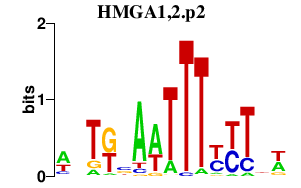
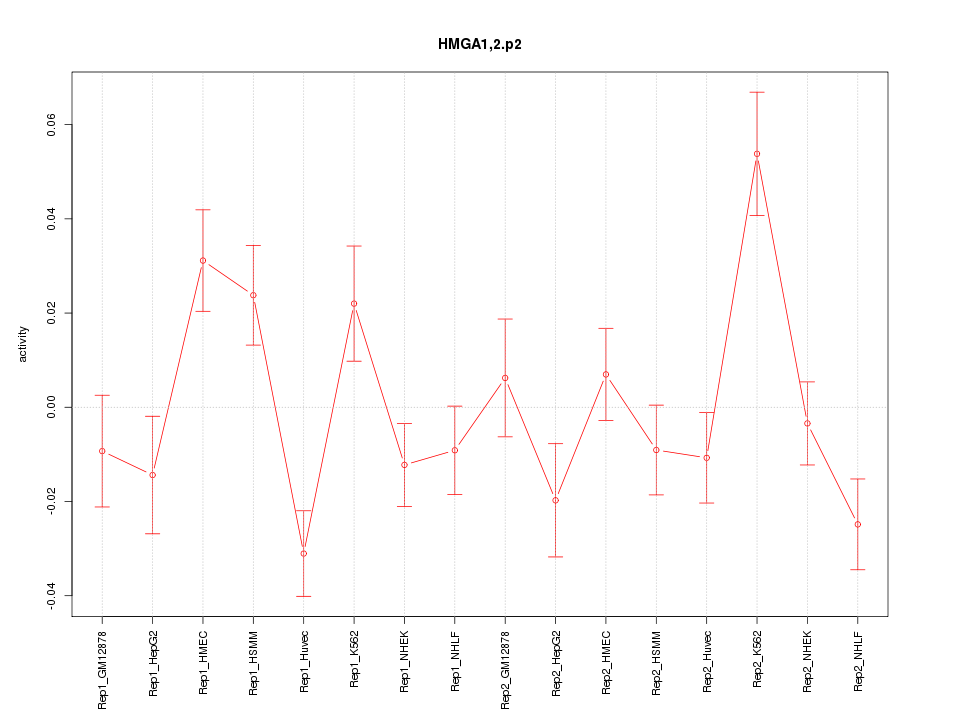
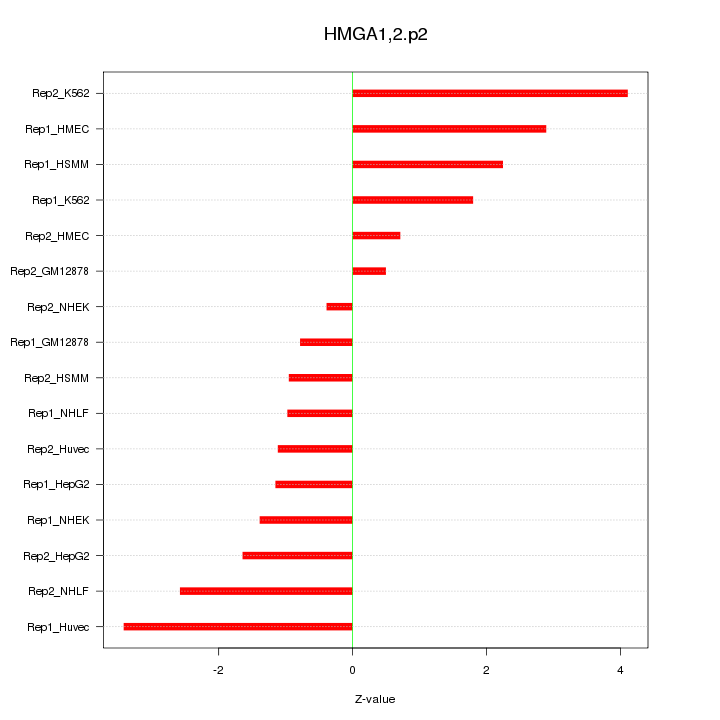
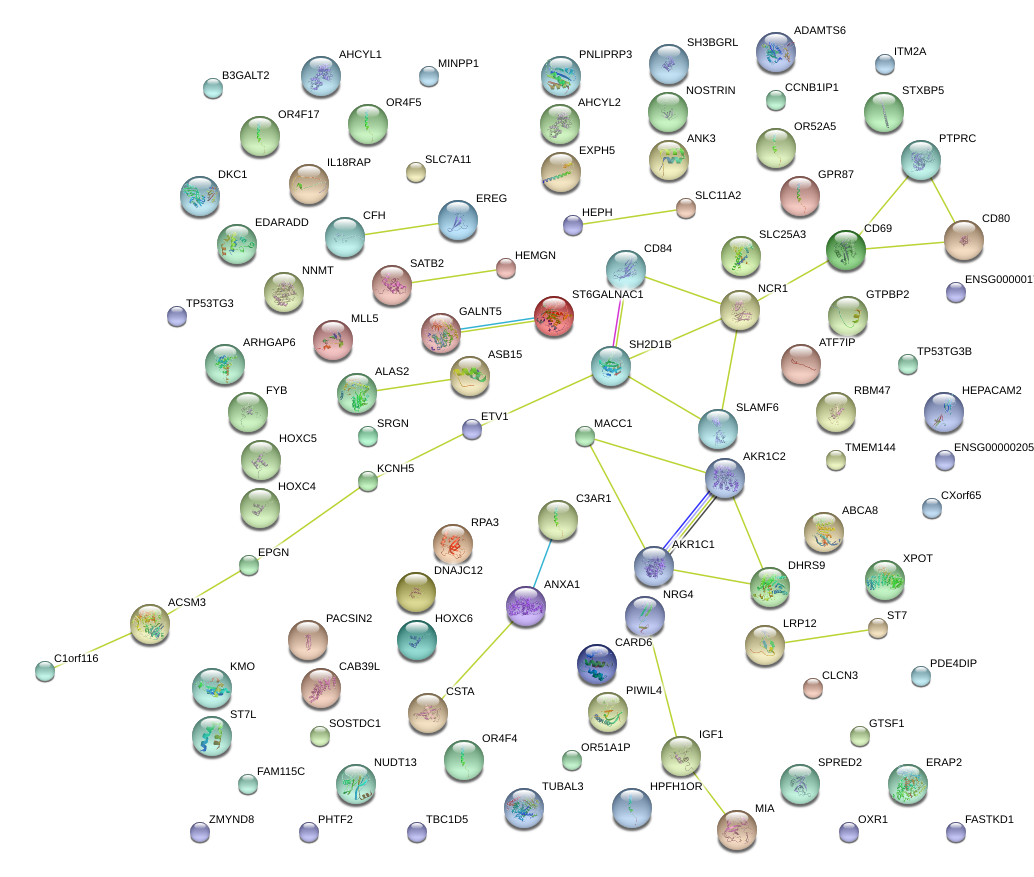
|
chr7_-_16471817
|
2.583
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr10_+_89257569
|
2.191
|
NM_001178118
|
MINPP1
|
multiple inositol-polyphosphate phosphatase 1
|
|
chr1_-_143708467
|
2.060
|
NM_001195260
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr4_+_75449720
|
1.999
|
NM_001432
|
EREG
|
epiregulin
|
|
chr1_+_196874867
|
1.905
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr7_+_77307382
|
1.881
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr1_-_158815850
|
1.873
|
|
CD84
|
CD84 molecule
|
|
chr1_-_158815895
|
1.816
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr1_+_194887630
|
1.790
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr1_-_205272619
|
1.790
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr7_+_123036312
|
1.744
|
NM_080928
|
ASB15
|
ankyrin repeat and SOCS box containing 15
|
|
chr1_+_239762179
|
1.743
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr4_+_159350850
|
1.731
|
NM_018342
|
TMEM144
|
transmembrane protein 144
|
|
chr7_-_7724762
|
1.726
|
NM_002947
|
RPA3
|
replication protein A3, 14kDa
|
|
chr1_+_234624266
|
1.716
|
NM_145861
|
EDARADD
|
EDAR-associated death domain
|
|
chr4_+_170817787
|
1.714
|
|
CLCN3
|
chloride channel 3
|
|
chr1_+_196874759
|
1.684
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr7_-_112545840
|
1.671
|
|
LOC401397
|
hypothetical LOC401397
|
|
chr7_-_112545847
|
1.658
|
|
LOC401397
|
hypothetical LOC401397
|
|
chr1_+_196874839
|
1.652
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr2_+_157822355
|
1.564
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr7_+_143043871
|
1.467
|
NM_001130026
|
FAM115C
|
family with sequence similarity 115, member C
|
|
chr9_+_74956601
|
1.465
|
|
ANXA1
|
annexin A1
|
|
chr6_+_147568795
|
1.458
|
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr3_-_120761092
|
1.450
|
NM_005191
|
CD80
|
CD80 molecule
|
|
chr3_-_17757402
|
1.414
|
NM_014744
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chr7_+_104468614
|
1.372
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr4_-_40211324
|
1.359
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr17_-_72151314
|
1.316
|
NM_018414
|
ST6GALNAC1
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1
|
|
chr9_-_99740243
|
1.313
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr7_+_116447499
|
1.311
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr6_-_43703057
|
1.301
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr12_+_97513644
|
1.293
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr8_+_107807415
|
1.271
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr11_-_5110383
|
1.254
|
NM_001005160
|
OR52A5
|
olfactory receptor, family 52, subfamily A, member 5
|
|
chrX_-_70243362
|
1.252
|
NM_001025265
|
CXorf65
|
chromosome X open reading frame 65
|
|
chr13_-_48873735
|
1.243
|
NM_030925
|
CAB39L
|
calcium binding protein 39-like
|
|
chr6_-_68655708
|
1.241
|
|
|
|
|
chr6_-_43703024
|
1.233
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr4_+_22608252
|
1.232
|
|
|
|
|
chr10_+_70517863
