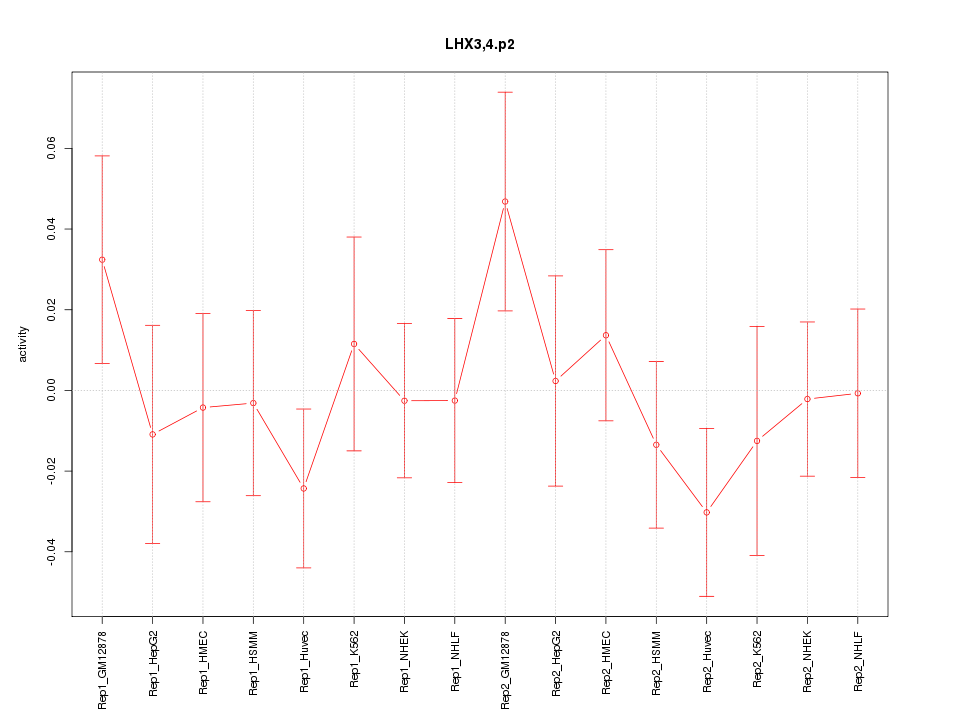
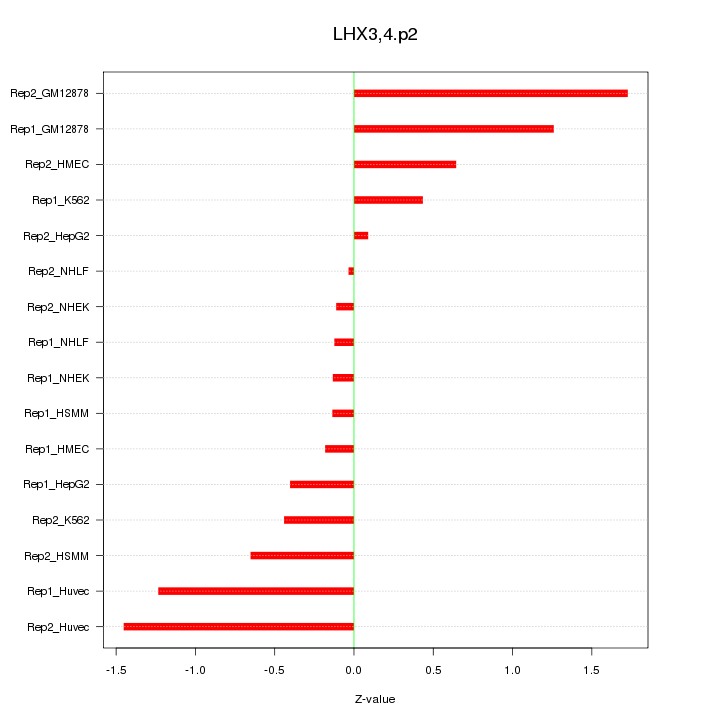
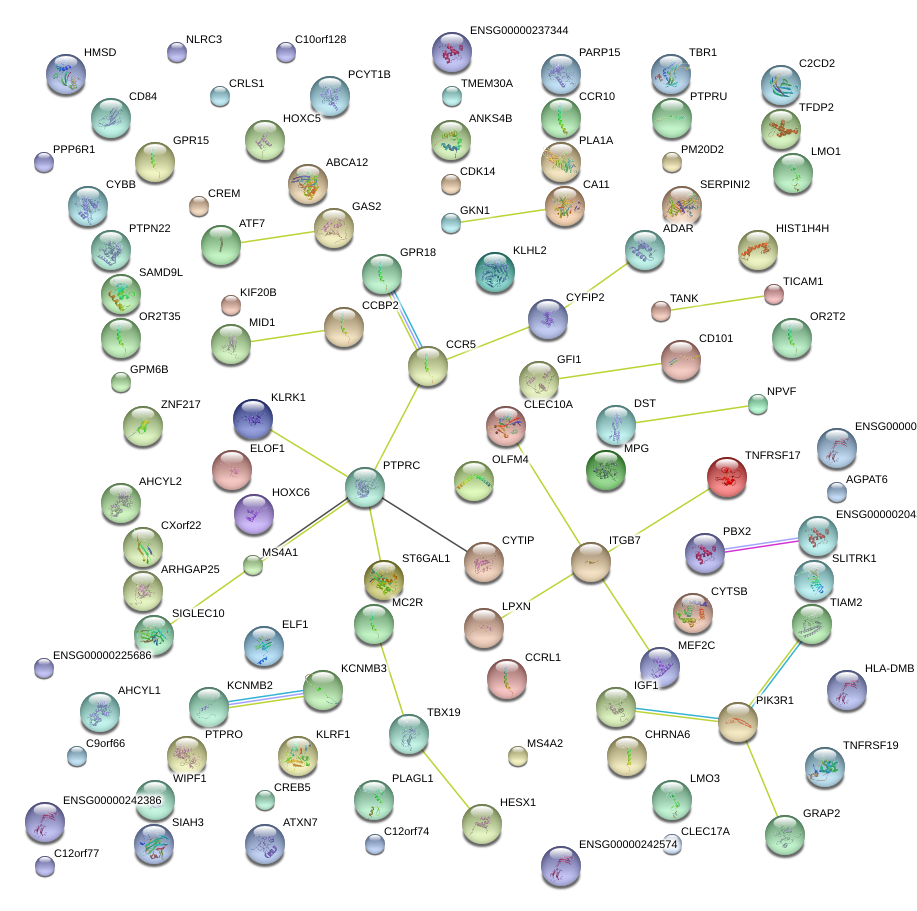
|
chr5_-_88155360
|
2.771
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr19_-_56612653
|
2.023
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr2_-_175170674
|
1.451
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr17_+_19917420
|
1.290
|
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr13_+_23042722
|
1.234
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr16_+_11966464
|
1.197
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr18_+_59767567
|
1.148
|
NM_001123366
|
HMSD
|
histocompatibility (minor) serpin domain containing
|
|
chr1_+_196874839
|
1.126
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr6_-_32265489
|
1.093
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr12_-_101396459
|
1.036
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_114215768
|
0.970
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chr3_-_57209319
|
0.965
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr19_+_14554895
|
0.948
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr1_-_92725017
|
0.876
|
NM_005263
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr20_-_51633042
|
0.870
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr7_-_25234582
|
0.870
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr7_+_90177082
|
0.829
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr5_+_67622242
|
0.821
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr17_-_38087366
|
0.796
|
NM_016602
|
CCR10
|
chemokine (C-C motif) receptor 10
|
|
chr22_+_38627129
|
0.782
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr8_-_42742729
|
0.743
|
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr22_+_38627031
|
0.739
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr1_+_166516901
|
0.722
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr6_+_31538934
|
0.715
|
NM_006674
|
HCP5
|
HLA complex P5
|
|
chr3_+_99733432
|
0.707
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr2_-_158008763
|
0.675
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr11_+_22646229
|
0.635
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr6_-_144427340
|
0.631
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr11_+_59979857
|
0.625
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr10_-_50066386
|
0.602
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr2_-_215711313
|
0.592
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr7_-_92615593
|
0.589
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr3_-_143230124
|
0.580
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr13_-_83354425
|
0.552
|
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr7_+_128802719
|
0.552
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr12_-_51887266
|
0.551
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr1_+_196874867
|
0.551
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr12_+_91620749
|
0.539
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr5_+_156628929
|
0.529
