|
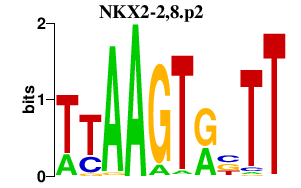
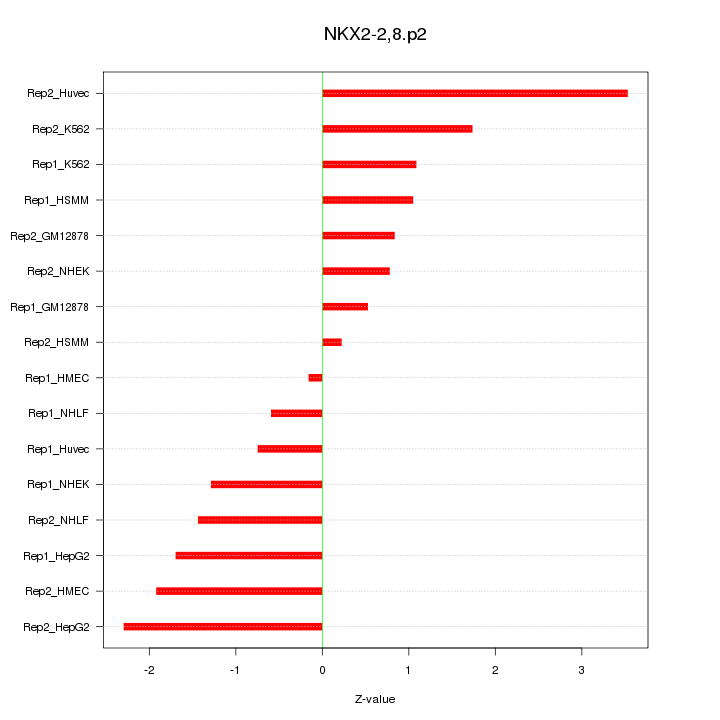
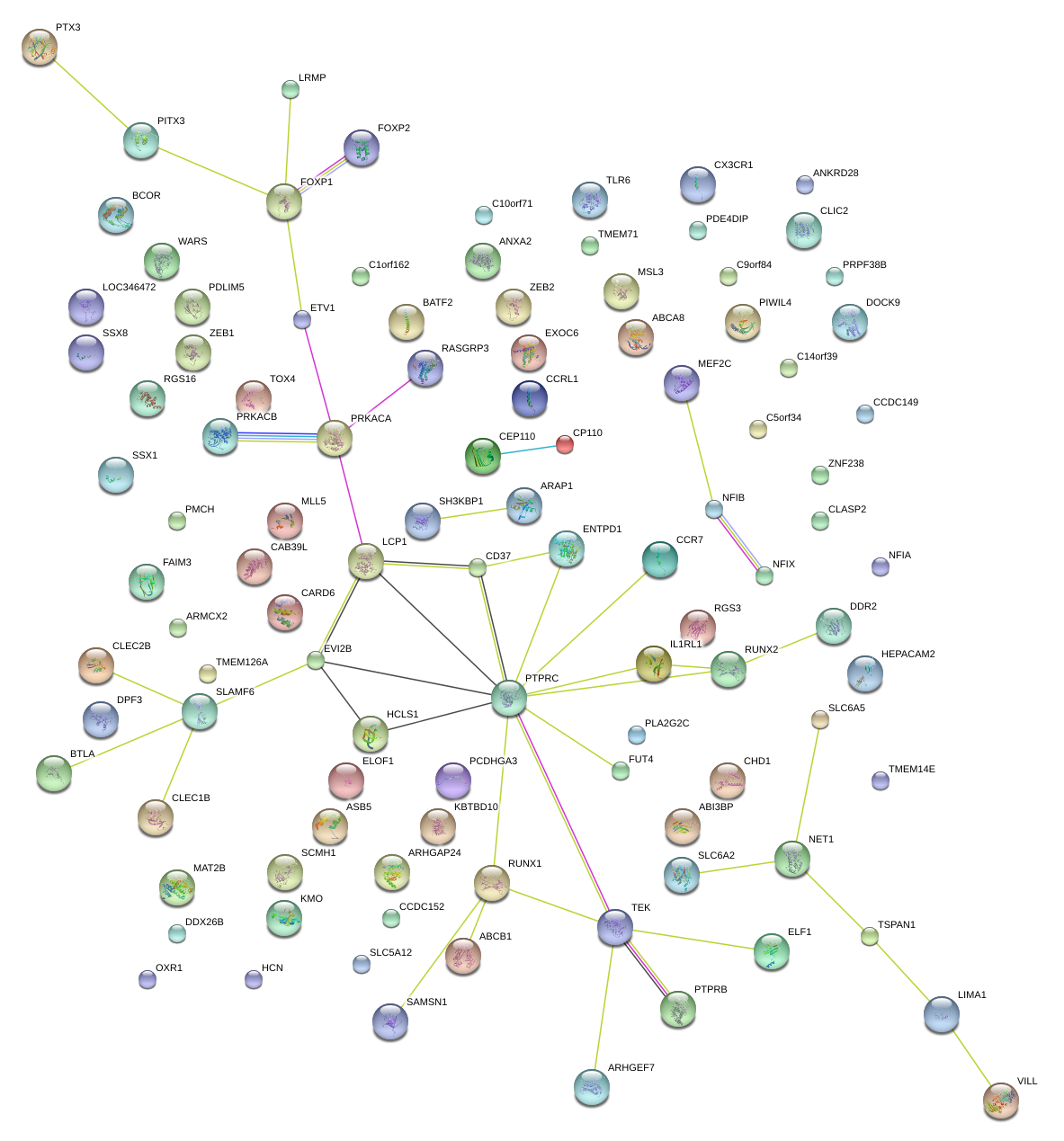
chr13_-_40454188
|
4.983
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr3_-_15814678
|
2.547
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr3_-_122862391
|
2.403
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr3_-_122862447
|
2.272
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr2_+_102320148
|
2.218
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr12_-_101115677
|
2.123
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr13_-_40454360
|
1.879
|
NM_001145353
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr21_-_14840506
|
1.831
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr12_-_9913199
|
1.750
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr2_-_144991556
|
1.673
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr13_+_110653407
|
1.529
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr10_+_50177192
|
1.460
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr15_-_58477407
|
1.435
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr1_+_242281175
|
1.340
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr9_+_115303527
|
1.255
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_122923864
|
1.226
|
|
CEP110
|
centrosomal protein 110kDa
|
|
chr5_+_40877053
|
1.216
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr11_+_65027721
|
1.174
|
|
|
|
|
chrX_+_134482212
|
1.133
|
NM_182540
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr4_-_24590867
|
1.102
|
NM_173463
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr4_+_86918870
|
1.090
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chrX_-_154216929
|
1.054
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr2_+_170074457
|
1.040
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr5_+_162862808
|
1.040
|
NM_182796
|
MAT2B
|
methionine adenosyltransferase II, beta
|
|
chr1_+_239762179
|
1.031
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr1_+_242281199
|
1.029
|
|
ZNF238
|
zinc finger protein 238
|
|
chrX_+_11687655
|
1.014
|
NM_006800
|
MSL3
|
male-specific lethal 3 homolog (Drosophila)
|
|
chr10_+_97505398
|
1.012
|
NM_001164178
NM_001164181
NM_001164179
NM_001164182
NM_001164183
NM_001776
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1
|
|
chr21_-_35343065
|
0.995
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_-_41399632
|
0.994
|
NM_001031694
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr14_-_99911409
|
0.991
|
|
WARS
|
tryptophanyl-tRNA synthetase
|
|
chr4_+_86918882
|
0.987
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr17_-_64462976
|
0.958
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr3_+_38010081
|
0.950
|
NM_015873
|
VILL
|
villin-like
|
|
chr3_-_71262426
|
0.945
|
|
FOXP1
|
forkhead box P1
|
|
chr1_+_160868822
|
0.927
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr3_-_71262405
|
0.888
|
|
FOXP1
|
forkhead box P1
|
|
chr14_-_99911528
|
0.881
|
|
WARS
|
tryptophanyl-tRNA synthetase
|
|
chr1_-_143708467
|
0.874
|
NM_001195260
