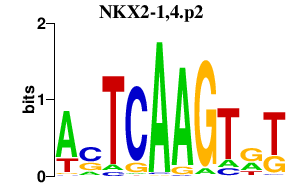
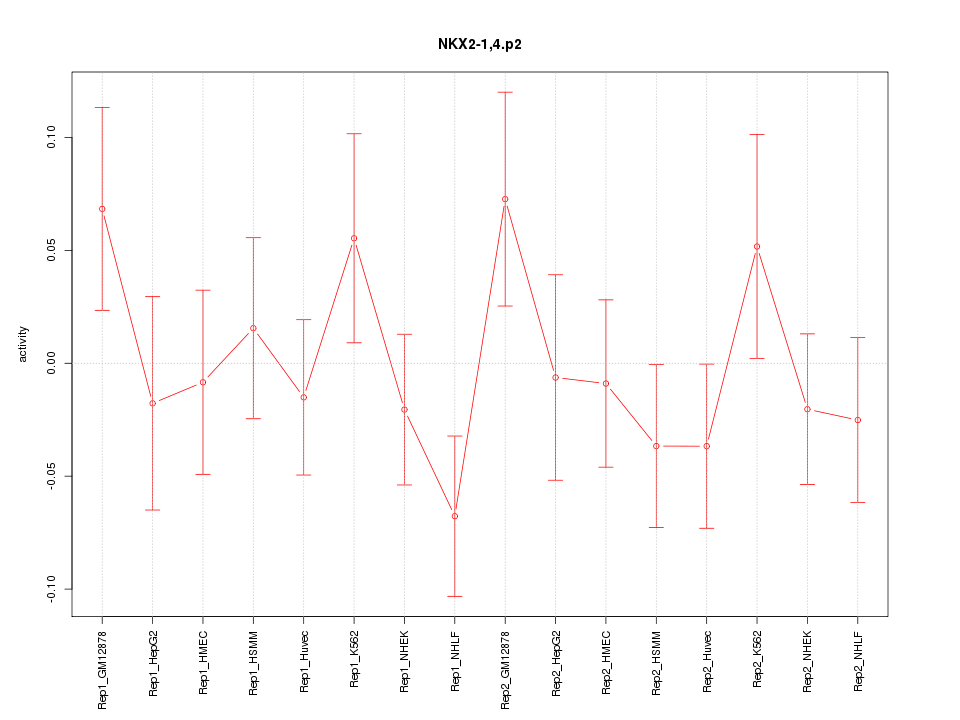
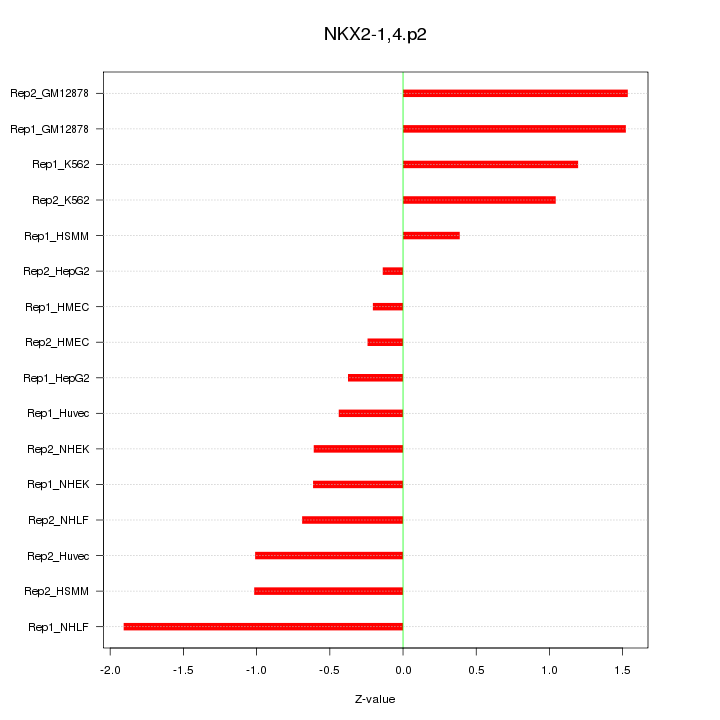
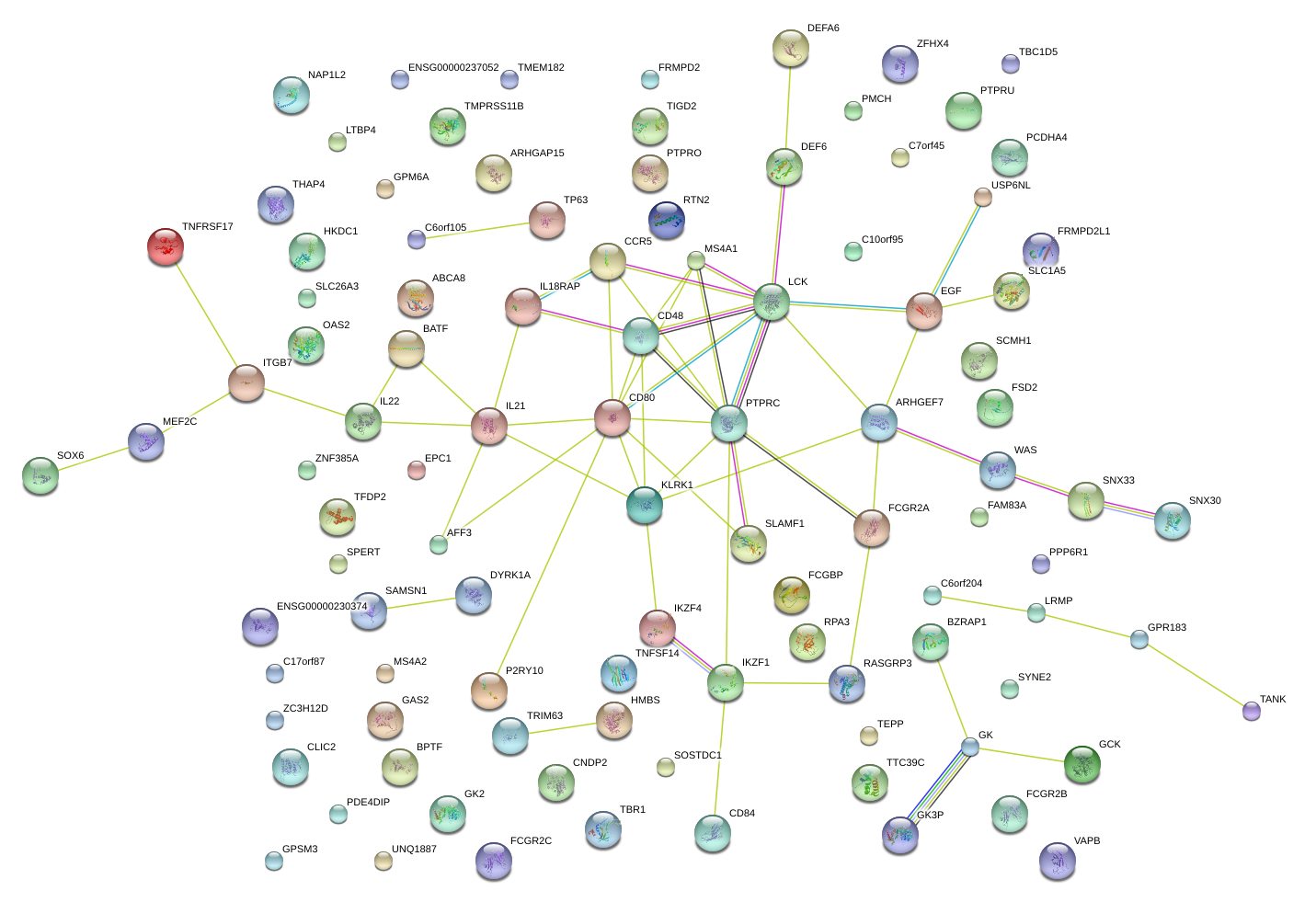
|
chr16_+_11966464
|
2.667
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chrX_+_78087484
|
2.586
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr21_-_14840506
|
2.582
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr19_-_6621078
|
2.357
|
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14
|
|
chrX_-_154216929
|
1.877
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr3_-_143230124
|
1.707
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr19_-_6621598
|
1.706
|
NM_003807
NM_172014
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14
|
|
chr5_-_88155360
|
1.496
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_+_159817752
|
1.389
|
NM_201563
|
FCGR2C
|
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr1_-_143706196
|
1.287
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_+_159899528
|
1.203
|
NM_001002273
NM_001002274
NM_001002275
NM_001190828
NM_004001
|
FCGR2B
FCGR2C
|
Fc fragment of IgG, low affinity IIb, receptor (CD32)
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr2_+_161701711
|
1.158
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr4_+_111053488
|
1.152
|
NM_001178130
NM_001178131
NM_001963
|
EGF
|
epidermal growth factor
|
|
chr1_+_32489426
|
1.085
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr10_-_11614279
|
1.010
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr1_-_143706300
|
0.942
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr19_-_50688310
|
0.931
|
NM_206901
|
RTN2
|
reticulon 2
|
|
chr1_-_143706265
|
0.914
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chrX_+_48427103
|
0.909
|
NM_000377
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr10_-_104201234
|
0.897
|
NM_024886
|
C10orf95
|
chromosome 10 open reading frame 95
|
|
chr19_-_60458915
|
0.884
|
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1
|
|
chr19_-_51979838
|
0.864
|
NM_001145145
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5
|
|
chr2_-_100125402
|
0.863
|
NM_002285
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr11_+_59979857
|
0.853
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr1_+_180836050
|
0.848
|
|
|
|
|
chr11_-_16380967
|
0.715
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr16_+_56567839
|
0.712
|
NM_199046
NM_199456
|
TEPP
|
testis, prostate and placenta expressed
|
|
chr10_-_32707549
|
0.695
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr15_-_81271859
|
0.693
|
NM_001007122
|
FSD2
|
fibronectin type III and SPRY domain containing 2
|
|
chr4_-_68793928
|
0.662
|
NM_182502
|
TMPRSS11B
|
transmembrane protease, serine 11B
|
|
chr1_-_158948176
|
0.652
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr12_-_51887266
|
0.651
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr7_+_50318801
|
0.648
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr3_-_120761092
|
0.601
|
NM_005191
|
CD80
|
CD80 molecule
|
|
chr12_-_119733062
|
0.590
|
|
SPPL3
|
signal peptide peptidase 3
|
|
chr7_+_129634935
|
0.579
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr2_+_143603368
|
0.550
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr10_+_70650064
