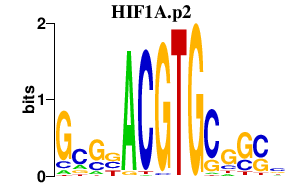
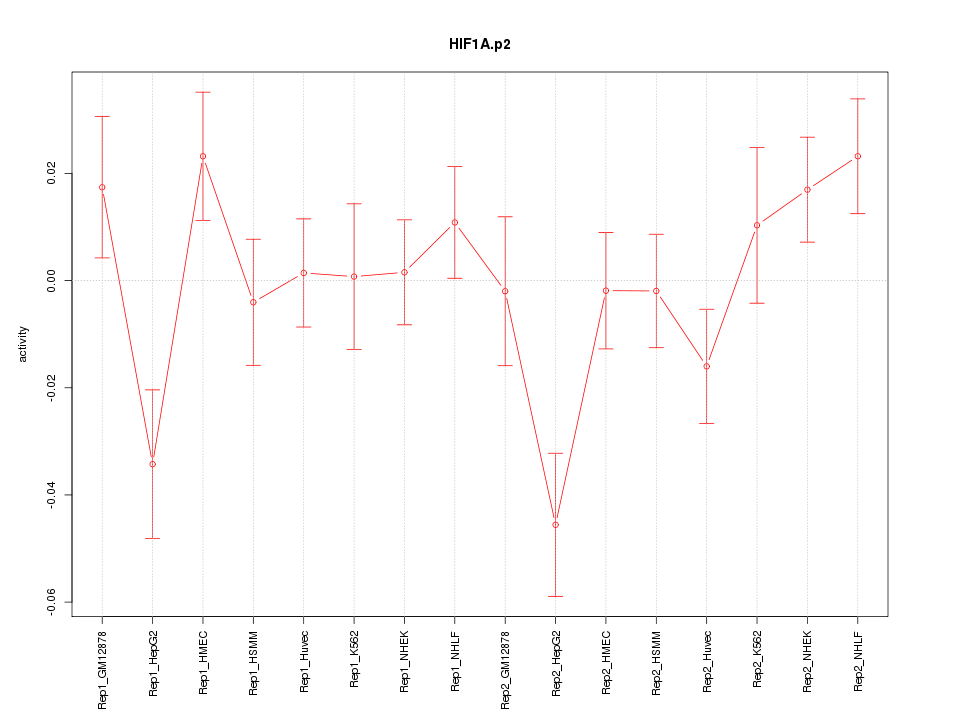
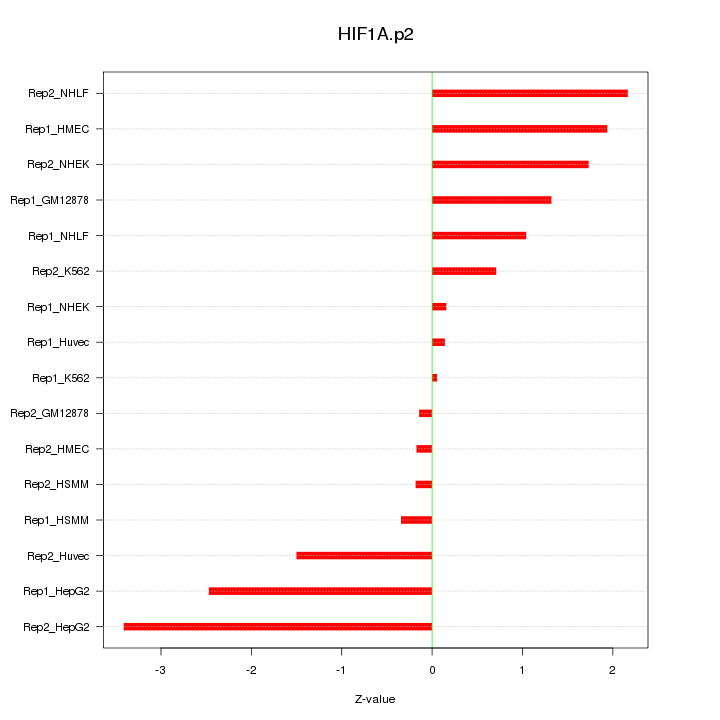
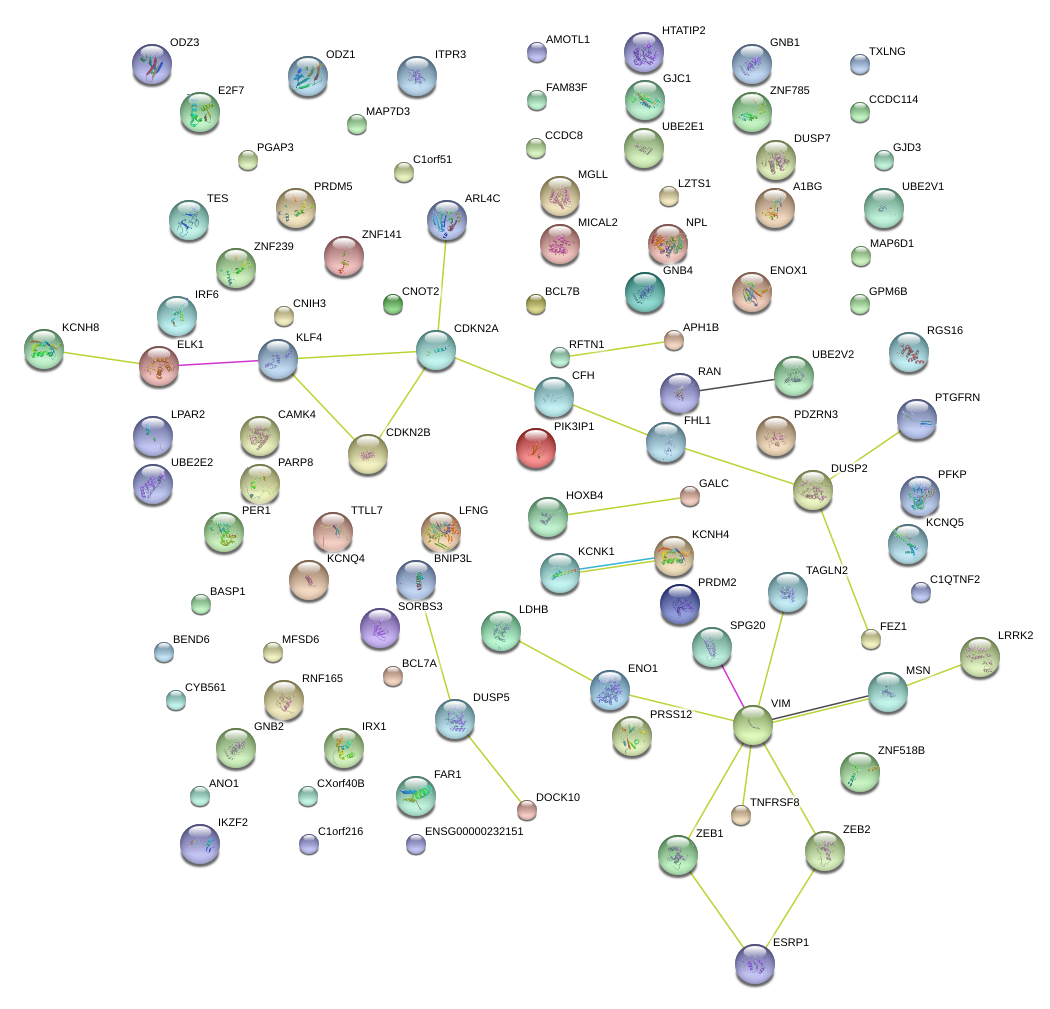
|
chr22_+_38720881
|
2.033
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr10_-_88720908
|
1.811
|
|
LOC100133190
|
hypothetical LOC100133190
|
|
chr13_-_43259032
|
1.798
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr1_+_231816372
|
1.409
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr10_+_112247614
|
1.319
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr1_+_13899272
|
1.260
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr3_-_180651874
|
1.231
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr11_+_20341835
|
1.224
|
NM_001098520
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr11_+_12088639
|
1.206
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr11_+_20341806
|
1.172
|
NM_006410
NM_001098521
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr11_+_20341900
|
1.126
|
NM_001098522
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr3_-_180652024
|
1.124
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr4_+_321607
|
1.095
|
|
ZNF141
|
zinc finger protein 141
|
|
chr19_-_53516962
|
1.089
|
|
CCDC114
|
coiled-coil domain containing 114
|
|
chr4_+_321587
|
1.077
|
NM_003441
|
ZNF141
|
zinc finger protein 141
|
|
chr6_+_73388713
|
1.054
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr1_+_231816540
|
1.052
|
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr11_+_94141151
|
0.985
|
NM_130847
|
AMOTL1
|
angiomotin like 1
|
|
chr10_+_3099708
|
0.962
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr6_+_33697023
|
0.956
|
NM_002224
|
ITPR3
|
inositol 1,4,5-triphosphate receptor, type 3
|
|
chr5_+_49998677
|
0.945
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr10_-_43390014
|
0.933
|
NM_001099282
NM_001099283
NM_001099284
|
ZNF239
|
zinc finger protein 239
|
|
chr3_-_129024665
|
0.926
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr19_-_51608665
|
0.899
|
NM_032040
|
CCDC8
|
coiled-coil domain containing 8
|
|
chr1_+_148521852
|
0.894
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr12_+_129922589
|
0.889
|
|
RAN
|
RAN, member RAS oncogene family
|
|
chr4_-_10068055
|
0.876
|
NM_053042
|
ZNF518B
|
zinc finger protein 518B
|
|
chrX_+_16714452
|
0.869
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr3_-_129023850
|
0.857
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr12_+_129922570
|
0.857
|
|
RAN
|
RAN, member RAS oncogene family
|
|
chr21_+_25856311
|
0.852