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr8_+_107807415
|
0.872
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr19_+_54530458
|
0.870
|
|
CD37
|
CD37 molecule
|
|
chr4_+_95595419
|
0.857
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr19_+_54530485
|
0.844
|
NM_001040031
NM_001774
|
CD37
|
CD37 molecule
|
|
chr9_-_113561613
|
0.839
|
NM_001080551
|
C9orf84
|
chromosome 9 open reading frame 84
|
|
chr10_+_94584449
|
0.833
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr19_+_54530497
|
0.832
|
|
CD37
|
CD37 molecule
|
|
chr1_+_111818111
|
0.826
|
NM_174896
|
C1orf162
|
chromosome 1 open reading frame 162
|
|
chr13_-_98428244
|
0.813
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr7_-_92686773
|
0.804
|
NM_198151
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr17_-_26665144
|
0.804
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr5_+_42792676
|
0.783
|
NM_001134848
|
CCDC152
|
coiled-coil domain containing 152
|
|
chr11_+_65023218
|
0.777
|
|
|
|
|
chr3_+_158637273
|
0.773
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chrX_-_100801472
|
0.769
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr19_+_54530512
|
0.765
|
|
CD37
|
CD37 molecule
|
|
chr1_-_20374273
|
0.763
|
NM_001105572
|
PLA2G2C
|
phospholipase A2, group IIC
|
|
chr12_-_69317441
|
0.757
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr14_-_72430545
|
0.756
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr2_-_144994349
|
0.753
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chrX_+_47999740
|
0.734
|
NM_005635
|
SSX1
|
synovial sarcoma, X breakpoint 1
|
|
chr1_-_180840016
|
0.734
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr11_+_65023038
|
0.721
|
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding)
|
|
chr14_-_99911679
|
0.716
|
NM_004184
NM_213646
|
WARS
|
tryptophanyl-tRNA synthetase
|
|
chr11_+_93916653
|
0.702
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr13_-_45654327
|
0.687
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr12_-_48902692
|
0.685
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chrX_-_19727775
|
0.681
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr3_+_133798770
|
0.675
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr5_-_88155360
|
0.674
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_+_84382575
|
0.672
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_-_102194938
|
0.667
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr19_+_54530546
|
0.656
|
|
CD37
|
CD37 molecule
|
|
chr17_-_35975227
|
0.655
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr3_-_33675484
|
0.652
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr5_-_98290117
|
0.647
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr1_+_196874839
|
0.643
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr9_-_41944687
|
0.638
|
|
MGC21881
|
hypothetical locus MGC21881
|
|
chr3_+_188197029
|
0.636
|
|
|
|
|
chr1_-_197173159
|
0.633
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr7_+_104468614
|
0.632
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr4_-_177427252
|
0.631