|
0.546
|
NM_025130
|
HKDC1
|
hexokinase domain containing 1
|
|
chr6_-_32268267
|
0.533
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr17_-_5078773
|
0.521
|
NM_207103
|
C17orf87
|
chromosome 17 open reading frame 87
|
|
chr12_-_101115677
|
0.517
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr21_+_37714470
|
0.516
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr18_+_70317548
|
0.510
|
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr6_-_119137923
|
0.505
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr3_+_190831909
|
0.496
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr2_+_143603439
|
0.487
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr5_-_88004891
|
0.486
|
|
LOC645323
|
hypothetical LOC645323
|
|
chrX_-_100920493
|
0.471
|
|
GK
|
glycerol kinase
|
|
chr1_-_158815850
|
0.466
|
|
CD84
|
CD84 molecule
|
|
chr19_-_45132372
|
0.448
|
NM_003890
|
FCGBP
|
Fc fragment of IgG binding protein
|
|
chr1_-_178100204
|
0.447
|
|
|
|
|
chr14_+_63752424
|
0.446
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr17_+_63338500
|
0.443
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chrX_+_48427137
|
0.440
|
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr17_-_53761119
|
0.438
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr19_+_45797413
|
0.429
|
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr2_-_242205556
|
0.428
|
NM_001164356
|
THAP4
|
THAP domain containing 4
|
|
chr1_-_158815895
|
0.413
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr13_-_98757634
|
0.411
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr7_-_107230867
|
0.403
|
NM_000111
|
SLC26A3
|
solute carrier family 26, member 3
|
|
chr2_+_102744921
|
0.402
|
NM_144632
|
TMEM182
|
transmembrane protein 182
|
|
chr3_-_17757402
|
0.395
|
NM_014744
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chr20_+_56455866
|
0.392
|
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr12_+_111900650
|
0.383
|
NM_001032731
NM_002535
NM_016817
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa
|
|
chr12_+_25096755
|
0.381
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr14_+_75058518
|
0.381
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr1_-_41398126
|
0.381
|
NM_001172222
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr3_+_46386636
|
0.377
|
NM_000579
NM_001100168
|
CCR5
|
chemokine (C-C motif) receptor 5
|
|
chr12_+_54700955
|
0.376
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr8_+_77928372
|
0.372
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr9_+_114671635
|
0.364
|
|
SNX30
|
sorting nexin family member 30
|
|
chr1_-_158883666
|
0.361
|
NM_003037
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1
|
|
chr5_+_140166842
|
0.360
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr12_-_51887318
|
0.351
|
|
ITGB7
|
integrin, beta 7
|
|
chr4_-_176970372
|
0.351
|
|
GPM6A
|
glycoprotein M6A
|
|
chr17_-_64462976
|
0.350
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr6_+_35373632
|
0.349
|
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr4_-_176970407
|
0.347
|
|
GPM6A
|
glycoprotein M6A
|
|
chr1_+_196874867
|
0.344
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr13_+_110637173
|
0.344
|
NM_001113513
