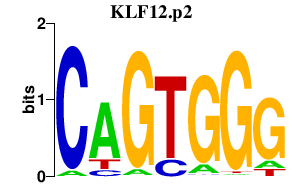
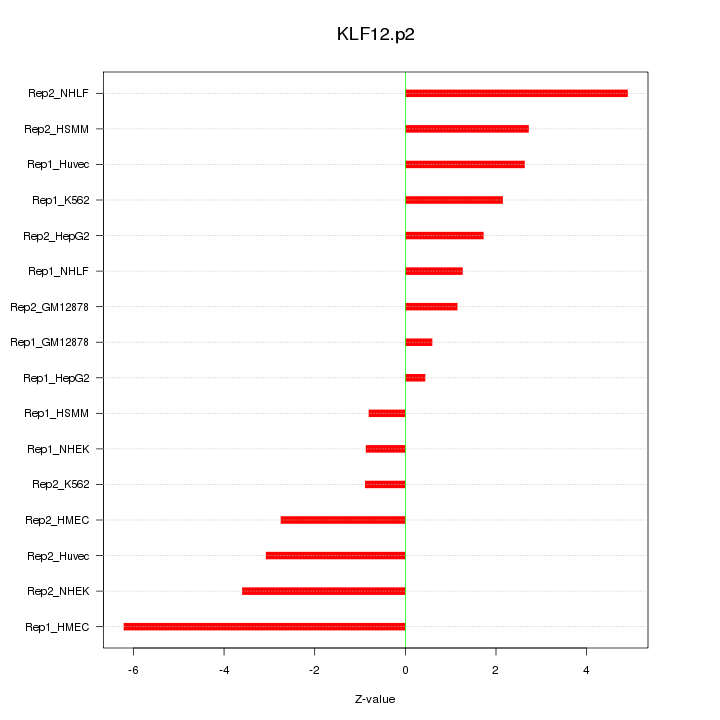
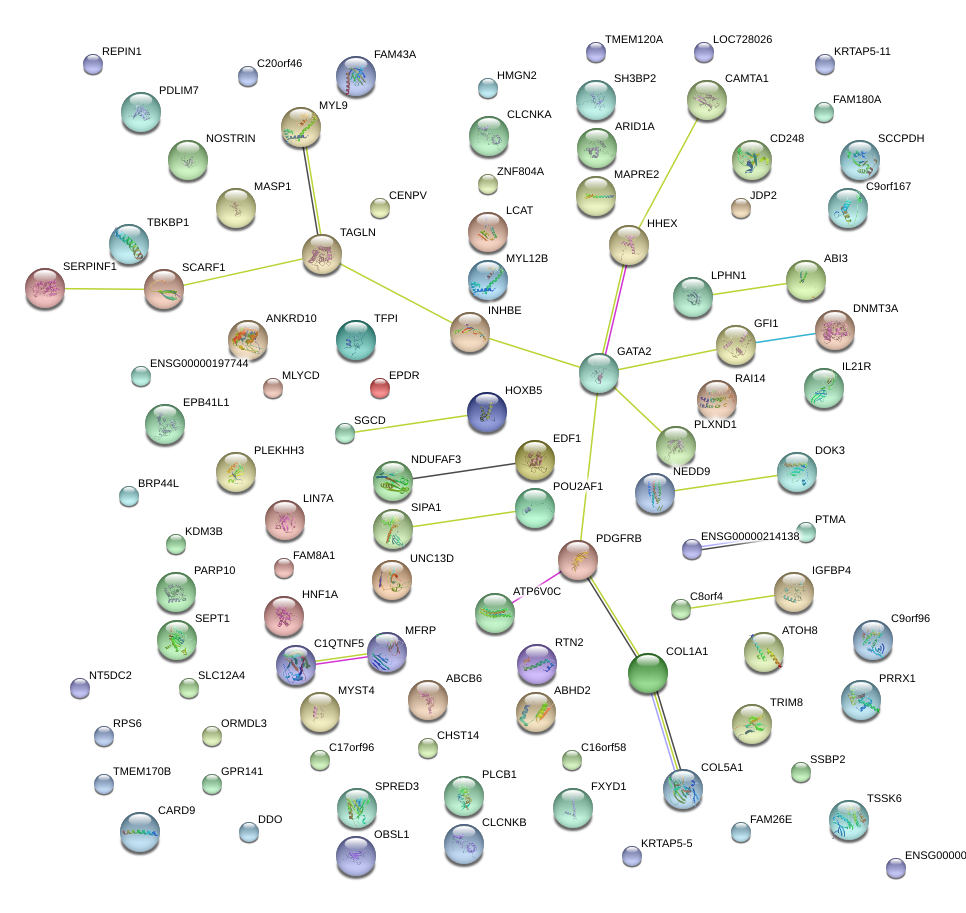
|
chr12_-_79855580
|
2.545
|
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr12_-_79855618
|
2.436
|
NM_004664
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr15_+_38550451
|
2.151
|
NM_130468
|
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14
|
|
chr1_+_168899936
|
2.041
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr9_+_136673464
|
1.880
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr10_+_94439658
|
1.780
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr11_-_65841077
|
1.759
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr16_-_66535923
|
1.747
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr2_+_85834111
|
1.667
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr9_+_136673534
|
1.490
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr19_-_14121728
|
1.450
|
|
LPHN1
|
latrophilin 1
|
|
chr20_+_8060762
|
1.430
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr11_-_118716244
|
1.414
|
|
MFRP
|
membrane frizzled-related protein
|
|
chr16_-_66535457
|
1.333
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr2_-_25328452
|
1.308
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr4_+_2783764
|
1.263
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr9_+_139292094
|
1.261
|
NM_017723
|
C9orf167
|
chromosome 9 open reading frame 167
|
|
chr5_-_81082614
|
1.249
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr2_-_220144254
|
1.243
|
|
OBSL1
|
obscurin-like 1
|
|
chr8_+_40130143
|
1.222
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr17_-_35331435
|
1.186
|
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr11_+_1607580
|
1.157
|
NM_001001480
|
KRTAP5-5
|
keratin associated protein 5-5
|
|
chr7_+_149699185
|
1.135
|
NM_014374
|
REPIN1
|
replication initiator 1
|
|
chr1_+_26671488
|
1.132
|
NM_005517
|
HMGN2
|
high-mobility group nucleosomal binding domain 2
|
|
chr16_+_2503915
|
1.122
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr10_-_105410840
|
1.110
|
|
|
|
|
chr17_+_35853177
|
1.110
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr12_+_119900931
|
1.104
|
NM_000545
|
HNF1A
|
HNF1 homeobox A
|
|
chr20_+_34603311
|
1.098
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr16_+_2503676
|
1.094
|
NM_001198569
NM_001694
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr17_-_1495742
|
1.083
|
NM_003693
NM_145350
NM_145352
|
SCARF1
|
scavenger receptor class F, member 1
|
|
chr3_-_130761967
|
1.075
|
|
PLXND1
|
plexin D1
|
|
chr11_+_65164767
|
1.068
|
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr1_+_26895108
|
1.062
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr3_-_129690056
|
1.059
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr17_-_44026042
|
1.056
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr5_-_149515331
|
1.048
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr17_-_38081967
|
1.045
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr11_+_65164963
|
1.025
|
|
|
|
|
chr16_+_82490230
|
1.007
|
NM_012213
|