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr11_-_72111027
|
0.623
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr3_-_153541468
|
0.621
|
NM_001123228
|
TMEM14E
|
transmembrane protein 14E
|
|
chr7_+_113853790
|
0.620
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr2_+_33514896
|
0.618
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr1_+_196874759
|
0.613
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr8_-_133841988
|
0.611
|
NM_001145153
NM_144649
|
TMEM71
|
transmembrane protein 71
|
|
chr9_+_66293049
|
0.606
|
|
MGC21881
|
hypothetical locus MGC21881
|
|
chr5_+_140772691
|
0.606
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr1_+_61642287
|
0.605
|
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_196874867
|
0.605
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chrX_-_19727828
|
0.600
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr5_-_43550891
|
0.595
|
NM_198566
|
C5orf34
|
chromosome 5 open reading frame 34
|
|
chr12_+_25096755
|
0.559
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr13_-_48873735
|
0.559
|
NM_030925
|
CAB39L
|
calcium binding protein 39-like
|
|
chr11_-_26700121
|
0.557
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr17_-_26665194
|
0.541
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr7_-_13992351
|
0.540
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr1_+_169747788
|
0.532
|
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr1_-_205161821
|
0.531
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr12_-_120725210
|
0.531
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr7_-_87180499
|
0.530
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr3_-_113701073
|
0.527
|
NM_001085357
NM_181780
|
BTLA
|
B and T lymphocyte associated
|
|
chr4_-_38534802
|
0.526
|
NM_006068
|
TLR6
|
toll-like receptor 6
|
|
chr17_-_26665213
|
0.526
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr14_-_60022487
|
0.525
|
NM_174978
|
C14orf39
|
chromosome 14 open reading frame 39
|
|
chr17_-_26665220
|
0.524
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chrX_-_39921525
|
0.523
|
NM_001123383
NM_001123384
|
BCOR
|
BCL6 corepressor
|
|
chr1_-_158759606
|
0.522
|
NM_001184714
NM_001184715
NM_001184716
NM_052931
|
SLAMF6
|
SLAM family member 6
|
|
chr9_+_27099138
|
0.520
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr10_+_5478521
|
0.514
|
NM_005863
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr17_+_35425139
|
0.512
|
NM_000759
NM_001178147
NM_172219
NM_172220
|
CSF3
|
colony stimulating factor 3 (granulocyte)
|
|
chr2_-_63518589
|
0.506
|
NM_001042692
|
WDPCP
|
WD repeat containing planar cell polarity effector
|
|
chr1_+_149299504
|
0.504
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr10_-_64698831
|
0.493
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr4_-_74307637
|
0.490
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr1_-_35143570
|
0.488
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr1_+_190394214
|
0.488
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr1_+_234625338
|
0.488
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chr12_+_115455579