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr17_-_7996409
|
0.850
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chrX_-_47394808
|
0.847
|
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr8_+_95722448
|
0.840
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr2_-_235070431
|
0.836
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr4_+_183302105
|
0.814
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr1_-_158161877
|
0.814
|
|
TAGLN2
|
transgelin 2
|
|
chr11_+_13646806
|
0.810
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chrX_-_47394937
|
0.806
|
NM_001114123
NM_005229
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr2_-_225615556
|
0.802
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr1_-_158161902
|
0.802
|
NM_003564
|
TAGLN2
|
transgelin 2
|
|
chrX_-_135161185
|
0.784
|
NM_001173517
NM_024597
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr11_-_124871258
|
0.777
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr1_-_208046011
|
0.775
|
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr19_-_19599973
|
0.772
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr11_+_13646727
|
0.770
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chrX_-_13866366
|
0.767
|
|
|
|
|
chr19_-_63550712
|
0.763
|
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr7_+_115637795
|
0.758
|
NM_015641
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr5_+_17270781
|
0.758
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr1_+_12046020
|
0.750
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr9_-_109291575
|
0.748
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_-_35957030
|
0.715
|
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr3_-_185025989
|
0.710
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr17_-_58877276
|
0.710
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr11_+_69602055
|
0.706
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr5_+_110587546
|
0.700
|
NM_001744
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV
|
|
chr12_-_21701985
|
0.694
|
NM_001174097
|
LDHB
|
lactate dehydrogenase B
|
|
chrX_+_148430424
|
0.692
|
NM_001171907
NM_001171908
NM_178124
|
CXorf40A
|
chromosome X open reading frame 40A
|
|
chr5_-_159730196
|
0.690
|
NM_031908
|
C1QTNF2
|
C1q and tumor necrosis factor related protein 2
|
|
chr9_-_109291866
|
0.687
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_+_222870573
|
0.687
|
|
CNIH3
|
cornichon homolog 3 (Drosophila)
|
|
chr12_+_68923043
|
0.684
|
NM_014515
|
CNOT2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr12_-_21702036
|
0.683
|