MLYCD
|
malonyl-CoA decarboxylase
|
|
chr19_-_19487237
|
1.006
|
NM_032037
|
TSSK6
|
testis-specific serine kinase 6
|
|
chr16_+_27320983
|
0.997
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr12_+_119900677
|
0.984
|
|
HNF1A
|
HNF1 homeobox A
|
|
chr20_+_34206070
|
0.983
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr18_+_30810889
|
0.977
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr13_-_110365336
|
0.977
|
|
ANKRD10
|
ankyrin repeat domain 10
|
|
chr8_-_145132582
|
0.970
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr6_+_11646472
|
0.967
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr5_+_88220846
|
0.959
|
|
|
|
|
chr1_-_92724181
|
0.948
|
NM_001127216
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr7_-_135083906
|
0.938
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr9_-_138387903
|
0.929
|
|
CARD9
|
caspase recruitment domain family, member 9
|
|
chr5_+_137716169
|
0.927
|
NM_016604
|
KDM3B
|
lysine (K)-specific demethylase 3B
|
|
chr17_-_44026321
|
0.924
|
|
HOXB5
|
homeobox B5
|
|
chr2_+_185171480
|
0.910
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr19_+_43572679
|
0.908
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr19_+_40322231
|
0.902
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr9_-_138387926
|
0.899
|
NM_052813
NM_052814
|
CARD9
|
caspase recruitment domain family, member 9
|
|
chr2_-_188127294
|
0.897
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr17_+_44643067
|
0.891
|
|
ABI3
|
ABI family, member 3
|
|
chr5_+_155686333
|
0.889
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr9_+_135232937
|
0.883
|
|
C9orf96
|
chromosome 9 open reading frame 96
|
|
chr2_+_185171537
|
0.879
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr2_+_185171682
|
0.875
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr5_+_34692239
|
0.874
|
|
RAI14
|
retinoic acid induced 14
|
|
chr17_-_34084682
|
0.872
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr3_+_49033621
|
0.872
|
NM_199073
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr5_-_176868483
|
0.864
|
|
DOK3
|
docking protein 3
|
|
chr1_-_32479785
|
0.863
|
|
MTMR9LP
|
myotubularin related protein 9-like, pseudogene
|
|
chr6_+_116939500
|
0.862
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr6_+_17708483
|
0.854
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr5_-_81082699
|
0.853
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr17_+_1612092
|
0.846
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr12_+_56135353
|
0.845
|
NM_031479
|
INHBE
|
inhibin, beta E
|
|
chr5_+_88221013
|
0.828
|
|
RPS6
|
ribosomal protein S6
|
|
chr17_-_16197549
|
0.823
|
|
CENPV
|
centromere protein V
|
|
chr2_-_219791638
|
0.818
|
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr1_+_16247870
|
0.811
|
NM_001165945
|
CLCNKB
|
chloride channel Kb
|
|
chr6_-_11490487
|
0.804
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr16_-_30301357
|
0.794
|
|
SEPT1
|
septin 1
|
|
chr2_-_188127430
|
0.793
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr17_+_1612031
|
0.781
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr2_-_25390266