|
1.223
|
|
SRGN
|
serglycin
|
|
chr9_+_74956614
|
1.203
|
|
ANXA1
|
annexin A1
|
|
chr2_+_169637255
|
1.196
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr14_-_62638336
|
1.189
|
NM_172376
|
KCNH5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5
|
|
chr11_-_107969554
|
1.176
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chrX_+_65300803
|
1.170
|
NM_001130860
NM_014799
|
HEPH
|
hephaestin
|
|
chr10_+_70517823
|
1.164
|
NM_002727
|
SRGN
|
serglycin
|
|
chr16_-_32596378
|
1.162
|
NM_001099687
|
TP53TG3B
|
TP53 target 3B
|
|
chr12_+_52696908
|
1.154
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr11_-_5130174
|
1.153
|
NM_012375
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1
|
|
chr19_+_45973139
|
1.152
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr10_-_5035967
|
1.141
|
NM_001135241
|
AKR1C1
AKR1C2
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chrX_+_153644210
|
1.139
|
NM_001142463
NM_001363
|
DKC1
|
dyskeratosis congenita 1, dyskerin
|
|
chr7_-_20223467
|
1.135
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr7_+_116447527
|
1.133
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr11_+_113671744
|
1.131
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr15_-_74091817
|
1.128
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr1_+_234625338
|
1.125
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chr16_+_33111656
|
1.122
|
NM_001099687
|
TP53TG3B
|
TP53 target 3B
|
|
chr4_-_139382672
|
1.119
|
NM_014331
|
SLC7A11
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 11
|
|
chr20_-_45410033
|
1.105
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr12_-_53153613
|
1.101
|
|
GTSF1
|
gametocyte specific factor 1
|
|
chrX_-_78509662
|
1.066
|
NM_001171581
NM_004867
|
ITM2A
|
integral membrane protein 2A
|
|
chr4_+_75393053
|
1.063
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr11_+_93940121
|
1.061
|
NM_152431
|
PIWIL4
|
piwi-like 4 (Drosophila)
|
|
chr2_+_102401580
|
1.058
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr9_+_74956494
|
1.041
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr2_+_169367345
|
1.036
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr5_+_40877053
|
1.027
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr5_-_39238818
|
1.017
|
|
FYB
|
FYN binding protein
|
|
chr1_-_160648429
|
1.014
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr19_+_60109319
|
1.014
|
NM_001145457
NM_001145458
NM_004829
|
NCR1
|
natural cytotoxicity triggering receptor 1
|
|
chr3_+_123526700
|
1.012
|
NM_005213
|
CSTA
|
cystatin A (stefin A)
|
|
chr3_-_152517204
|
1.008
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr16_+_33168191
|
0.999
|
NM_001099687
|
TP53TG3B
|
TP53 target 3B
|
|
chr5_+_96237399
|
0.985
|
NM_001130140
NM_022350
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr22_-_41685827
|
0.985
|
NM_001184971
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr14_-_19867372
|
0.985
|
NM_182852
NM_182851
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr2_-_65447267
|
0.982
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr12_-_101396459
|
0.964
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr2_-_65447370
|
0.956
