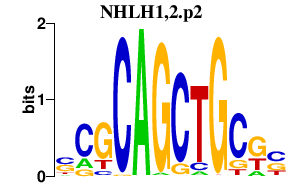
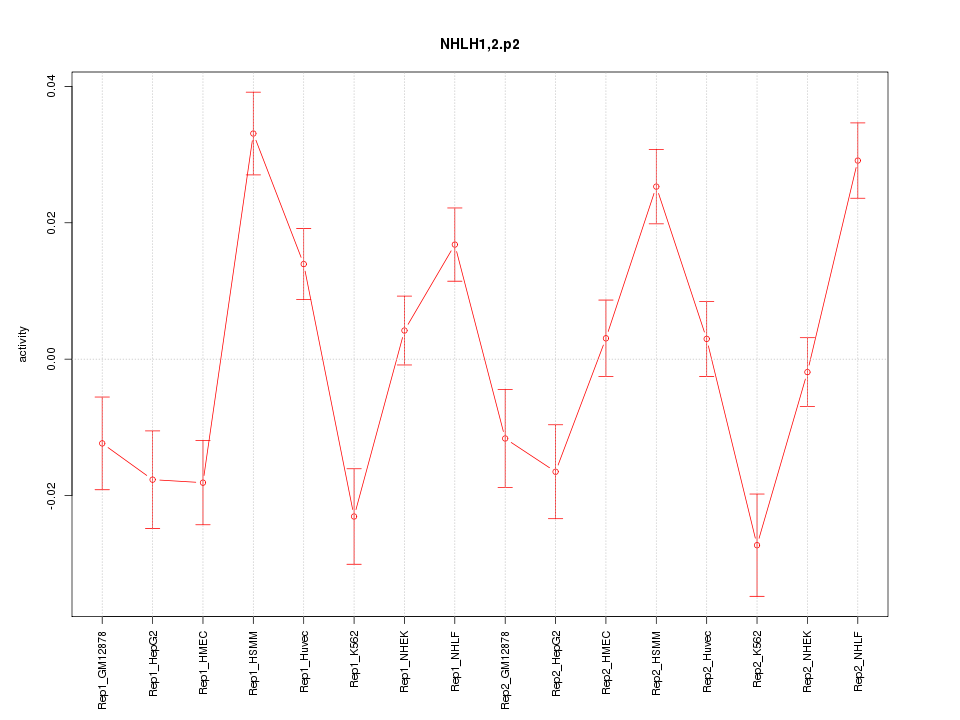
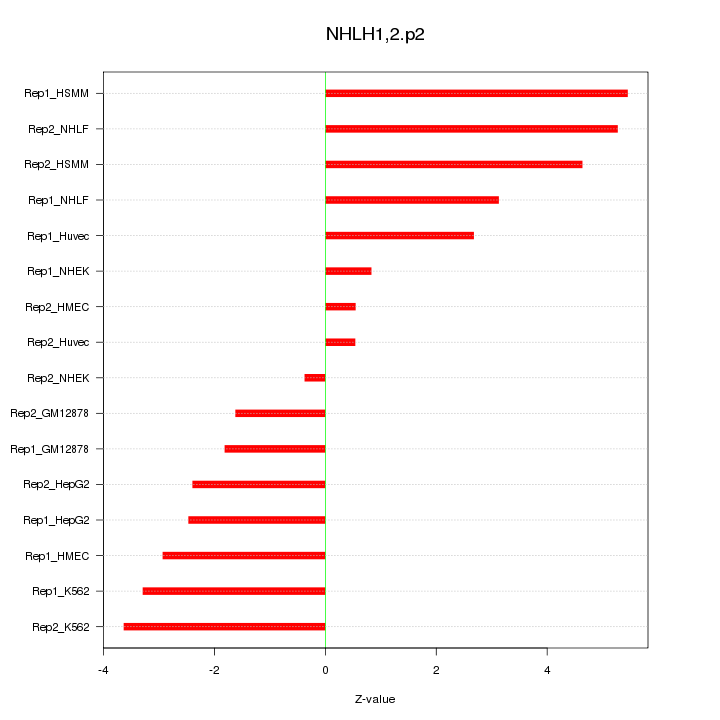
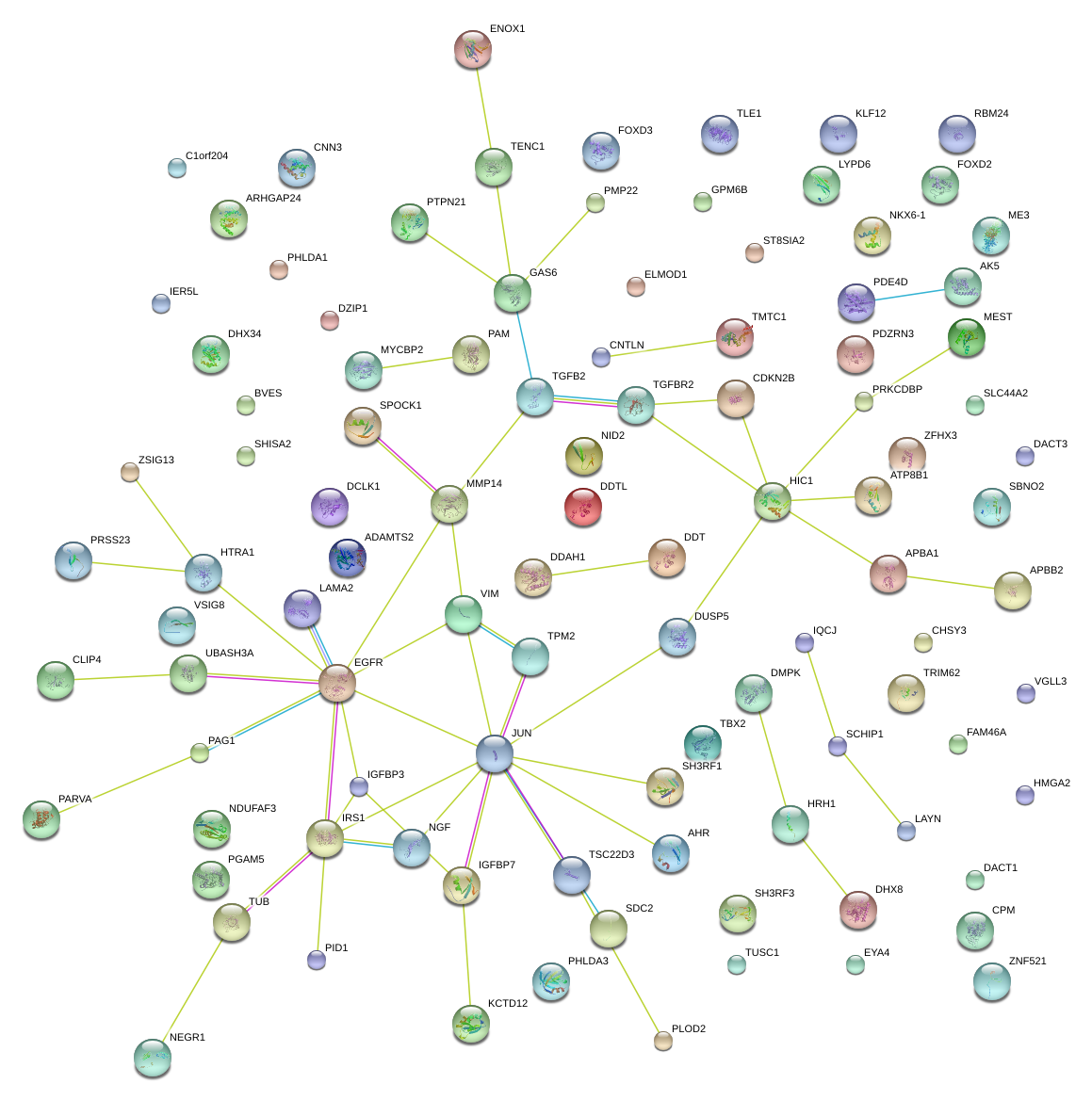
|
chr2_+_109112428
|
8.359
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr7_+_17304707
|
6.231
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr9_-_83493415
|
4.432
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr12_-_74710741
|
4.404
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr3_-_73756668
|
4.339
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr2_-_229844198
|
4.278
|
NM_001100818
NM_017933
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr9_+_17124974
|
4.125
|
NM_001114395
NM_017738
|
CNTLN
|
centlein, centrosomal protein
|
|
chr11_-_6298284
|
3.862
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr13_-_25523164
|
3.604
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr4_-_40911212
|
3.430
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr19_+_10597167
|
3.209
|
NM_020428
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr3_-_87122936
|
3.150
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr3_-_87122958
|
3.023
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr9_-_21549696
|
2.978
|
|
LOC554202
|
hypothetical LOC554202
|
|
chr7_-_45927263
|
2.928
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr13_-_43259032
|
2.878
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chrX_-_106846253
|
2.839
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr13_+_113546841
|
2.809
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr17_+_56832492
|
2.762
|
|
TBX2
|
T-box 2
|
|
chr7_+_129919330
|
2.738
|
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr5_-_58370693
|
2.721
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_+_29191711
|
2.712
|
NM_024692
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr5_+_129268352
|
2.704
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr7_+_55054416
|
2.703
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr5_-_72779585
|
2.697
|
|
FOXD1
|
forkhead box D1
|
|
chr15_+_90738108
|
2.619
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr19_-_1083109
|
2.608
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr6_+_129245978
|
2.572
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chrX_-_13866451
|
2.558
|
|
GPM6B
|
glycoprotein M6B
|
|
chr18_-_21185968
|
2.549
|
|
ZNF521
|
zinc finger protein 521
|
|
chr13_+_113546943
|
2.533
|
|
GAS6
|
growth arrest-specific 6
|
|
chr13_+_113546897
|
2.529
|
|
GAS6
|
growth arrest-specific 6
|
|
chr6_+_133604203
|
2.507
|
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr14_+_22375753
|
2.489
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr1_-_59020875
|
2.453
|
|
JUN
|
jun proto-oncogene
|
|
chr1_+_77520315
|
2.428
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr9_-_71476917
|
2.426
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr10_+_112247614
|
2.424
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr8_+_97575370