|
LDHB
|
lactate dehydrogenase B
|
|
chrX_+_135057202
|
0.682
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr12_-_21702021
|
0.679
|
|
LDHB
|
lactate dehydrogenase B
|
|
chrX_+_64804235
|
0.669
|
NM_002444
|
MSN
|
moesin
|
|
chr10_+_17312654
|
0.669
|
|
VIM
|
vimentin
|
|
chr12_-_75983329
|
0.660
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr15_+_46957556
|
0.657
|
NM_014335
|
EID1
|
EP300 interacting inhibitor of differentiation 1
|
|
chr1_-_35956979
|
0.651
|
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr3_+_23219707
|
0.649
|
NM_152653
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2 (UBC4/5 homolog, yeast)
|
|
chr7_+_2525922
|
0.647
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr12_+_38905053
|
0.644
|
NM_198578
|
LRRK2
|
leucine-rich repeat kinase 2
|
|
chr16_-_30504309
|
0.644
|
NM_152458
|
ZNF785
|
zinc finger protein 785
|
|
chr12_-_21702000
|
0.641
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr5_+_3649167
|
0.640
|
NM_024337
|
IRX1
|
iroquois homeobox 1
|
|
chr6_+_56927665
|
0.640
|
NM_152731
|
BEND6
|
BEN domain containing 6
|
|
chr3_-_16530132
|
0.638
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr1_-_173428455
|
0.638
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr6_+_56927882
|
0.632
|
|
BEND6
|
BEN domain containing 6
|
|
chrX_-_148430250
|
0.630
|
|
LOC100131434
|
hypothetical LOC100131434
|
|
chr17_-_44010543
|
0.627
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chrX_-_13866451
|
0.626
|
|
GPM6B
|
glycoprotein M6B
|
|
chr13_-_35818491
|
0.623
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr18_+_42167904
|
0.619
|
|
RNF165
|
ring finger protein 165
|
|
chr13_-_35818777
|
0.615
|
NM_001142295
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr3_-_16529996
|
0.612
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr3_-_73756668
|
0.609
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr2_-_96174502
|
0.602
|
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr9_-_21984414
|
0.597
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr3_+_23219490
|
0.594
|
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2 (UBC4/5 homolog, yeast)
|
|
chr4_-_119493322
|
0.593
|
NM_003619
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin)
|
|
chr1_+_222870798
|
0.591
|
NM_152495
|
CNIH3
|
cornichon homolog 3 (Drosophila)
|
|
chr13_-_35818396
|
0.587
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr14_-_87529222
|
0.587
|
|
GALC
|
galactosylceramidase
|
|
chr8_+_26296439
|
0.586
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr7_-_72609957
