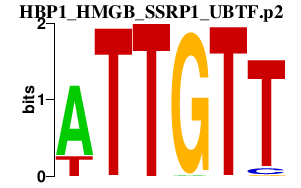
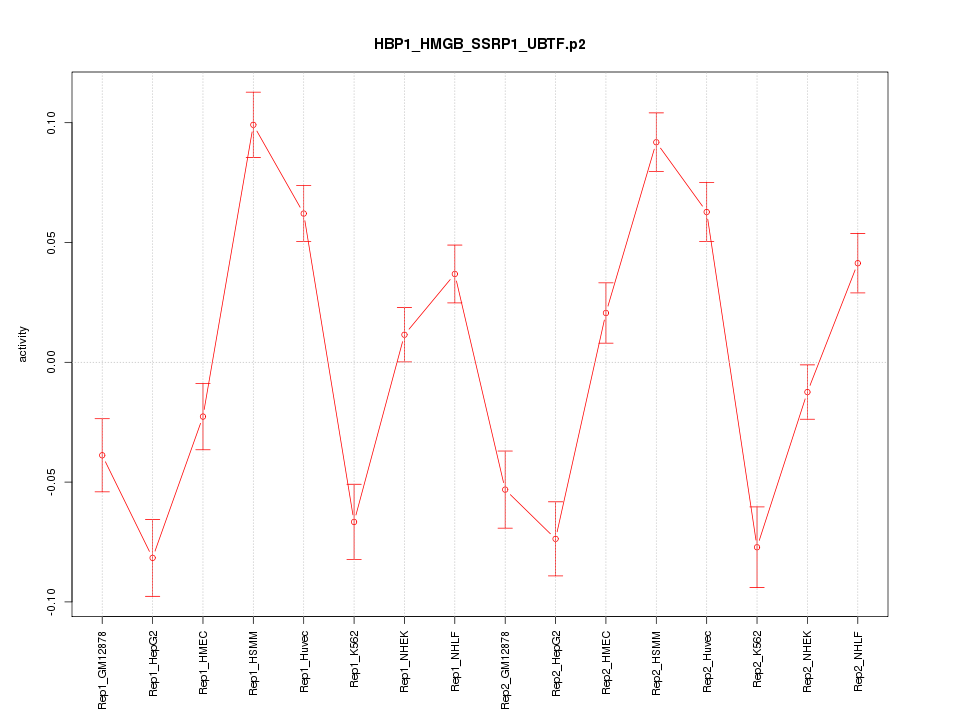
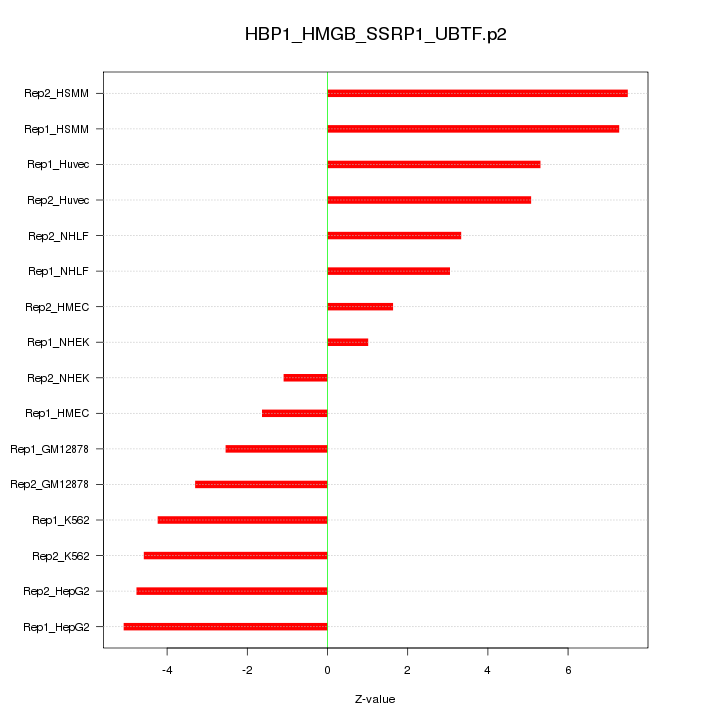
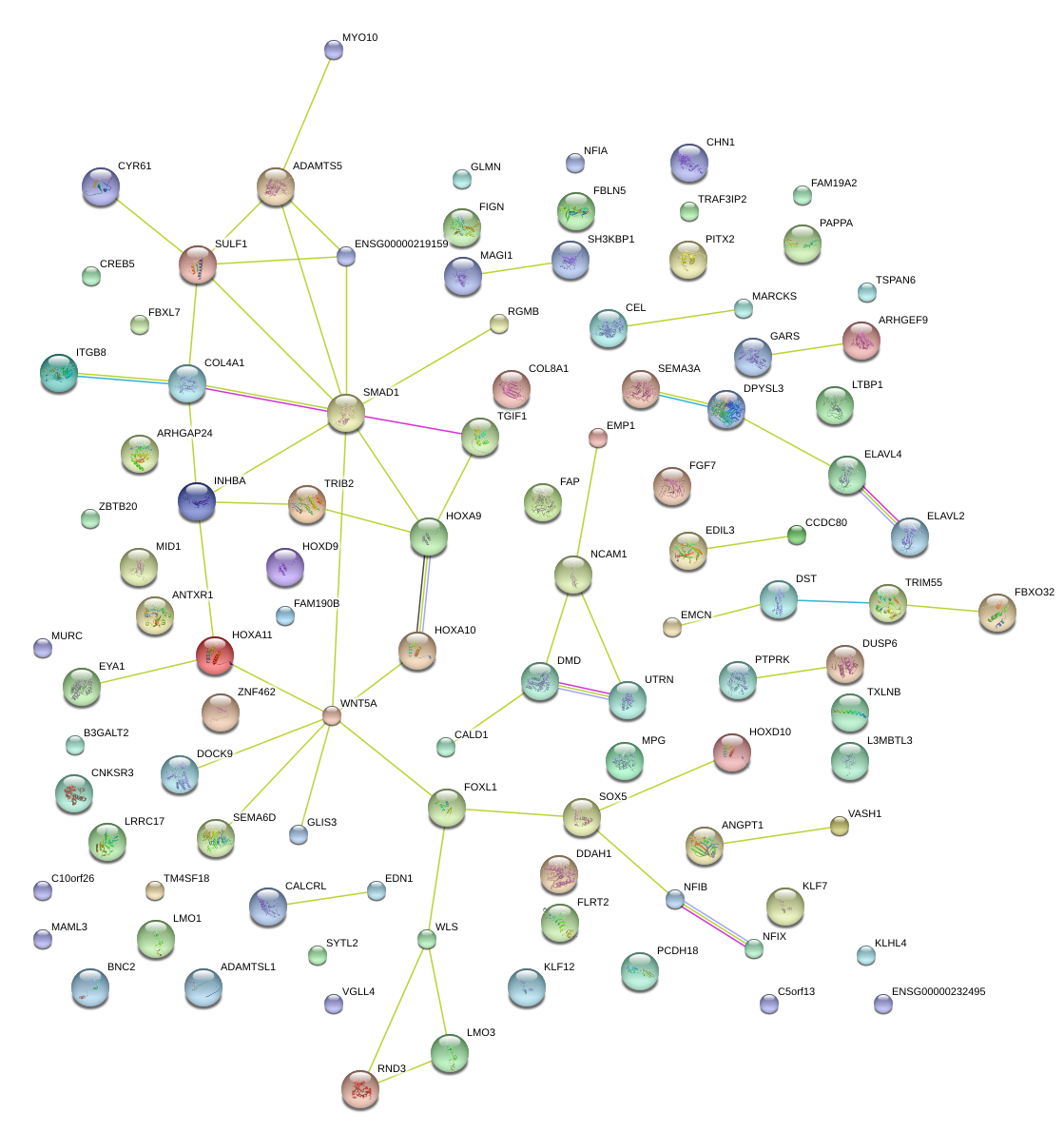
|
chr7_+_134114957
|
17.805
|
|
CALD1
|
caldesmon 1
|
|
chr7_+_134114912
|
17.231
|
|
CALD1
|
caldesmon 1
|
|
chr7_-_27186400
|
16.686
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr6_+_12398514
|
16.008
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr11_-_85107569
|
15.474
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr11_-_85108030
|
13.797
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr13_-_73605914
|
13.631
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr16_+_85169615
|
12.871
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr13_-_98428244
|
12.738
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr5_-_111119846
|
12.688
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_-_72436519
|
12.380
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr13_-_73606362
|
11.495
|
|
KLF12
|
Kruppel-like factor 12
|
|
chr8_-_108579270
|
11.308
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr13_-_109757442
|
10.750
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr2_-_162808123
|
10.700
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr4_+_86918882
|
10.666
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_-_197173159
|
10.546
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr6_-_111995213
|
10.499
|
NM_001164283
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr5_-_16989371
|
10.276
|
NM_012334
|
MYO10
|
myosin X
|
|
chr7_-_27171673
|
10.155
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr5_-_16989290
|
9.983
|
|
MYO10
|
myosin X
|
|
chr2_-_56266408
|
9.773
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr9_+_18464090
|
9.616
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr9_+_18464138
|
9.433
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr2_+_69093641
|
9.289
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr9_+_108665168
|
8.958
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr6_-_56824672
|
8.909
|
|
DST
|
dystonin
|
|
chr10_+_104525877
|
8.837
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr5_+_98132898
|
8.612
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr3_-_115825742
|
8.518
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr8_+_67201684
|
8.497
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr12_+_13240983
|
8.137
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr8_-_124622434
|
8.091
|
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr10_+_86174665
|
8.078
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr12_+_13240987
|
7.983
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr3_-_116272911
|
7.979
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_24606616
|
7.917
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr14_+_85066265
|
7.857
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr8_-_72437020
|
7.848
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr7_+_28692122
|
7.826
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_+_15553736
|
7.770
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr2_+_12775814
|
7.697
|
|
|
|
|
chr7_+_102340579
|
7.641
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr8_+_70567634
|
7.626
|
|
SULF1
|
sulfatase 1
|
|
chr14_+_85066207
|
7.623
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr8_+_70567581
|
7.524
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr7_+_20337249
|
7.379
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr14_-_91483325
|
7.377
|
|
FBLN5
|
fibulin 5
|
|
chr15_+_47502666
|
7.366