|
2.376
|
|
SDC2
|
syndecan 2
|
|
chr1_+_68070573
|
2.368
|
|
|
|
|
chr1_-_115682374
|
2.360
|
NM_002506
|
NGF
|
nerve growth factor (beta polypeptide)
|
|
chr14_-_88090641
|
2.358
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr2_-_227371698
|
2.358
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr11_+_7997366
|
2.318
|
|
TUB
|
tubby homolog (mouse)
|
|
chr1_-_85703321
|
2.244
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr13_-_95094920
|
2.225
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr1_-_158091296
|
2.212
|
|
C1orf204
|
chromosome 1 open reading frame 204
|
|
chr10_+_17311307
|
2.211
|
|
VIM
|
vimentin
|
|
chr6_+_3696224
|
2.191
|
|
|
|
|
chr10_+_17311283
|
2.190
|
|
VIM
|
vimentin
|
|
chr10_+_17311347
|
2.187
|
|
VIM
|
vimentin
|
|
chr13_-_76358370
|
2.179
|
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr9_-_35679799
|
2.175
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr12_+_51730101
|
2.167
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr17_-_15106579
|
2.160
|
|
PMP22
|
peripheral myelin protein 22
|
|
chr17_-_15106597
|
2.150
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr12_-_29827952
|
2.149
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr11_+_110916629
|
2.148
|
|
LAYN
|
layilin
|
|
chr6_-_82518576
|
2.141
|
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr14_-_51605487
|
2.139
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr22_-_22652018
|
2.136
|
NM_001355
|
DDT
|
D-dopachrome tautomerase
|
|
chr10_+_17311242
|
2.133
|
|
VIM
|
vimentin
|
|
chr19_-_50977485
|
2.127
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr9_-_35679902
|
2.120
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr9_-_25668230
|
2.115
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr14_-_51605461
|
2.113
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr6_-_105691675
|
2.110
|
|
BVES
|
blood vessel epicardial substance
|
|
chr10_+_17311282
|
2.097
|
|
VIM
|
vimentin
|
|
chr3_+_11171213
|
2.096
|
NM_001098212
|
HRH1
|
histamine receptor H1
|
|
chr8_-_82186832
|
2.075
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr5_-_178704934
|
2.074
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr18_-_53621277
|
2.071
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr9_-_21999270
|
2.063
|
NM_004936
NM_078487
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4)
|
|
chr11_+_86189138
|
2.021
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr11_+_12355600
|
2.020
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr5_-_136862130
|
2.010
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr3_-_147361465
|
1.986
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr1_+_216586199
|
1.957
|
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr10_+_124211353
|
1.946
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr1_-_199704773
|
1.942
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr10_+_99463446
|
1.931
