|
chr4_-_152366970
|
2.008
|
NM_001009555
NM_001128923
|
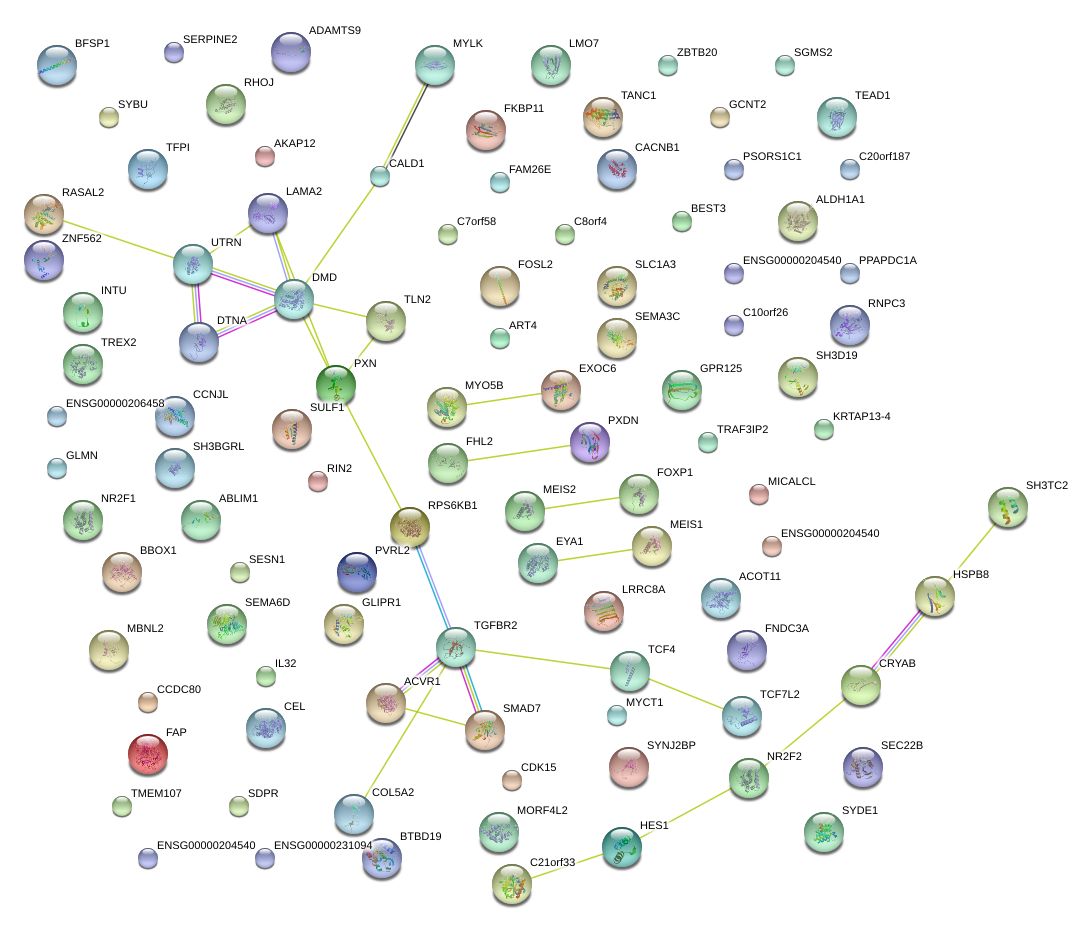
SH3D19
|
SH3 domain containing 19
|
|
chr10_+_94584449
|
1.801
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr20_+_19818209
|
1.567
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chrX_-_33139349
|
1.553
|
NM_004006
|
DMD
|
dystrophin
|
|
chr7_+_134114957
|
1.549
|
|
CALD1
|
caldesmon 1
|
|
chr7_+_134114701
|
1.382
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr6_+_151688327
|
1.370
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr11_+_12265022
|
1.312
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr14_+_62740878
|
1.312
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr10_-_116434102
|
1.255
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr2_-_188127294
|
1.248
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr10_-_116434364
|
1.226
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr2_-_188127430
|
1.192
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr2_-_192419896
|
1.153
|
|
SDPR
|
serum deprivation response
|
|
chr7_+_134114912
|
1.121
|
|
CALD1
|
caldesmon 1
|
|
chr19_+_15079141
|
1.080
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr13_+_48582389
|
1.043
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr7_-_80386258
|
1.001
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr3_-_64648703
|
0.965
|
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9
|
|
chr3_-_115585178
|
0.944
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr20_-_17487480
|
0.936
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr12_+_74160779
|
0.917
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr5_-_148422799
|
0.916
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr7_-_80386602
|
0.876
|
NM_006379
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr12_-_119172339
|
0.848
|
NM_025157
|
PXN
|
paxillin
|
|
chr12_-_14887648
|
0.830
|
NM_021071
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group)
|
|
chr3_-_113842350
|
0.827
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr2_-_189752575
|
0.823
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr12_-_68369322
|
0.813
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr10_+_122206455
|
0.811
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr3_+_195336625
|
0.795
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr10_+_104525877
|
0.791
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr4_-_22126437
|
0.790
|
NM_145290
|
GPR125
|
G protein-coupled receptor 125
|
|
chr2_+_202379442
|
0.785
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr6_+_116939500
|
0.782
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr8_-_110689396
|
0.762
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr2_+_66516196
|
0.759
|
|
MEIS1
|
Meis homeobox 1
|
|
chr2_-_224611568
|
0.736
|
NM_001136530
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr3_+_30623376
|
0.735
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr6_+_129245978
|
0.678
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr4_+_108965167
|
0.654
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr2_-_192420168
|
0.643
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr9_+_130719034
|
0.634
|
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr12_+_118100977
|
0.628
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr2_-_162808123
|
0.617
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr19_+_50041424
|
0.612
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr4_+_128773529
|
0.611
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr7_+_120416680
|
0.608
|
NM_001105533
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr6_+_31190505
|
0.608
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr3_-_113842553
|
0.606
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_54786394
|
0.603
|
NM_015547
NM_147161