|
0.781
|
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr17_+_43127628
|
0.780
|
NM_014726
|
TBKBP1
|
TBK1 binding protein 1
|
|
chr17_-_71352090
|
0.778
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr5_+_34692124
|
0.775
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr2_-_220144427
|
0.775
|
|
OBSL1
|
obscurin-like 1
|
|
chr15_+_87432384
|
0.774
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr2_+_169367345
|
0.771
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr5_-_176857158
|
0.769
|
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr2_-_220144511
|
0.765
|
NM_001173408
NM_001173431
NM_015311
|
OBSL1
|
obscurin-like 1
|
|
chr17_-_45620911
|
0.765
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr11_+_116575249
|
0.762
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr19_-_50688310
|
0.762
|
NM_206901
|
RTN2
|
reticulon 2
|
|
chr3_-_52542748
|
0.759
|
NM_001134231
|
NT5DC2
|
5'-nucleotidase domain containing 2
|
|
chr9_-_138880530
|
0.758
|
NM_003792
NM_153200
|
EDF1
|
endothelial differentiation-related factor 1
|
|
chr7_+_37926943
|
0.749
|
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr11_-_118716736
|
0.749
|
|
C1QTNF5
MFRP
|
C1q and tumor necrosis factor related protein 5
membrane frizzled-related protein
|
|
chr6_-_166716353
|
0.747
|
|
BRP44L
|
brain protein 44-like
|
|
chr20_+_34603290
|
0.744
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr16_+_2503994
|
0.741
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr3_-_188492163
|
0.735
|
NM_001031849
NM_001879
NM_139125
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor)
|
|
chr2_+_232281491
|
0.734
|
|
PTMA
|
prothymosin, alpha
|
|
chr20_-_1113116
|
0.731
|
NM_018354
|
C20orf46
|
chromosome 20 open reading frame 46
|
|
chr10_+_76256305
|
0.728
|
NM_012330
|
MYST4
|
MYST histone acetyltransferase (monocytic leukemia) 4
|
|
chr16_+_2503978
|
0.728
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr2_-_219791902
|
0.728
|
NM_005689
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr1_+_244954153
|
0.724
|
|
SCCPDH
|
saccharopine dehydrogenase (putative)
|
|
chr3_+_195888701
|
0.721
|
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr7_-_75461814
|
0.720
|
|
TMEM120A
|
transmembrane protein 120A
|
|
chr1_+_6767968
|
0.719
|
NM_001195563
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr11_-_110755112
|
0.714
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr17_+_1612056
|
0.709
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr10_+_104394185
|
0.706
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr19_+_50963484
|
0.704
|
|
|
|
|
chr14_+_74964261
|
0.700
|
NM_001135047
|
JDP2
|
Jun dimerization protein 2
|
|
chr3_+_49034050
|
0.697
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr16_-_31427130
|
0.693
|
NM_022744
|
C16orf58
|
chromosome 16 open reading frame 58
|
|
chr6_-_110843432
|
0.692
|
NM_003649
NM_004032
|
DDO
|
D-aspartate oxidase
|
|
chr13_-_110365416
|
0.692
|
NM_017664
|
ANKRD10
|
ankyrin repeat domain 10
|
|
chr17_+_35751796
|
0.689
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr2_+_42128521