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr11_+_22646229
|
0.342
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr2_+_161980865
|
0.341
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr4_+_90252990
|
0.332
|
NM_145715
|
TIGD2
|
tigger transposable element derived 2
|
|
chr12_-_53064565
|
0.327
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chr7_-_7724762
|
0.327
|
NM_002947
|
RPA3
|
replication protein A3, 14kDa
|
|
chr7_-_16471817
|
0.322
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr12_+_15590552
|
0.316
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr10_-_49152918
|
0.312
|
NM_001018071
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr2_+_102401580
|
0.307
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr11_+_118463906
|
0.304
|
NM_001024382
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr6_-_11887265
|
0.304
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr13_+_45174446
|
0.304
|
NM_152719
|
SPERT
|
spermatid associated
|
|
chrX_-_72351381
|
0.300
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr6_-_149847809
|
0.296
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr1_+_196874759
|
0.294
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr2_+_33554824
|
0.294
|
NM_001139488
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr8_+_124264054
|
0.290
|
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr4_-_123761615
|
0.289
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr1_-_26266571
|
0.288
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chr17_+_20048890
|
0.287
|
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr9_-_99740243
|
0.286
|
NM_197978
|
HEMGN
|
hemogen
|
|
chrX_+_48529905
|
0.282
|
NM_002049
|
GATA1
|
GATA binding protein 1 (globin transcription factor 1)
|
|
chr17_-_10393664
|
0.281
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr1_-_89364273
|
0.280
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr10_-_76538859
|
0.276
|
NM_001007271
NM_001007272
NM_001007273
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr2_-_175170674
|
0.273
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr14_-_94669547
|
0.273
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chrX_-_72351392
|
0.272
|
NM_021963
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr19_-_53950458
|
0.272
|
NM_000148
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group)
|
|
chr15_-_53349775
|
0.271
|
NM_183234
NM_004580
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr6_-_149847718
|
0.271
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr12_+_54132264
|
0.266
|
NM_054105
|
OR6C2
|
olfactory receptor, family 6, subfamily C, member 2
|
|
chr11_+_118259684
|
0.264
|
NM_001716
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr8_+_124263932
|
0.263
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr17_-_26665144
|
0.263
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr2_+_178893216
|
0.257
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr12_-_110410730
|
0.245
|
|
ATXN2
|
ataxin 2
|
|
chr5_-_142795269
|
0.240
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr9_-_5958367
|
0.237
|
|
|
|
|
chr3_-_180460372
|
0.236
|
NM_171829
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr7_+_143457083
|
0.234
|