|
ACOT11
|
acyl-CoA thioesterase 11
|
|
chr8_+_40130143
|
0.589
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr2_+_28469850
|
0.576
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr6_+_153060722
|
0.572
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr11_-_111287657
|
0.571
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr11_+_27019084
|
0.567
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr6_-_111995213
|
0.565
|
NM_001164283
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chrX_-_102828238
|
0.563
|
NM_001142418
NM_001142423
NM_001142424
NM_001142425
NM_001142426
NM_001142429
NM_001142430
NM_012286
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr15_+_60838053
|
0.560
|
|
TLN2
|
talin 2
|
|
chr2_+_159533329
|
0.557
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chrX_+_80344253
|
0.555
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr3_+_195336631
|
0.553
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr5_+_36644187
|
0.551
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr11_+_12652427
|
0.551
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr18_-_51219880
|
0.549
|
|
TCF4
|
transcription factor 4
|
|
chr18_-_45630154
|
0.545
|
|
MYO5B
|
myosin VB
|
|
chr2_-_105421661
|
0.539
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr2_-_158439868
|
0.536
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr8_+_70541412
|
0.529
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr6_-_109521969
|
0.528
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr9_-_74757735
|
0.527
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr15_+_94670160
|
0.516
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr15_+_45263517
|
0.515
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr13_+_75232797
|
0.506
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr16_+_3055639
|
0.502
|
NM_001012634
|
IL32
|
interleukin 32
|
|
chr3_-_113842652
|
0.498
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr16_+_3055640
|
0.497
|
|
IL32
|
interleukin 32
|
|
chr1_+_143807876
|
0.496
|
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr21_+_30724442
|
0.491
|
NM_181600
|
KRTAP13-4
|
keratin associated protein 13-4
|
|
chr18_-_44723116
|
0.486
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr1_+_176577228
|
0.485
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr6_+_10663934
|
0.483
|
NM_001491
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr1_+_45046740
|
0.482
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr18_+_30427279
|
0.478
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chrX_+_80344215
|
0.475
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr3_-_124893875
|
0.473
|
|
MYLK
|
myosin light chain kinase
|
|
chr14_-_69953433
|
0.470
|
NM_018373
|
SYNJ2BP
|
synaptojanin 2 binding protein
|
|
chr5_-_159672060
|
0.468
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr13_+_96672541
|
0.467
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr3_-_71262405
|
0.466
|
|
FOXP1
|
forkhead box P1
|
|
chr8_-_72436832
|
0.465
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr4_-_159313167
|
0.465
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr6_+_46728590
|
0.461
|
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr12_+_10351683
|
0.460
|
NM_002262
NM_007334
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr12_+_49604782
|
0.452
|
NM_014033
|
METTL7A
|
methyltransferase like 7A
|
|
chr4_-_75183700
|
0.448
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr16_+_3055313
|
0.448
|
NM_001012631
NM_001012633
NM_001012718
NM_004221
|
IL32
|
interleukin 32
|
|
chr11_-_5419358
|
0.445
|
NM_001005288
|
OR51I1
|
olfactory receptor, family 51, subfamily I, member 1
|
|
chr2_+_28469864
|
0.441
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr4_-_75123193
|
0.440
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr1_-_151848039