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr3_-_55496370
|
7.353
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr6_+_114285209
|
7.216
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr15_+_45797977
|
7.215
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr9_-_23816062
|
7.211
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr2_-_207738858
|
7.203
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr4_-_111777651
|
7.128
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr2_+_33213111
|
6.906
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr9_-_14303509
|
6.879
|
|
NFIB
|
nuclear factor I/B
|
|
chr11_+_112337199
|
6.850
|
NM_000615
NM_001076682
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr1_-_191422346
|
6.830
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr6_-_56824371
|
6.760
|
|
DST
|
dystonin
|
|
chr6_-_139654807
|
6.664
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr1_+_85819004
|
6.642
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr5_+_15553539
|
6.631
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr7_-_27171648
|
6.597
|
|
HOXA9
|
homeobox A9
|
|
chr1_-_85816520
|
6.569
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr3_+_148593911
|
6.510
|
|
|
|
|
chr3_-_194118321
|
6.495
|
|
MB21D2
|
Mab-21 domain containing 2
|
|
chrX_+_86659370
|
6.483
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr6_+_130381408
|
6.452
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr3_-_55496607
|
6.449
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr4_-_138673078
|
6.434
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr6_-_128883125
|
6.430
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr2_+_12775336
|
6.427
|
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr9_-_16717887
|
6.426
|
|
BNC2
|
basonuclin 2
|
|
chr21_-_27261276
|
6.417
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr8_-_72436832
|
6.389
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr7_-_27171600
|
6.359
|
|
HOXA9
|
homeobox A9
|
|
chr9_-_14303899
|
6.318
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_-_68470585
|
6.308
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr7_-_41709191
|
6.307
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr5_+_15553288
|
6.298
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr3_-_11585264
|
6.245
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr3_+_100840129
|
6.171
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chrX_-_99778340
|
6.155
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr9_+_102380156
|
5.866
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr9_-_14304036
|
5.853
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr12_-_16649211
|
5.830
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr9_+_126460511
|
5.779
|
|
|
|
|
chr3_-_150533948
|
5.776
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr4_+_86918870
|
5.722
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr5_-_83715947
|
5.713
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr2_-_164300758
|
5.684
|
NM_018086
|
FIGN
|
fidgetin
|
|
chr9_-_4289915
|
5.667
|
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr4_+_146623405
|
5.599
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr3_-_65999548
|
5.583
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr5_-_146813343
|
5.582
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr2_-_188021117
|
5.562
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr2_-_151052391
|
5.467
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr4_-_141293572
|
5.460
|
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr14_+_76298693