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr10_-_69267774
|
0.955
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr12_+_52697160
|
0.951
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr2_-_200033445
|
0.920
|
|
SATB2
|
SATB homeobox 2
|
|
chr19_+_61624
|
0.916
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr12_-_49708279
|
0.911
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr2_-_170138558
|
0.909
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr3_-_42818221
|
0.902
|
|
|
|
|
chr12_-_8110268
|
0.894
|
|
C3AR1
|
complement component 3a receptor 1
|
|
chr10_+_118177371
|
0.883
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chrX_-_55074125
|
0.879
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr17_-_64462976
|
0.876
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr12_-_8110196
|
0.870
|
NM_004054
|
C3AR1
|
complement component 3a receptor 1
|
|
chr16_+_20682812
|
0.868
|
NM_005622
NM_202000
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr7_+_128802719
|
0.867
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr1_-_158759606
|
0.865
|
NM_001184714
NM_001184715
NM_001184716
NM_052931
|
SLAMF6
|
SLAM family member 6
|
|
chr10_+_74540215
|
0.864
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chr12_+_63090008
|
0.858
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr7_-_13992351
|
0.851
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr12_+_14429499
|
0.850
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr5_-_64813459
|
0.845
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chrX_+_80343958
|
0.844
|
NM_003022
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chrX_-_11194015
|
0.843
|
NM_013423
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr1_-_191422346
|
0.842
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr10_-_5436781
|
0.838
|
NM_001171864
NM_024803
|
TUBAL3
|
tubulin, alpha-like 3
|
|
chr7_-_92693716
|
0.836
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr10_-_62002672
|
0.831
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr12_-_9804682
|
0.823
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr7_+_141054621
|
0.819
|
NM_001105558
|
WEE2
|
WEE1 homolog 2 (S. pombe)
|
|
chr15_-_70350609
|
0.815
|
NM_020214
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6
|
|
chr10_+_5976354
|
0.801
|
NM_032807
|
FBXO18
|
F-box protein, helicase, 18
|
|
chr7_-_104696678
|
0.801
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr12_+_18305740
|
0.800
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr8_+_26206648
|
0.796
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr19_-_14750347
|
0.794
|
NM_013447
NM_152916
NM_152917
NM_152918
NM_152919
NM_152920
NM_152921
|
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2
|
|
chr1_+_50344550
|
0.783
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr4_-_100070808
|
0.781
|
NM_001130679
NM_001968
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr9_+_129893948
|
0.779
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr7_-_135263737
|
0.777
|
|
MTPN
LUZP6
|
myotrophin
leucine zipper protein 6
|
|
chr13_-_35686665
|
0.777
|
NM_017826
|
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2
|
|
chr5_-_147996769
|
0.773
|
NM_001040169