|
0.487
|
|
NCRNA00173
|
non-protein coding RNA 173
|
|
chr9_+_102380156
|
0.484
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr4_-_71751205
|
0.483
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr1_-_156923111
|
0.482
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr17_+_72827654
|
0.480
|
|
SEPT9
|
septin 9
|
|
chr11_+_65023556
|
0.471
|
|
|
|
|
chr1_+_23994914
|
0.469
|
|
|
|
|
chr9_+_263025
|
0.467
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr7_+_838663
|
0.466
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr2_+_102401580
|
0.460
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr8_+_124264054
|
0.457
|
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr6_-_29081015
|
0.454
|
NM_001010877
|
ZNF311
|
zinc finger protein 311
|
|
chr1_+_144198658
|
0.453
|
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr1_-_67039518
|
0.449
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr17_-_31281843
|
0.448
|
NM_001034836
NM_001163120
NM_001163121
NM_145654
|
RDM1
|
RAD52 motif 1
|
|
chr8_-_17599438
|
0.440
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr1_-_41398126
|
0.438
|
NM_001172222
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr7_-_13997574
|
0.433
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr1_-_103269356
|
0.427
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr18_-_65766068
|
0.423
|
|
CD226
|
CD226 molecule
|
|
chr1_+_165564904
|
0.423
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_-_115060956
|
0.418
|
NM_002524
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr2_-_183095308
|
0.416
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr7_+_23686268
|
0.411
|
NM_001127364
NM_001127365
NM_199136
|
C7orf46
|
chromosome 7 open reading frame 46
|
|
chr6_-_47030492
|
0.410
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr4_-_145046109
|
0.406
|
NM_002102
NM_198682
|
GYPE
|
glycophorin E (MNS blood group)
|
|
chr22_+_38627031
|
0.406
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr3_+_25444837
|
0.406
|
NM_000965
|
RARB
|
retinoic acid receptor, beta
|
|
chr21_+_29322100
|
0.405
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr17_+_72827227
|
0.405
|
|
SEPT9
|
septin 9
|
|
chr12_-_47869067
|
0.404
|
NM_006009
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr1_-_224993498
|
0.402
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|
|
chr3_+_25444753
|
0.402
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr1_+_103840900
|
0.400
|
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3
|
|
chr1_+_45251007
|
0.398
|
|
UROD
|
uroporphyrinogen decarboxylase
|
|
chr12_-_47869134
|
0.398
|
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr6_+_30799130
|
0.397
|
|
TUBB
|
tubulin, beta
|
|
chr7_-_111216180
|
0.396
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr22_+_38627129
|
0.394
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr7_-_107667728
|
0.393
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chrX_-_70248022
|
0.390
|
NM_000206
|
IL2RG
|
interleukin 2 receptor, gamma
|
|
chr5_+_54491702
|
0.388