NM_001001659
|
OR2A14
|
olfactory receptor, family 2, subfamily A, member 14
|
|
chr8_-_82536312
|
0.234
|
NM_001080526
|
FABP9
|
fatty acid binding protein 9, testis
|
|
chr2_-_88946767
|
0.234
|
|
|
|
|
chr10_+_86121526
|
0.233
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chrX_-_137649180
|
0.232
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr17_-_26665213
|
0.229
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_-_26665220
|
0.227
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr12_-_52980914
|
0.227
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr10_+_114125003
|
0.224
|
NM_203380
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr1_+_61642287
|
0.223
|
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_172683834
|
0.222
|
NM_005684
|
GPR52
|
G protein-coupled receptor 52
|
|
chr8_+_9990471
|
0.222
|
NM_001135671
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr12_-_54392328
|
0.222
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr12_+_117067214
|
0.218
|
|
PEBP1
|
phosphatidylethanolamine binding protein 1
|
|
chr12_+_130004404
|
0.217
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr17_+_65009986
|
0.217
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr1_-_246503738
|
0.215
|
NM_001004695
|
OR2T33
|
olfactory receptor, family 2, subfamily T, member 33
|
|
chrX_+_70682204
|
0.214
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr11_+_62195710
|
0.213
|
|
C11orf83
|
chromosome 11 open reading frame 83
|
|
chr10_-_69125932
|
0.212
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr1_+_144124547
|
0.209
|
NM_145277
NM_202004
NM_213652
NM_213653
|
HFE2
|
hemochromatosis type 2 (juvenile)
|
|
chr3_-_57209319
|
0.209
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr10_-_108448990
|
0.208
|
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chr12_-_53064723
|
0.208
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr19_+_16691786
|
0.207
|
NM_001007525
|
NWD1
|
NACHT and WD repeat domain containing 1
|
|
chr1_+_151498802
|
0.206
|
NM_000427
|
LOR
|
loricrin
|
|
chr14_+_75058591
|
0.205
|
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr16_+_29982941
|
0.202
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr11_+_63775567
|
0.195
|
NM_000932
NM_001184883
|
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific)
|
|
chrX_-_48101044
|
0.191
|
NM_021014
NM_175711
|
SSX3
|
synovial sarcoma, X breakpoint 3
|
|
chr1_+_38033666
|
0.189
|
NM_152496
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr17_-_3407780
|
0.189
|
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr8_-_7207899
|
0.188
|
NM_001164457
|
ZNF705G
|
zinc finger protein 705G
|
|
chr6_+_31782618
|
0.186
|
NM_001003693
|
LY6G6F
|
lymphocyte antigen 6 complex, locus G6F
|
|
chrX_-_72351348
|
0.186
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr1_-_43168247
|
0.185
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr12_-_7739591
|
0.183
|
NM_020634
|
GDF3
|
growth differentiation factor 3
|
|
chr5_-_147996769
|
0.182
|
NM_001040169
NM_001040172
NM_001040173
NM_199453
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr13_-_74799898
|
0.181
|
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr1_-_42574116
|
0.180
|
NM_001198850
|
FOXJ3
|
forkhead box J3