|
5.414
|
|
VASH1
|
vasohibin 1
|
|
chr7_+_27190688
|
5.384
|
|
|
|
|
chr9_+_117955890
|
5.380
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr4_-_111777584
|
5.337
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr7_-_83662152
|
5.268
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chrX_-_62891066
|
5.252
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr3_-_11660395
|
5.250
|
NM_001128219
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr4_-_101658094
|
5.247
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chrX_-_19598912
|
5.204
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr14_+_76297885
|
5.144
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr18_+_3441589
|
5.142
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr7_-_27190882
|
5.116
|
|
HOXA11
|
homeobox A11
|
|
chrX_-_10548458
|
5.100
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr2_-_175578156
|
5.095
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr7_+_134114701
|
5.068
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr5_-_83716346
|
5.046
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr3_-_113842133
|
5.021
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chrX_-_33139349
|
4.970
|
NM_004006
|
DMD
|
dystrophin
|
|
chr18_-_21184824
|
4.941
|
|
|
|
|
chr11_+_112337299
|
4.926
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr11_+_112337491
|
4.925
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr14_-_91483757
|
4.913
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chr12_-_88270375
|
4.897
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr3_-_113841967
|
4.889
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_+_93388951
|
4.847
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr6_-_134540666
|
4.804
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_+_148705421
|
4.794
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr3_-_115960807
|
4.768
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_-_83052606
|
4.767
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chrX_+_77889861
|
4.761
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr8_-_6407882
|
4.734
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr1_-_85703198
|
4.705
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr8_-_29262240
|
4.698
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr11_+_131286513
|
4.697
|
|
NTM
|
neurotrimin
|
|
chr3_-_113842237
|
4.693
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr15_-_68781662
|
4.596
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr3_-_113842255
|
4.594
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr14_+_22375747
|
4.593
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr7_-_47588160
|
4.593
|
|
TNS3
|
tensin 3
|
|
chr3_+_174598937
|
4.579
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr14_+_22375753
|
4.515
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr7_-_47588266
|
4.485
|
|
TNS3
|
tensin 3
|
|
chr1_-_177378801
|
4.472
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr12_+_13240868
|
4.466
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr11_+_131285747
|
4.443
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr2_+_85834111
|
4.439
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chrX_+_135079461
|
4.404
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr3_-_145049853
|
4.402
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr11_+_101488380
|
4.391
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr4_-_141294682
|
4.373
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr13_+_110653407
|
4.357
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr1_-_95165089