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr2_+_161981138
|
0.526
|
|
TBR1
|
T-box, brain, 1
|
|
chr1_-_158815895
|
0.524
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr3_+_46386636
|
0.520
|
NM_000579
NM_001100168
|
CCR5
|
chemokine (C-C motif) receptor 5
|
|
chr12_-_52281050
|
0.501
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr12_+_52697160
|
0.498
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr3_-_168674502
|
0.492
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr1_+_117345894
|
0.489
|
NM_004258
|
CD101
|
CD101 molecule
|
|
chrX_-_13744933
|
0.486
|
|
GPM6B
|
glycoprotein M6B
|
|
chr9_-_205892
|
0.484
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chrX_+_37524190
|
0.473
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr12_-_51887318
|
0.466
|
|
ITGB7
|
integrin, beta 7
|
|
chr2_+_161980865
|
0.458
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr2_-_65013070
|
0.446
|
|
LOC400958
|
hypothetical LOC400958
|
|
chr2_+_161981046
|
0.429
|
|
TBR1
|
T-box, brain, 1
|
|
chr10_+_91488704
|
0.422
|
|
KIF20B
|
kinesin family member 20B
|
|
chr19_-_53841064
|
0.416
|
NM_001217
|
CA11
|
carbonic anhydrase XI
|
|
chr11_-_58099884
|
0.412
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr12_+_15590811
|
0.407
|
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr13_+_52500830
|
0.405
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr1_-_152845399
|
0.404
|
NM_001193495
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chrX_+_37524250
|
0.403
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chrX_-_10761729
|
0.397
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr7_+_37689903
|
0.393
|
|
|
|
|
chr4_+_166350620
|
0.393
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr16_-_3567354
|
0.393
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr3_+_63928459
|
0.392
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr7_+_90176647
|
0.392
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr21_-_42219849
|
0.387
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr1_+_196874759
|
0.384
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr13_-_45323757
|
0.369
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr18_-_13905534
|
0.368
|
NM_000529
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone)
|
|
chrX_+_55951280
|
0.367
|
|
|
|
|
chr2_+_161701711
|
0.365
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr19_-_60458915
|
0.365
|
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1
|
|
chr6_-_76022565
|
0.361
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr3_+_188222337
|
0.358
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr13_-_40491405
|
0.356
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr12_-_16652202
|
0.353
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr6_-_56824371
|
0.348
|
|
DST
|
dystonin
|
|
chrX_-_24600661
|
0.348
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr13_-_98708549
|
0.346
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr3_+_120799411
|
0.345
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr7_+_28418668