|
0.687
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr17_-_1342700
|
0.685
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr17_-_1029758
|
0.681
|
NM_021962
|
ABR
|
active BCR-related gene
|
|
chr5_+_133870175
|
0.679
|
|
|
|
|
chr22_+_18499354
|
0.678
|
NM_001185024
NM_013373
|
ZDHHC8
|
zinc finger, DHHC-type containing 8
|
|
chr19_-_51081147
|
0.677
|
NM_015649
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr3_+_52543023
|
0.676
|
|
|
|
|
chr17_-_59273988
|
0.673
|
|
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2
|
|
chr17_+_23723180
|
0.672
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr3_-_129689394
|
0.671
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr7_-_75461868
|
0.669
|
|
TMEM120A
|
transmembrane protein 120A
|
|
chr20_-_2769245
|
0.668
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr20_+_2769338
|
0.668
|
NM_022575
NM_080413
|
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae)
|
|
chr11_-_70971556
|
0.660
|
NM_001005405
|
KRTAP5-11
|
keratin associated protein 5-11
|
|
chr17_-_36451189
|
0.660
|
NM_030967
|
KRTAP1-3
KRTAP1-1
|
keratin associated protein 1-3
keratin associated protein 1-1
|
|
chr19_-_11169457
|
0.658
|
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr22_-_41372620
|
0.657
|
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chr5_-_176857207
|
0.653
|
NM_005451
NM_203352
NM_213636
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr17_-_1336073
|
0.651
|
|
MYO1C
|
myosin IC
|
|
chr13_-_27967215
|
0.651
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr17_+_23723220
|
0.648
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr11_+_67563030
|
0.644
|
NM_006019
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr17_+_23723113
|
0.642
|
NM_015077
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr3_-_185578621
|
0.641
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr17_-_71352017
|
0.640
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr17_-_44642934
|
0.640
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr1_-_182273451
|
0.636
|
NM_015101
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr20_-_2769120
|
0.634
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr2_-_73152323
|
0.634
|
NM_144579
|
SFXN5
|
sideroflexin 5
|
|
chr20_-_2769789
|
0.631
|
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr9_+_135233047
|
0.624
|
NM_153710
|
C9orf96
|
chromosome 9 open reading frame 96
|
|
chr19_-_3928516
|
0.622
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr3_+_171619346
|
0.621
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr6_-_111911106
|
0.616
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr1_+_1994943
|
0.615
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chr10_-_44200458
|
0.615
|
NM_000609
NM_001033886
NM_001178134
NM_199168
|
CXCL12
|
chemokine (C-X-C motif) ligand 12
|
|
chr14_-_22891263
|
0.612
|
NM_020372
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr11_-_118471313
|
0.611
|
|
H2AFX
|
H2A histone family, member X
|
|
chr6_+_32003380
|
0.610
|
|
C2
|
complement component 2
|
|
chr7_-_99871981