|
0.440
|
|
S100A16
|
S100 calcium binding protein A16
|
|
chr2_+_222871109
|
0.438
|
NM_153038
|
CCDC140
|
coiled-coil domain containing 140
|
|
chr16_+_3055642
|
0.437
|
NM_001012632
NM_001012636
|
IL32
|
interleukin 32
|
|
chr12_+_49605010
|
0.436
|
|
METTL7A
|
methyltransferase like 7A
|
|
chr10_+_53744043
|
0.432
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr6_-_56615544
|
0.430
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr16_+_3055602
|
0.429
|
NM_001012635
|
IL32
|
interleukin 32
|
|
chr3_+_46514488
|
0.427
|
NM_031440
|
RTP3
|
receptor (chemosensory) transporter protein 3
|
|
chr13_-_32658215
|
0.424
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_-_75641587
|
0.423
|
NM_001135032
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr3_+_113061218
|
0.421
|
NM_001134439
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chrX_-_43717864
|
0.418
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr2_+_69093641
|
0.418
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr16_+_68516401
|
0.417
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr3_+_184453907
|
0.415
|
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
|
|
chr18_+_3441589
|
0.413
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr12_+_13240868
|
0.413
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_-_78217278
|
0.412
|
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr3_-_71262426
|
0.412
|
|
FOXP1
|
forkhead box P1
|
|
chr3_-_15814678
|
0.409
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr4_-_101658094
|
0.406
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr6_+_134800533
|
0.405
|
|
LOC154092
|
hypothetical LOC154092
|
|
chr2_-_75641535
|
0.405
|
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr6_-_161614962
|
0.403
|
NM_020133
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
|
|
chr5_-_94443300
|
0.400
|
NM_001002796
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chrX_+_86659370
|
0.399
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr21_-_34820916
|
0.397
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr17_+_21249040
|
0.394
|
NM_001194958
|
KCNJ18
|
potassium inwardly-rectifying channel, subfamily J, member 18
|
|
chr3_+_113061296
|
0.393
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr6_+_146906520
|
0.392
|
NM_006834
|
RAB32
|
RAB32, member RAS oncogene family
|
|
chr3_+_69871252
|
0.389
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr13_+_96672705
|
0.388
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr4_+_77575276
|
0.387
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr1_+_20788027
|
0.387
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr4_-_74307637
|
0.385
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr18_+_54013575
|
0.381
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr10_+_71232281
|
0.379
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr3_+_113061330
|
0.376
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr5_+_140454382
|
0.372
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr1_+_150749943
|
0.372
|
NM_178438
|
LCE5A
|
late cornified envelope 5A
|
|
chr5_-_92942680
|
0.371
|
|
FLJ42709
|
hypothetical LOC441094
|
|
chr6_-_109522207
|
0.369
|
|
SESN1
|
sestrin 1
|
|
chrX_+_135106578
|
0.366
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr20_+_29656744
|
0.365
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr14_-_51605695
|
0.361
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr5_-_94442941
|
0.360
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chrX_-_2892288
|
0.359
|
NM_000047
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chr14_+_88360690
|
0.356
|
NM_144596
NM_198309
NM_198310
|
TTC8
|
tetratricopeptide repeat domain 8
|
|
chr22_-_34564611
|
0.353
|
|
RPL41P5
|
ribosomal protein L41 pseudogene 5
|
|
chr3_-_113841448
|
0.352