|
NM_031484
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr4_-_57671307
|
1.910
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr16_-_71650034
|
1.904
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr3_+_49034050
|
1.903
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr17_+_1906262
|
1.900
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr13_-_73606362
|
1.892
|
|
KLF12
|
Kruppel-like factor 12
|
|
chr5_+_102229612
|
1.882
|
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr14_-_51605515
|
1.879
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr1_-_95165089
|
1.877
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr9_-_130980359
|
1.864
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr1_-_72520735
|
1.861
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr12_-_67613213
|
1.845
|
NM_001005502
NM_198320
|
CPM
|
carboxypeptidase M
|
|
chr12_+_64504835
|
1.828
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr4_+_86968069
|
1.818
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr3_+_30623376
|
1.799
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr13_-_35603466
|
1.799
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr6_+_17389752
|
1.794
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr7_+_129919157
|
1.792
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr6_-_105691235
|
1.790
|
NM_007073
|
BVES
|
blood vessel epicardial substance
|
|
chr10_+_99463468
|
1.788
|
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr11_+_106967283
|
1.783
|
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr2_+_149895274
|
1.776
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr7_+_129919134
|
1.775
|
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr3_+_160964574
|
1.762
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr14_+_58174489
|
1.755
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr1_-_33420213
|
1.750
|
NM_018207
|
TRIM62
|
tripartite motif containing 62
|
|
chr11_-_86060812
|
1.749
|
NM_001014811
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chrX_-_13866366
|
1.744
|
|
|
|
|
chr1_+_31658549
|
1.738
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr7_-_130891861
|
1.729
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr6_-_82519093
|
1.721
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr1_+_238321794
|
1.720
|
NM_020066
|
FMN2
|
formin 2
|
|
chr19_-_63301402
|
1.714
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr21_+_45318861
|
1.711
|
NM_001112
NM_001160230
NM_015833
NM_015834
|
ADARB1
|
adenosine deaminase, RNA-specific, B1
|
|
chr16_+_27320983
|
1.704
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr11_+_133444175
|
1.686
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr1_-_84237316
|
1.685
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr5_-_178705036
|
1.684
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr4_+_158216631
|
1.675
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr18_-_21186107