|
4.285
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr3_+_29297806
|
4.272
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr11_+_113436438
|
4.252
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr7_-_27108826
|
4.225
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr1_-_224993498
|
4.221
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|
|
chr13_-_32678142
|
4.208
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr7_+_20336849
|
4.179
|
|
ITGB8
|
integrin, beta 8
|
|
chr5_+_15553768
|
4.175
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr5_+_36642424
|
4.172
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr7_-_27153892
|
4.146
|
NM_024014
|
HOXA6
|
homeobox A6
|
|
chr2_-_152299229
|
4.114
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr12_-_47868563
|
4.110
|
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr6_+_21774624
|
4.091
|
|
FLJ22536
|
hypothetical locus LOC401237
|
|
chr13_+_109757593
|
4.086
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr5_+_61910318
|
4.086
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr3_-_194118627
|
4.067
|
NM_178496
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr12_-_8706533
|
4.067
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr4_-_7991992
|
4.042
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr11_+_112337411
|
4.036
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr1_-_199742589
|
4.019
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr20_+_19818209
|
4.005
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr5_-_121441910
|
3.930
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr19_-_45016587
|
3.927
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr3_-_15814678
|
3.921
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr4_-_74307637
|
3.901
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr7_+_28415495
|
3.890
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr9_+_81377289
|
3.881
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr7_-_27149750
|
3.872
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr5_-_39400319
|
3.858
|
NM_001737
|
C9
|
complement component 9
|
|
chr15_-_81667169
|
3.834
|
NM_016073
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr5_-_121441866
|
3.819
|
|
LOX
|
lysyl oxidase
|
|
chr14_+_62740878
|
3.819
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr9_+_81377419
|
3.816
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chrX_-_106846253
|
3.810
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_68070573
|
3.806
|
|
|
|
|
chr4_-_140696740
|
3.776
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chrX_+_15428820
|
3.775
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr4_-_152366970
|
3.753
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr12_-_16650687
|
3.752
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr19_+_13767207
|
3.741
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr1_+_162795538
|
3.729
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr12_-_16650697
|
3.708
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_-_121441805
|
3.702
|
|
LOX
|
lysyl oxidase
|
|
chr7_+_134226727
|
3.681
|
|
CALD1
|
caldesmon 1
|
|
chr13_-_32658215
|
3.672
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr6_+_31190505
|
3.666
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr15_-_53996597
|
3.654
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr3_+_114414064
|
3.602
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr4_-_138673014
|
3.590
|
|
PCDH18
|
protocadherin 18
|
|
chr18_-_51406359
|
3.583
|
|
TCF4
|
transcription factor 4
|