|
0.608
|
NM_145030
|
C7orf47
|
chromosome 7 open reading frame 47
|
|
chr14_-_22358752
|
0.607
|
NM_001126106
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr15_-_80123935
|
0.606
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr6_+_19945578
|
0.605
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr16_-_30301634
|
0.605
|
NM_052838
|
SEPT1
|
septin 1
|
|
chr4_+_72271218
|
0.602
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chrX_-_154146930
|
0.602
|
NM_171998
|
RAB39B
|
RAB39B, member RAS oncogene family
|
|
chr7_+_23496661
|
0.601
|
|
RPS2P32
|
ribosomal protein S2 pseudogene 32
|
|
chr5_+_34692352
|
0.600
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr5_-_176869463
|
0.600
|
NM_024872
|
DOK3
|
docking protein 3
|
|
chr17_-_59131194
|
0.599
|
|
LIMD2
|
LIM domain containing 2
|
|
chr17_-_59131210
|
0.592
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr16_+_65756179
|
0.590
|
|
HSF4
|
heat shock transcription factor 4
|
|
chr1_+_149286796
|
0.589
|
NM_017860
|
C1orf56
|
chromosome 1 open reading frame 56
|
|
chr10_+_111755715
|
0.583
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr15_+_62230962
|
0.582
|
NM_024798
|
SNX22
|
sorting nexin 22
|
|
chr17_-_71352182
|
0.581
|
NM_199242
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr15_-_72952648
|
0.581
|
NM_005697
|
SCAMP2
|
secretory carrier membrane protein 2
|
|
chr5_-_32480586
|
0.580
|
NM_016107
|
ZFR
|
zinc finger RNA binding protein
|
|
chr5_-_176869943
|
0.578
|
NM_001144875
NM_001144876
|
DOK3
|
docking protein 3
|
|
chr7_+_90177082
|
0.578
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr1_+_26220895
|
0.578
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr10_-_102269580
|
0.576
|
NM_015490
|
SEC31B
|
SEC31 homolog B (S. cerevisiae)
|
|
chr8_-_145119627
|
0.576
|
NM_201378
|
PLEC
|
plectin
|
|
chr1_-_182273114
|
0.576
|
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr16_+_66129481
|
0.573
|
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr9_-_90983501
|
0.573
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr5_-_32480486
|
0.570
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr1_-_42978320
|
0.570
|
NM_001123395
NM_001185117
NM_148960
|
CLDN19
|
claudin 19
|
|
chr17_+_12633504
|
0.570
|
NM_014859
|
ARHGAP44
|
Rho GTPase activating protein 44
|
|
chr19_+_47521600
|
0.567
|
NM_001410
|
MEGF8
|
multiple EGF-like-domains 8
|
|
chr1_-_53380709
|
0.567
|
NM_006671
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7
|
|
chr2_+_232281470
|
0.567
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr16_+_31132842
|
0.566
|
NM_001008274
|
TRIM72
|
tripartite motif containing 72
|
|
chr19_+_10689772
|
0.565
|
|
DNM2
|
dynamin 2
|
|
chr17_+_71772875
|
0.564
|
NM_182565
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr6_-_166716449
|
0.564
|
NM_016098
|
BRP44L
|
brain protein 44-like
|
|
chr6_-_109882116
|
0.562
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr3_-_121883647
|
0.562
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr19_+_6723705
|
0.561
|
NM_005428
|
VAV1
|
vav 1 guanine nucleotide exchange factor
|