4
|
|
chr8_+_19215279
|
0.979
|
NM_001174160
NM_022071
NM_001174159
|
SH2D4A
|
SH2 domain containing 4A
|
|
chr12_-_6356783
|
0.972
|
NM_001159575
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr9_+_12764988
|
0.968
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr1_-_56817823
|
0.964
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr1_-_56817568
|
0.962
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr13_-_35818491
|
0.953
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr9_-_36266930
|
0.948
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr9_+_129900637
|
0.941
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr12_-_27126721
|
0.938
|
NM_001080406
|
C12orf71
|
chromosome 12 open reading frame 71
|
|
chr3_+_190990142
|
0.929
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr2_-_151052391
|
0.926
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr2_-_189752575
|
0.921
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr5_+_140546839
|
0.920
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr9_-_14769530
|
0.919
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr1_+_161308318
|
0.907
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr13_-_35818396
|
0.906
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr11_+_36572615
|
0.900
|
NM_138787
|
C11orf74
|
chromosome 11 open reading frame 74
|
|
chr8_-_130868315
|
0.900
|
NM_031415
|
GSDMC
|
gasdermin C
|
|
chr3_+_139550368
|
0.899
|
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr1_+_144236398
|
0.894
|
|
ITGA10
|
integrin, alpha 10
|
|
chr13_-_47885018
|
0.890
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr3_+_156280772
|
0.890
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_-_58497913
|
0.888
|
NM_003500
|
ACOX2
|
acyl-CoA oxidase 2, branched chain
|
|
chr3_-_150533948
|
0.886
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chrX_+_102498035
|
0.880
|
NM_001006612
NM_001006613
NM_001006614
NM_016303
|
WBP5
|
WW domain binding protein 5
|
|
chr7_+_120416103
|
0.872
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr11_+_114054409
|
0.870
|
NM_182495
|
FAM55B
|
family with sequence similarity 55, member B
|
|
chr20_+_44771532
|
0.869
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr12_-_8706533
|
0.868
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr16_+_68516401
|
0.854
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr6_-_80303822
|
0.850
|
NM_001122769
NM_181714
|
LCA5
|
Leber congenital amaurosis 5
|
|
chr12_-_8706637
|
0.847
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr9_+_129900590
|
0.842
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr18_-_20105900
|
0.842
|
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr2_+_36777336
|
0.842
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr5_-_151046638
|
0.841
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr7_+_120415958
|
0.839
|
NM_024913
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr1_-_23393808
|
0.835
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr1_-_159474503
|
0.834
|
NM_001077469
NM_001077470
NM_001077471
NM_001077472
NM_001077473
NM_001077474
NM_001077475
NM_001077476
NM_001077477
NM_001077478
NM_001077479
NM_001077480
NM_001077481
NM_001077482
NM_005122
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3
|
|
chr18_-_21184824
|
0.834
|
|
|
|
|
chr10_-_99151003
|
0.828
|
NM_001145114
NM_015179
|
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae)
|
|
chr17_-_17816425
|
0.827
|
NM_001033551
NM_001082968
|
TOM1L2
|
target of myb1-like 2 (chicken)
|
|
chr1_-_56817802
|
0.827
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr3_-_150569574
|
0.825
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr1_-_24342158
|
0.824
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr2_+_54411574
|
0.818
|
NM_001100396
|
C2orf73
|
chromosome 2 open reading frame 73
|
|
chr1_-_199732323
|
0.815
|
NM_001193572
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr6_+_134800533
|
0.810
|
|
LOC154092
|
hypothetical LOC154092
|
|
chr12_-_8706746
|
0.805
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr1_-_229071957
|
0.802
|
NM_001136494
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr1_+_144236346
|
0.797
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr1_+_20788027
|
0.795
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr11_+_112337199
|
0.792
|
NM_000615
NM_001076682
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr2_+_174968701
|
0.789
|
NM_001193528
NM_024583
|
SCRN3
|
secernin 3
|
|
chr3_-_113842652
|
0.782
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr12_+_12935222
|
0.781
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr3_+_42176662
|
0.780
|
NM_014965
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr8_+_12847521
|
0.758
|
NM_020844
|
KIAA1456
|
KIAA1456
|
|
chr9_-_8723893
|
0.758
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_-_105835525
|
0.757
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr7_-_13997574
|
0.757
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr3_-_113842553
|
0.755
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr16_+_54072969
|
0.755
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr6_+_36752244
|
0.754
|
NM_078467
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr2_-_200033445
|
0.754
|
|
SATB2
|
SATB homeobox 2
|
|
chr2_+_36777406
|
0.749
|
|
VIT
|
vitrin
|
|
chr13_-_47885248
|
0.746
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr3_+_62280435
|
0.745
|
NM_020685
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr2_-_165768798
|
0.737
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr5_+_178419938
|
0.736
|
NM_014594
|
ZNF354C
|
zinc finger protein 354C
|
|
chr12_+_79625538
|
0.735
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr17_+_3326045
|
0.730
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr5_+_125786717
|
0.729
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr11_+_834445
|
0.727
|
NM_001025237
|
TSPAN4
|
tetraspanin 4
|
|
chr1_-_56817717
|
0.727
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_+_12774264
|
0.720
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr15_-_70288071
|
0.718
|
|
PKM2
|
pyruvate kinase, muscle
|
|
chr6_+_52393763
|
0.716
|
NM_001172420
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr15_-_37999519
|
0.716
|
|
|
|
|
chr6_+_108988797
|
0.714
|
|
FOXO3
|
forkhead box O3
|
|
chr13_-_73605914
|
0.708
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr21_+_36429079
|
0.708
|
NM_001236
|
CBR3
|
carbonyl reductase 3
|
|
chr2_+_189547358
|
0.705
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr17_-_15495534
|
0.698
|
|
TRIM16
|
tripartite motif containing 16
|
|
chr9_-_74757735
|
0.696
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr2_+_174968751
|
0.695
|
|
SCRN3
|
secernin 3
|
|
chr2_+_202379442
|
0.692
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr4_+_187227302
|
0.684
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr17_+_1612056
|
0.679
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr3_-_15814678
|
0.677
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr9_+_130722994
|
0.675
|
NM_001100876
NM_174933
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr20_-_29751755
|
0.675
|
|
|
|
|
chr17_-_36469869
|
0.672
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr2_+_135526304
|
0.671
|
NM_001172435
NM_012233
|
RAB3GAP1
|
RAB3 GTPase activating protein subunit 1 (catalytic)
|
|
chr5_-_58688428
|
0.670
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_-_40537002
|
0.667
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr10_+_11087345
|
0.663
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr15_+_97010219
|
0.662
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr13_+_96672705
|
0.654
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr8_+_107807415
|
0.653
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|