|
0.342
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr20_+_5935897
|
0.342
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr2_+_68815416
|
0.339
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr3_+_123779122
|
0.334
|
NM_001113523
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr6_-_33016561
|
0.328
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr1_+_246682699
|
0.326
|
NM_001004136
|
OR2T2
|
olfactory receptor, family 2, subfamily T, member 2
|
|
chr10_+_35466715
|
0.321
|
NM_183011
NM_183012
|
CREM
|
cAMP responsive element modulator
|
|
chr6_+_155579788
|
0.320
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr12_-_25041639
|
0.318
|
NM_001101339
|
C12orf77
|
chromosome 12 open reading frame 77
|
|
chr12_+_9871343
|
0.316
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr3_+_179759181
|
0.311
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr3_+_123385219
|
0.311
|
NM_000388
|
CASR
|
calcium-sensing receptor
|
|
chr17_-_9635325
|
0.309
|
NM_001105571
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr13_+_47948586
|
0.308
|
|
RB1
|
retinoblastoma 1
|
|
chr10_+_18729518
|
0.308
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr20_-_18425811
|
0.307
|
NM_006606
|
RBBP9
|
retinoblastoma binding protein 9
|
|
chr4_+_123311207
|
0.305
|
NM_015312
|
KIAA1109
|
KIAA1109
|
|
chr1_-_149681265
|
0.305
|
NM_207171
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chrX_+_107174855
|
0.302
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chrX_+_70682204
|
0.297
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr7_-_132371208
|
0.296
|
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma
|
|
chr22_-_38619739
|
0.294
|
NM_152512
|
ENTHD1
|
ENTH domain containing 1
|
|
chr5_+_166644420
|
0.287
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr14_-_105610144
|
0.285
|
|
IGHA1
IGHG1
IGHG3
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr1_-_230717850
|
0.282
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr12_+_52696908
|
0.279
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr5_+_6799100
|
0.279
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|
|
chr10_+_63478974
|
0.277
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr5_-_1935764
|
0.277
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chrX_+_99726445
|
0.276
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr6_+_31691745
|
0.275
|
NM_032955
|
AIF1
|
allograft inflammatory factor 1
|
|
chr6_-_138231006
|
0.273
|
|
|
|
|
chrX_+_85404626
|
0.273
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr9_+_92603832
|
0.268
|
NM_001135052
NM_003177
|
SYK
|
spleen tyrosine kinase
|
|
chr8_-_102872372
|
0.268
|
|
NCALD
|
neurocalcin delta
|
|
chr1_+_242283129
|
0.266
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chrX_+_115215985
|
0.266
|
NM_000686
|
AGTR2
|
angiotensin II receptor, type 2
|
|
chr4_+_88790477
|
0.264
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr4_+_38740246
|
0.263
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr14_-_46882144
|
0.259
|
NM_182830