NM_001040172
NM_001040173
NM_199453
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr6_-_119137923
|
0.763
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr11_-_5247948
|
0.762
|
NM_005330
|
HBE1
|
hemoglobin, epsilon 1
|
|
chr10_+_4995597
|
0.752
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr1_-_21310462
|
0.750
|
NM_003760
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr15_-_41195994
|
0.749
|
|
RPS3A
|
ribosomal protein S3A
|
|
chr12_-_642786
|
0.747
|
NM_016533
|
NINJ2
|
ninjurin 2
|
|
chr19_-_43608641
|
0.747
|
NM_001146202
NM_001146203
NM_001146204
NM_001146205
NM_001146206
NM_001146207
NM_170604
|
RASGRP4
|
RAS guanyl releasing protein 4
|
|
chr7_+_91762227
|
0.746
|
|
|
|
|
chrX_-_141120722
|
0.746
|
NM_016249
|
MAGEC2
|
melanoma antigen family C, 2
|
|
chr4_-_74704964
|
0.746
|
NM_177532
NM_201431
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr11_+_123491313
|
0.746
|
NM_001130142
NM_014622
NM_198315
|
VWA5A
|
von Willebrand factor A domain containing 5A
|
|
chr10_-_921694
|
0.743
|
NM_015155
|
LARP4B
|
La ribonucleoprotein domain family, member 4B
|
|
chr12_-_69434650
|
0.738
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr20_+_57027703
|
0.737
|
NM_030773
|
TUBB1
|
tubulin, beta 1
|
|
chr3_-_158703821
|
0.733
|
NM_001167911
NM_001167912
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr1_+_557269
|
0.731
|
|
|
|
|
chr1_+_242283129
|
0.726
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr12_-_53153639
|
0.719
|
NM_144594
|
GTSF1
|
gametocyte specific factor 1
|
|
chr19_+_10258642
|
0.716
|
NM_001039132
NM_001544
NM_022377
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)
|
|
chr8_-_116749382
|
0.713
|
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr3_-_15814678
|
0.709
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr6_-_49820030
|
0.706
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr7_-_91632525
|
0.704
|
NM_001161528
|
LOC401387
|
leucine-rich repeat and death domain-containing protein
|
|
chr6_-_49712483
|
0.704
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr11_+_113280727
|
0.699
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr13_+_107753764
|
0.697
|
|
|
|
|
chr19_-_12858956
|
0.691
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr7_-_92615593
|
0.690
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr3_+_142175329
|
0.687
|
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr4_+_159351092
|
0.681
|
|
TMEM144
|
transmembrane protein 144
|
|
chr2_-_174475528
|
0.679
|
|
|
|
|
chr10_+_4995685
|
0.677
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr8_-_62764839
|
0.676
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr16_-_53520208
|
0.673
|
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding)
|
|
chr11_+_4859624
|
0.669
|
NM_001004759
|
OR51T1
|
olfactory receptor, family 51, subfamily T, member 1
|
|
chr10_-_50066386
|
0.666
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chrX_-_80344096
|
0.666
|
NM_030763
|
HMGN5
|
high-mobility group nucleosome binding domain 5
|
|
chr1_-_26058434
|
0.665
|
NM_024037
|
C1orf135
|
chromosome 1 open reading frame 135
|
|
chr6_-_50039776
|
0.661
|
NM_001037499
|
DEFB114
|
defensin, beta 114
|
|