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_+_102340579
|
0.352
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr8_-_13018123
|
0.352
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr7_-_83662152
|
0.352
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr1_-_199657468
|
0.351
|
NM_003281
|
TNNI1
|
troponin I type 1 (skeletal, slow)
|
|
chr3_+_52504410
|
0.350
|
|
STAB1
|
stabilin 1
|
|
chr22_+_39583001
|
0.349
|
NM_022098
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative
|
|
chr15_-_72981595
|
0.346
|
|
C15orf17
|
chromosome 15 open reading frame 17
|
|
chr7_+_105304445
|
0.344
|
|
LOC100506768
CDHR3
|
hypothetical LOC100506768
cadherin-related family member 3
|
|
chr9_-_13269562
|
0.344
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chr21_-_35181956
|
0.340
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_-_46728188
|
0.337
|
NM_016593
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1
|
|
chr14_-_22610572
|
0.337
|
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr8_-_37847095
|
0.334
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr4_+_120167264
|
0.331
|
|
SYNPO2
|
synaptopodin 2
|
|
chr11_-_104344473
|
0.329
|
NM_001225
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr7_-_92615593
|
0.329
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr1_-_45045543
|
0.326
|
NM_001013632
|
TCTEX1D4
|
Tctex1 domain containing 4
|
|
chr8_-_72436519
|
0.326
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr10_-_131652007
|
0.325
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr14_+_85066207
|
0.324
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr14_-_22610610
|
0.324
|
NM_001164816
NM_001164817
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr17_+_65677255
|
0.323
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr3_-_188946168
|
0.320
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr1_-_148175345
|
0.319
|
|
MTMR11
|
myotubularin related protein 11
|
|
chr11_+_65027721
|
0.318
|
|
|
|
|
chr6_+_10803141
|
0.316
|
NM_017906
|
PAK1IP1
|
PAK1 interacting protein 1
|
|
chr14_-_51605487
|
0.315
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr14_-_51605461
|
0.314
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr8_-_38505331
|
0.313
|
NM_207412
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr14_-_51605515
|
0.311
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr5_-_146813343
|
0.309
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr7_+_129771865
|
0.308
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr6_+_28342766
|
0.306
|
NM_001023560
NM_001111039
NM_152736
|
ZNF187
|
zinc finger protein 187
|
|
chr1_+_194887630
|
0.306
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr17_+_35425139
|
0.305
|
NM_000759
NM_001178147
NM_172219
NM_172220
|
CSF3
|
colony stimulating factor 3 (granulocyte)
|
|
chr1_-_148174722
|
0.304
|
NM_181873
|
MTMR11
|
myotubularin related protein 11
|
|
chr14_+_54104383
|
0.304
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr7_-_158304062
|
0.303
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr10_+_24795446
|
0.302
|
|
KIAA1217
|
KIAA1217
|
|
chr7_+_142790047
|
0.300
|
|
ZYX
|
zyxin
|
|
chr9_-_72673753
|
0.300
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr14_+_104218487
|
0.300
|
|
|
|
|
chr13_-_30634031
|
0.300
|
NM_006644
|
HSPH1
|
heat shock 105kDa/110kDa protein 1
|
|
chr12_+_11694172
|
0.299
|
|
ETV6
|
ets variant 6
|
|
chr13_+_47705493
|
0.299
|
|
ITM2B
|
integral membrane protein 2B
|
|
chr21_+_29439768
|
0.299
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chrX_-_21686079
|
0.298
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr18_+_18003401
|
0.297
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr6_+_158358067
|
0.296
|
NM_001178088
|
SYNJ2
|
synaptojanin 2
|
|
chr12_-_84754289
|
0.296
|
NM_005447
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|