|
0.585
|
NM_001197244
NM_001707
|
BCL7B
|
B-cell CLL/lymphoma 7B
|
|
chr2_+_190981257
|
0.581
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr12_-_21702046
|
0.576
|
NM_002300
|
LDHB
|
lactate dehydrogenase B
|
|
chr17_-_40263124
|
0.576
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr14_-_87529212
|
0.574
|
|
GALC
|
galactosylceramidase
|
|
chr1_-_180840016
|
0.574
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr2_-_213724577
|
0.572
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr2_-_96174865
|
0.570
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr1_+_117253882
|
0.560
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr13_-_35818603
|
0.559
|
NM_001142296
NM_015087
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chrX_+_64804259
|
0.559
|
|
MSN
|
moesin
|
|
chr4_-_122063118
|
0.558
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr22_-_30016707
|
0.555
|
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr1_-_84237316
|
0.553
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr1_-_8861278
|
0.552
|
|
ENO1
|
enolase 1, (alpha)
|
|
chrX_-_13866565
|
0.547
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr3_-_52065130
|
0.546
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr15_+_61356801
|
0.545
|
NM_001145646
NM_031301
|
APH1B
|
anterior pharynx defective 1 homolog B (C. elegans)
|
|
chr22_+_16020241
|
0.545
|
|
CECR4
|
cat eye syndrome chromosome region, candidate 4 (non-protein coding)
|
|
chr8_+_22479115
|
0.539
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr2_-_144991386
|
0.538
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_+_181025494
|
0.538
|
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chr8_+_49083522
|
0.537
|
NM_003350
|
UBE2V2
|
ubiquitin-conjugating enzyme E2 variant 2
|
|
chr12_+_52806103
|
0.535
|
|
LOC400043
|
hypothetical LOC400043
|
|
chr1_+_199975563
|
0.535
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr7_-_72609804
|
0.534
|
|
BCL7B
|
B-cell CLL/lymphoma 7B
|
|
chr4_-_143987053
|
0.532
|
NM_001101669
NM_003866
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa
|
|
chrX_-_148430357
|
0.532
|
|
LOC100131434
|
hypothetical LOC100131434
|
|
chr11_-_66092534
|
0.526
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr2_+_74502364
|
0.526
|
NM_032118
|
WDR54
|
WD repeat domain 54
|
|
chr17_-_8000362
|
0.517
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr15_-_81744469
|
0.516
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr6_-_121697305
|
0.513
|
NM_152730
|
C6orf170
|
chromosome 6 open reading frame 170
|
|
chr7_+_27102197