|
1.675
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr4_-_86106567
|
1.668
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr5_+_102229425
|
1.652
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr19_-_63301386
|
1.651
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr11_-_86061003
|
1.645
|
NM_006680
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr12_-_26166485
|
1.640
|
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr8_+_97575196
|
1.639
|
|
SDC2
|
syndecan 2
|
|
chr2_-_19421852
|
1.634
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr11_+_124539814
|
1.633
|
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr8_+_97575244
|
1.633
|
|
SDC2
|
syndecan 2
|
|
chr2_-_37752731
|
1.621
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr19_-_63301418
|
1.619
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr13_-_35603513
|
1.616
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr4_-_57671273
|
1.611
|
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chrX_-_13866565
|
1.611
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr1_-_143750988
|
1.611
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr13_-_35603432
|
1.611
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr14_-_37795290
|
1.611
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr13_-_73605914
|
1.610
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr17_+_56832255
|
1.601
|
|
TBX2
|
T-box 2
|
|
chr6_+_142664490
|
1.597
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr1_-_56817717
|
1.597
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr14_+_99220349
|
1.585
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr9_-_128924706
|
1.583
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr12_+_27824429
|
1.576
|
NM_020782
|
KLHDC5
|
kelch domain containing 5
|
|
chr13_-_22905822
|
1.574
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chrX_+_152699616
|
1.567
|
NM_001170760
NM_001170761
NM_014370
|
SRPK3
|
SRSF protein kinase 3
|
|
chr17_-_19711580
|
1.563
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr11_+_124539768
|
1.562
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr4_-_140696740
|
1.559
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr2_+_5750229
|
1.559
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr4_-_186934059
|
1.558
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr13_+_113546912
|
1.533
|
|
GAS6
|
growth arrest-specific 6
|
|
chr3_-_79151232
|
1.529
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr9_-_128924782
|
1.527
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr3_-_151171576
|
1.522
|
|
PFN2
|
profilin 2
|
|
chr11_-_65841077
|
1.511
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr2_+_191818124
|
1.507
|
NM_001130158
NM_012223
NM_001161819
|
MYO1B
|
myosin IB
|
|
chr2_-_219404755
|
1.506
|
NM_017431
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit
|
|
chr14_+_22375747
|
1.506
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr1_+_15145001