|
|
chrX_-_110542040
|
0.179
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chrX_+_41468351
|
0.178
|
NM_080817
|
GPR82
|
G protein-coupled receptor 82
|
|
chr3_+_68136056
|
0.178
|
NM_213609
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1
|
|
chr19_-_60360768
|
0.177
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr1_-_160648429
|
0.176
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr14_-_20694023
|
0.175
|
NM_001004731
|
OR5AU1
|
olfactory receptor, family 5, subfamily AU, member 1
|
|
chr1_-_9052207
|
0.175
|
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr10_-_6144119
|
0.175
|
NM_000417
|
IL2RA
|
interleukin 2 receptor, alpha
|
|
chr3_+_188397967
|
0.173
|
NM_153708
|
RTP1
|
receptor (chemosensory) transporter protein 1
|
|
chr1_-_156937065
|
0.172
|
NM_001005279
|
OR6K2
|
olfactory receptor, family 6, subfamily K, member 2
|
|
chr1_-_76170563
|
0.170
|
NM_080868
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chr4_-_77547481
|
0.167
|
NM_001042784
|
CCDC158
|
coiled-coil domain containing 158
|
|
chr2_-_157890400
|
0.167
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr3_-_180467483
|
0.166
|
NM_001163677
NM_171828
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr9_+_130485754
|
0.165
|
NM_001122821
|
SET
|
SET nuclear oncogene
|
|
chr1_-_240679380
|
0.165
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr1_+_154120170
|
0.165
|
|
SYT11
|
synaptotagmin XI
|
|
chr6_-_152681171
|
0.164
|
NM_015293
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chrX_-_48655978
|
0.163
|
|
PIM2
|
pim-2 oncogene
|
|
chr14_+_55148488
|
0.163
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr17_-_39630951
|
0.162
|
NM_001098833
NM_020218
|
ATXN7L3
|
ataxin 7-like 3
|
|
chr8_-_110726921
|
0.161
|
NM_001099751
NM_001099752
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr6_-_11219932
|
0.161
|
NM_207582
|
ERVFRDE1
|
HERV-FRD provirus ancestral Env polyprotein
|
|
chr5_-_156468614
|
0.159
|
NM_032782
|
HAVCR2
|
hepatitis A virus cellular receptor 2
|
|
chr12_-_48246488
|
0.157
|
NM_001012300
|
MCRS1
|
microspherule protein 1
|
|
chr11_-_5212168
|
0.155
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr18_-_59137488
|
0.153
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr10_-_75005421
|
0.153
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr1_+_149299530
|
0.152
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr6_+_155579788
|
0.152
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr4_-_185963859
|
0.151
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr1_-_100416358
|
0.149
|
NM_144620
|
LRRC39
|
leucine rich repeat containing 39
|
|
chr8_-_6901661
|
0.149
|
NM_021010
|
DEFA5
|
defensin, alpha 5, Paneth cell-specific
|
|
chr2_-_37397669
|
0.149
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr7_+_38984133
|
0.149
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr11_-_31789168
|
0.149
|
|
PAX6
|
paired box 6
|
|
chr21_-_35343065
|
0.148
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr2_+_38009405
|
0.148
|
NM_001170792
|
FAM82A1
|
family with sequence similarity 82, member A1
|
|
chr17_-_74244539
|
0.148
|
|
|
|
|
chr11_-_57643491
|
0.147
|
NM_001005211
|
OR9I1
|
olfactory receptor, family 9, subfamily I, member 1
|