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr7_+_149696263
|
0.388
|
|
REPIN1
|
replication initiator 1
|
|
chr2_+_173394560
|
0.388
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr1_+_149299307
|
0.385
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr7_-_92693716
|
0.381
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr10_-_50066386
|
0.379
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr19_+_46121963
|
0.377
|
|
CYP2B7P1
|
cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1
|
|
chr2_+_143603368
|
0.373
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr5_-_169340321
|
0.372
|
NM_001129891
|
FAM196B
|
family with sequence similarity 196, member B
|
|
chr5_+_159781454
|
0.367
|
|
PTTG1
|
pituitary tumor-transforming 1
|
|
chr12_-_91346054
|
0.366
|
NM_001025232
|
CLLU1OS
|
chronic lymphocytic leukemia up-regulated 1 opposite strand
|
|
chr8_+_144522389
|
0.364
|
NM_052924
|
RHPN1
|
rhophilin, Rho GTPase binding protein 1
|
|
chr5_-_150501393
|
0.361
|
NM_001193544
|
ANXA6
|
annexin A6
|
|
chr12_+_25096472
|
0.359
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr15_-_91078307
|
0.357
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr12_-_88573949
|
0.356
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr4_+_134292630
|
0.354
|
|
PCDH10
|
protocadherin 10
|
|
chrX_+_122923279
|
0.352
|
|
STAG2
|
stromal antigen 2
|
|
chr1_-_180622083
|
0.352
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr4_-_71751070
|
0.351
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr8_+_17866373
|
0.350
|
|
PCM1
|
pericentriolar material 1
|
|
chr1_-_115060857
|
0.350
|
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chrX_+_142794838
|
0.349
|
NM_001012989
|
UBE2NL
|
ubiquitin-conjugating enzyme E2N-like
|
|
chr1_-_177378801
|
0.349
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr1_+_149299490
|
0.348
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_+_53253204
|
0.347
|
|
SCP2
|
sterol carrier protein 2
|
|
chr1_-_74971679
|
0.345
|
NM_001130042
NM_001130043
NM_001134759
NM_001889
|
CRYZ
|
crystallin, zeta (quinone reductase)
|
|
chr1_-_200396328
|
0.344
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr7_+_115650240
|
0.344
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr12_+_4700012
|
0.343
|
NM_017417
|
GALNT8
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 8 (GalNAc-T8)
|
|
chr1_+_165565006
|
0.342
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr19_+_43847049
|
0.339
|
|
|
|
|
chr10_-_61570665
|
0.338
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr18_-_29273979
|
0.338
|
NM_001105528
|
C18orf34
|
chromosome 18 open reading frame 34
|
|
chr5_-_96544694
|
0.336
|
|
RIOK2
|
RIO kinase 2 (yeast)
|
|
chr3_-_39209075
|
0.336
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr11_-_128217550
|
0.329
|
NM_000220
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr5_+_71538456
|
0.327
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr1_-_26266571
|
0.325
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chr10_+_70152216
|
0.324
|
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|