|
0.510
|
|
|
|
|
chr3_-_36961539
|
0.509
|
NM_014831
|
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr5_+_176493488
|
0.507
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr5_+_17270474
|
0.503
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr7_+_39739690
|
0.492
|
|
NCRNA00265
|
non-protein coding RNA 265
|
|
chr7_+_48041632
|
0.490
|
NM_001100159
|
C7orf57
|
chromosome 7 open reading frame 57
|
|
chr10_+_80740584
|
0.489
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr7_+_154720547
|
0.487
|
|
INSIG1
|
insulin induced gene 1
|
|
chr14_+_104523660
|
0.486
|
NM_174891
|
C14orf79
|
chromosome 14 open reading frame 79
|
|
chr1_-_146398501
|
0.484
|
|
LOC100286793
FLJ39739
|
hypothetical LOC100286793
hypothetical FLJ39739
|
|
chr19_-_11352850
|
0.477
|
|
EPOR
|
erythropoietin receptor
|
|
chr8_-_9046460
|
0.477
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr19_-_36531958
|
0.473
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr8_+_94998258
|
0.468
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr18_+_54682261
|
0.465
|
|
ZNF532
|
zinc finger protein 532
|
|
chr8_+_145718702
|
0.464
|
|
|
|
|
chr9_-_139316523
|
0.458
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr9_+_90193058
|
0.456
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr3_+_114414064
|
0.456
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr16_+_88169612
|
0.454
|
NM_014427
NM_153636
|
CPNE7
|
copine VII
|
|
chr6_+_1336077
|
0.452
|
|
|
|
|
chr5_+_148717806
|
0.449
|
|
PCYOX1L
|
prenylcysteine oxidase 1 like
|
|
chr9_+_132874324
|
0.448
|
NM_006059
|
LAMC3
|
laminin, gamma 3
|
|
chr19_+_43502380
|
0.447
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr8_-_72918697
|
0.446
|
|
MSC
|
musculin
|
|
chr8_-_124123772
|
0.445
|
NM_001134671
NM_024295
|
DERL1
|
Der1-like domain family, member 1
|
|
chr7_+_55054416
|
0.442
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr5_-_127447197
|
0.441
|
|
FLJ33630
|
hypothetical LOC644873
|
|
chr7_-_87687302
|
0.441
|
NM_003130
|
SRI
|
sorcin
|
|
chr1_+_182622987
|
0.441
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr22_-_22511173
|
0.438
|
NM_001002862
NM_001135751
NM_198440
|
DERL3
|
Der1-like domain family, member 3
|
|
chr8_+_98725505
|
0.437
|
NM_178812
|
MTDH
|
metadherin
|
|
chr3_-_81893308
|
0.437
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr18_+_40513889
|
0.437
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr5_+_176493233
|
0.435
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr1_-_68071635
|
0.435