chr21_-_38415273
|
0.658
|
NM_005867
|
DSCR4
|
Down syndrome critical region gene 4
|
|
chr3_-_95230143
|
0.657
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chr11_-_33139449
|
0.652
|
NM_001033505
NM_001033506
NM_001326
|
CSTF3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa
|
|
chr15_-_100280784
|
0.648
|
NM_001004195
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr10_+_47875229
|
0.648
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr20_-_30703219
|
0.647
|
NM_182584
|
C20orf203
|
chromosome 20 open reading frame 203
|
|
chr7_+_120416103
|
0.646
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr2_+_27046952
|
0.645
|
NM_012326
|
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3
|
|
chr4_-_74307637
|
0.645
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr7_-_112515035
|
0.645
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr10_-_11614279
|
0.644
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr20_+_56904042
|
0.642
|
|
GNAS
|
GNAS complex locus
|
|
chr19_+_52532210
|
0.640
|
NM_018485
|
GPR77
|
G protein-coupled receptor 77
|
|
chr22_-_39189369
|
0.632
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr12_-_8271460
|
0.623
|
NM_018088
|
FAM90A1
|
family with sequence similarity 90, member A1
|
|
chr11_+_10433241
|
0.619
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr1_-_47468029
|
0.618
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr10_+_74323344
|
0.616
|
NM_152635
|
OIT3
|
oncoprotein induced transcript 3
|
|
chr6_+_31782618
|
0.616
|
NM_001003693
|
LY6G6F
|
lymphocyte antigen 6 complex, locus G6F
|
|
chrX_+_47999740
|
0.609
|
NM_005635
|
SSX1
|
synovial sarcoma, X breakpoint 1
|
|
chr16_+_52037186
|
0.607
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr1_-_172199799
|
0.607
|
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chr11_-_125278296
|
0.607
|
NM_031307
|
PUS3
|
pseudouridylate synthase 3
|
|
chr6_-_42293564
|
0.605
|
|
MRPS10
|
mitochondrial ribosomal protein S10
|
|
chr2_+_170074457
|
0.605
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr18_-_27594696
|
0.598
|
NM_001034172
|
MCART2
|
mitochondrial carrier triple repeat 2
|
|
chrX_-_153698930
|
0.597
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr5_+_86712376
|
0.597
|
|
|
|
|
chr17_-_30909219
|
0.597
|
NM_001129820
|
SLFN14
|
schlafen family member 14
|
|
chr10_+_4995453
|
0.596
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr13_-_45654292
|
0.596
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr19_-_50688310
|
0.595
|
NM_206901
|
RTN2
|
reticulon 2
|
|
chr1_-_21249974
|
0.594
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr8_-_38324809
|
0.594
|
|
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1
|
|
chr6_+_14032655
|
0.593
|
NM_001165034
|
RNF182
|
ring finger protein 182
|
|
chr4_+_100957003
|
0.589
|
NM_014395
|
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides
|
|
chr6_-_123999640
|
0.586
|
NM_006073
|
TRDN
|
triadin
|
|
chrX_-_119095544
|
0.585
|
NM_001099685
NM_032498
|
RHOXF2B
RHOXF2
|
Rhox homeobox family, member 2B
Rhox homeobox family, member 2
|
|
chr2_-_232034638
|
0.584
|
|
NCL
|
nucleolin
|
|
chr8_-_13018123
|
0.582
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|