|
1.497
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr15_+_72006015
|
1.496
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr10_+_17311788
|
1.489
|
|
VIM
|
vimentin
|
|
chr9_+_133154896
|
1.486
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr17_-_53387493
|
1.485
|
|
CUEDC1
|
CUE domain containing 1
|
|
chr12_+_121233615
|
1.485
|
NM_001098519
|
LRRC43
|
leucine rich repeat containing 43
|
|
chr7_-_84654106
|
1.485
|
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr1_-_22136263
|
1.484
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr10_+_27833167
|
1.476
|
NM_021252
|
RAB18
|
RAB18, member RAS oncogene family
|
|
chr15_+_89228668
|
1.468
|
NM_001143785
NM_002005
|
FES
|
feline sarcoma oncogene
|
|
chr8_-_72918697
|
1.463
|
|
MSC
|
musculin
|
|
chr2_+_56265260
|
1.460
|
|
|
|
|
chr17_-_7434113
|
1.456
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr11_+_44543716
|
1.455
|
NM_001024844
NM_002231
|
CD82
|
CD82 molecule
|
|
chr1_-_72521079
|
1.455
|
|
|
|
|
chr9_-_128924819
|
1.454
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr3_+_151171755
|
1.444
|
|
LOC646903
|
hypothetical LOC646903
|
|
chr3_+_8518508
|
1.437
|
NM_014583
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr19_-_50977586
|
1.422
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr17_-_63965156
|
1.418
|
NM_017983
|
WIPI1
|
WD repeat domain, phosphoinositide interacting 1
|
|
chr3_+_49034494
|
1.417
|
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr7_+_27102508
|
1.411
|
|
|
|
|
chr14_+_62740878
|
1.409
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr6_-_119079712
|
1.409
|
NM_001042475
NM_206921
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr9_+_111582317
|
1.405
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr3_-_31997738
|
1.398
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chrX_+_73557809
|
1.393
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr5_-_136862317
|
1.387
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr15_-_58671929
|
1.370
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr9_-_16860664
|
1.362
|
|
BNC2
|
basonuclin 2
|
|
chr9_+_71848555
|
1.355
|
|
MAMDC2
|
MAM domain containing 2
|
|
chr19_-_50977592
|
1.351
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr5_-_54317102
|
1.345
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr5_-_112658200
|
1.341
|
|
MCC
|
mutated in colorectal cancers
|
|
chr7_-_27136876
|
1.341
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr12_-_94708477
|
1.338
|
|
NTN4
|
netrin 4
|
|
chr12_+_50587468
|
1.333
|
NM_000020
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr17_+_36515166
|
1.333
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr15_-_90999606
|
1.331
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr10_+_72642274
|
1.329
|
NM_170744
|
UNC5B
|
unc-5 homolog B (C. elegans)
|
|
chr3_+_100840129
|
1.325
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|