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr16_-_79395379
|
0.434
|
NM_152342
|
CDYL2
|
chromodomain protein, Y-like 2
|
|
chrX_+_101267315
|
0.434
|
NM_080390
|
TCEAL2
|
transcription elongation factor A (SII)-like 2
|
|
chr5_+_148717762
|
0.433
|
NM_024028
|
PCYOX1L
|
prenylcysteine oxidase 1 like
|
|
chr9_-_21984379
|
0.432
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr2_-_84961749
|
0.428
|
NM_001080824
|
C2orf89
|
chromosome 2 open reading frame 89
|
|
chr7_+_44802691
|
0.423
|
NM_021130
|
PPIA
|
peptidylprolyl isomerase A (cyclophilin A)
|
|
chr9_+_111582317
|
0.422
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr1_-_143232344
|
0.419
|
|
LOC728855
|
hypothetical LOC728855
|
|
chr8_-_124123707
|
0.418
|
|
DERL1
|
Der1-like domain family, member 1
|
|
chr1_-_158161898
|
0.418
|
|
TAGLN2
|
transgelin 2
|
|
chr6_-_116799548
|
0.417
|
|
LOC100287467
|
hypothetical LOC100287467
|
|
chrX_+_11686198
|
0.417
|
NM_078628
NM_078629
|
MSL3
|
male-specific lethal 3 homolog (Drosophila)
|
|
chr20_-_57015698
|
0.417
|
NM_001336
|
CTSZ
|
cathepsin Z
|
|
chr1_-_8861374
|
0.415
|
|
ENO1
|
enolase 1, (alpha)
|
|
chr3_-_109292444
|
0.414
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr9_+_81376666
|
0.412
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr22_+_48971067
|
0.410
|
|
TRABD
|
TraB domain containing
|
|
chr11_+_65357972
|
0.409
|
NM_152760
|
SNX32
|
sorting nexin 32
|
|
chr8_-_116750277
|
0.408
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr1_+_207915617
|
0.408
|
|
G0S2
|
G0/G1switch 2
|
|
chr1_-_143232464
|
0.408
|
|
|
|
|
chrX_+_64804318
|
0.408
|
|
MSN
|
moesin
|
|
chr11_+_32069183
|
0.407
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr14_-_87529600
|
0.405
|
NM_000153
NM_001037525
|
GALC
|
galactosylceramidase
|
|
chr13_-_98536640
|
0.405
|
NM_001130049
NM_015296
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr8_-_124123721
|
0.404
|
|
DERL1
|
Der1-like domain family, member 1
|
|
chr6_-_97391779
|
0.403
|
NM_001143957
NM_030784
|
GPR63
|
G protein-coupled receptor 63
|
|
chr5_+_148941228
|
0.403
|
NM_001001669
|
ARHGEF37
|
Rho guanine nucleotide exchange factor (GEF) 37
|
|
chr7_-_27102049
|
0.402
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr7_-_87687276
|
0.400
|
|
SRI
|
sorcin
|
|
chr2_-_217267742
|
0.398
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr8_-_101804114
|
0.396
|
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr1_-_35957314
|
0.396
|
NM_152374
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr2_+_222997456
|
0.396
|
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|