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr14_-_106249999
|
0.252
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr1_+_157067729
|
0.251
|
NM_002432
|
MNDA
|
myeloid cell nuclear differentiation antigen
|
|
chr16_+_2593351
|
0.248
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr5_+_140772691
|
0.247
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr1_+_173111306
|
0.244
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr2_-_32344192
|
0.244
|
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr1_+_111215343
|
0.240
|
NM_001040033
|
CD53
|
CD53 molecule
|
|
chr20_+_9146036
|
0.238
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr3_+_39399770
|
0.236
|
NM_017875
|
SLC25A38
|
solute carrier family 25, member 38
|
|
chrX_+_78087484
|
0.236
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr14_-_94306104
|
0.236
|
NM_173849
|
GSC
|
goosecoid homeobox
|
|
chr2_+_28892366
|
0.236
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr18_+_70352779
|
0.231
|
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family)
|
|
chr8_+_22266256
|
0.230
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr12_+_25096755
|
0.229
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr16_-_66075216
|
0.229
|
NM_001138
|
AGRP
|
agouti related protein homolog (mouse)
|
|
chr4_+_88748704
|
0.228
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr6_-_29635680
|
0.227
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr18_+_31488530
|
0.226
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr1_+_242281199
|
0.225
|
|
ZNF238
|
zinc finger protein 238
|
|
chr4_-_83566246
|
0.224
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr11_+_6854431
|
0.224
|
NM_207186
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4
|
|
chr11_-_117628211
|
0.223
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr14_+_19598043
|
0.223
|
NM_001004717
|
OR4L1
|
olfactory receptor, family 4, subfamily L, member 1
|
|
chr17_-_36528113
|
0.220
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr6_+_29963328
|
0.220
|
|
HLA-H
|
major histocompatibility complex, class I, H (pseudogene)
|
|
chr3_+_179736917
|
0.219
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chrX_-_72351392
|
0.219
|
NM_021963
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr4_-_83566045
|
0.218
|
|
|
|
|
chr6_+_50789215
|
0.218
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr1_-_89437098
|
0.217
|
NM_052941
|
GBP4
|
guanylate binding protein 4
|
|
chr6_+_152053323
|
0.214
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr1_+_78888064
|
0.214
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr12_-_101036407
|
0.212
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr13_-_102517196
|
0.212
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr10_+_18589589
|
0.212
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr11_-_20138445
|
0.211
|
NM_001029865
|
DBX1
|
developing brain homeobox 1
|
|
chr18_+_70352649
|
0.211
|
NM_032649
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family)
|
|
chr18_-_72857665
|
0.210
|
|
MBP
|
myelin basic protein
|
|
chr6_+_167445284
|
0.209
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr13_+_77007809
|
0.209
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr3_-_54937001
|
0.208
|
NM_020678
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1
|
|
chrX_-_102418329
|
0.208
|
NM_001012979
|
TCEAL5
|
transcription elongation factor A (SII)-like 5
|
|
chr17_-_9863929
|
0.207
|
|
GAS7
|
growth arrest-specific 7
|
|
chr18_+_27210737
|
0.206
|
NM_001134453
NM_177986
|
DSG4
|
desmoglein 4
|
|
chr17_-_3129017
|
0.206
|
NM_002551
|
OR3A2
|
olfactory receptor, family 3, subfamily A, member 2
|
|
chr3_+_103029523
|
0.206
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr6_-_56824672
|
0.204
|
|
DST
|
dystonin
|
|
chr15_-_70350609
|
0.204
|
NM_020214
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6
|
|
chr8_-_102872354
|
0.204
|
|
NCALD
|
neurocalcin delta
|
|
chr8_+_55691179
|
0.201
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chr13_-_83354474
|
0.199
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr7_-_122627235
|
0.198
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr5_-_160211623
|
0.197
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr12_+_14429499
|
0.197
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr6_+_31661934
|
0.195
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr4_-_116254381
|
0.195
|
NM_022569
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4
|
|
chr19_+_60962318
|
0.194
|
NM_001145014
|
RFPL4A
|
ret finger protein-like 4A
|
|
chr6_+_55147029
|
0.194
|
NM_001526
|
HCRTR2
|
hypocretin (orexin) receptor 2
|
|
chr9_+_124173049
|
0.193
|
NM_000962
NM_080591
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr15_+_46271158
|
0.191
|
NM_001145668
|
CTXN2
SLC12A1
|
cortexin 2
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr6_-_22405708
|
0.191
|
NM_000948
|
PRL
|
prolactin
|
|
chr12_-_56422120
|
0.191
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chrX_-_13745108
|
0.190
|
|
GPM6B
|
glycoprotein M6B
|
|
chr13_+_112349358
|
0.190
|
NM_207440
|
C13orf35
|
chromosome 13 open reading frame 35
|
|
chr8_-_7207899
|
0.190
|
NM_001164457
|
ZNF705G
|
zinc finger protein 705G
|
|
chrX_-_76824726
|
0.188
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr11_-_118405288
|
0.188
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr17_-_36376663
|
0.187
|
NM_213656
|
KRT39
|
keratin 39
|
|
chrX_-_13745134
|
0.187
|
|
GPM6B
|
glycoprotein M6B
|
|
chr2_-_99238001
|
0.186
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr12_+_52708460
|
0.185
|
NM_004503
|
HOXC6
|
homeobox C6
|
|
chr18_-_3835295
|
0.184
|
NM_001003809
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr2_-_80385227
|
0.184
|
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr8_+_32905027
|
0.184
|
|
|
|
|
chr8_+_67505091
|
0.183
|
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chrX_-_110542040
|
0.182
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chr13_-_40491491
|
0.181
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chrX_+_7147471
|
0.181
|
NM_000351
|
STS
|
steroid sulfatase (microsomal), isozyme S
|