|
chr4_+_169654742
|
2.220
|
NM_001166108
NM_016081
|
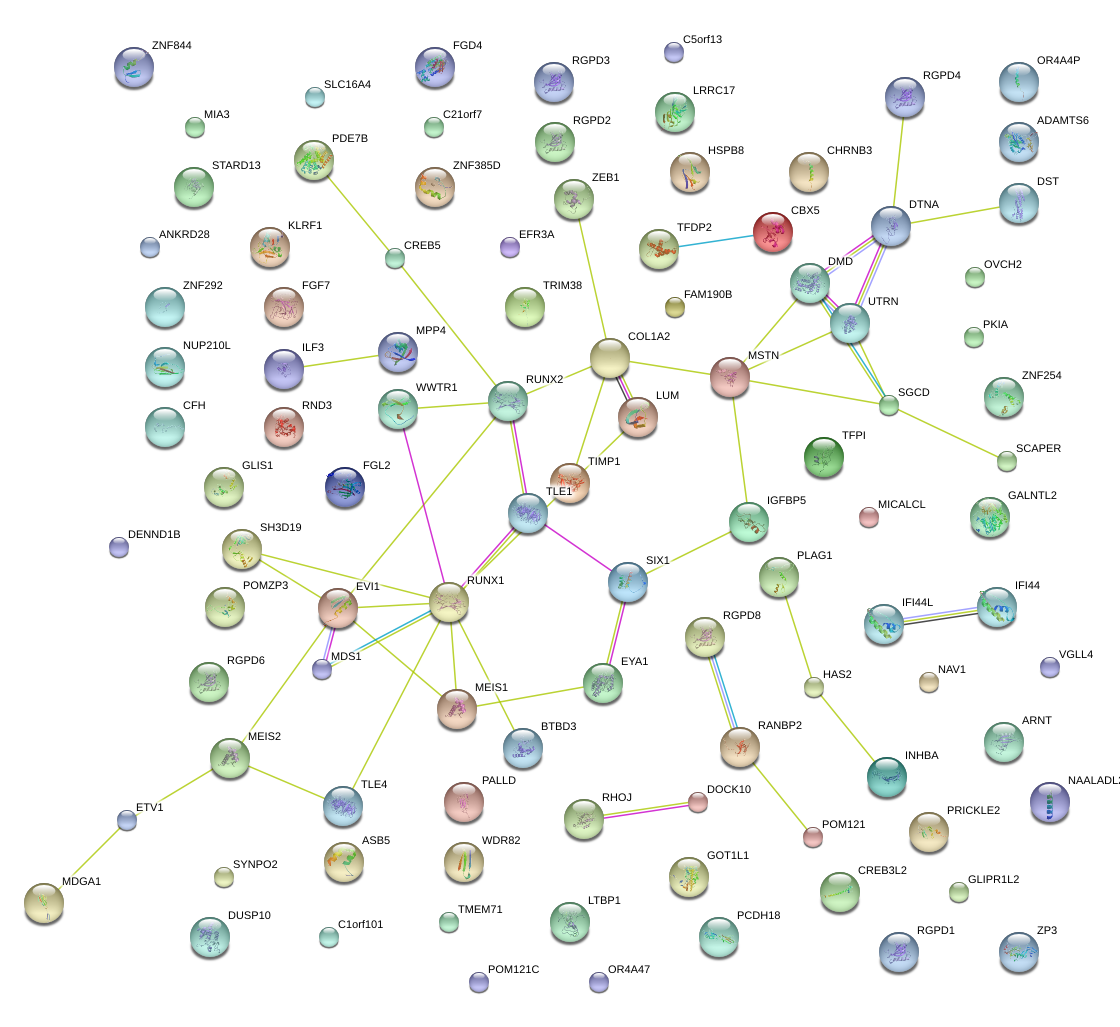
PALLD
|
palladin, cytoskeletal associated protein
|
|
chrX_-_33139349
|
1.484
|
NM_004006
|
DMD
|
dystrophin
|
|
chr8_-_122722674
|
1.291
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr12_+_32546243
|
1.204
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr21_-_35343065
|
1.018
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_194887630
|
0.999
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr21_-_35343448
|
0.996
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_-_143230124
|
0.974
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr5_+_155686333
|
0.962
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr9_+_81376666
|
0.937
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr2_-_217267742
|
0.923
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr21_-_35343456
|
0.919
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr4_-_138673078
|
0.903
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr7_+_93862137
|
0.894
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr21_-_35343500
|
0.861
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_-_64186151
|
0.852
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr3_-_15814678
|
0.830
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr1_-_53972464
|
0.825
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr21_-_35343331
|
0.817
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr9_+_81377419
|
0.815
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr3_+_16191159
|
0.808
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chr7_-_41709191
|
0.804
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr13_-_32658215
|
0.772
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr1_+_78858675
|
0.768
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr3_+_175641340
|
0.763
|
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr2_-_151052391
|
0.751
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr3_-_11585264
|
0.737
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr21_-_35343428
|
0.706
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr9_+_81377289
|
0.700
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr12_-_52939584
|
0.698
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr18_+_30544193
|
0.676
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_-_177427252
|
0.648
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr1_+_199884152
|
0.644
|
|
NAV1
|
neuron navigator 1
|
|
chr11_+_48466844
|
0.633
|
NM_001005512
|
OR4A47
|
olfactory receptor, family 4, subfamily A, member 47
|
|
chr4_-_138673014
|
0.610
|
|
PCDH18
|
protocadherin 18
|
|
chr12_-_52939636
|
0.606
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr3_-_21767819
|
0.598
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr2_-_225519974
|
0.591
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr5_-_111119846
|
0.590
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr19_+_24061717
|
0.581
|
NM_203282
|
ZNF254
|
zinc finger protein 254
|
|
chr14_+_62740878
|
0.569
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr12_-_90029328
|
0.558
|
|
LUM
|
lumican
|
|
chr6_-_37773609
|
0.548
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chr7_+_102340579
|
0.534
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr1_+_78888064
|
0.533
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr2_-_190635566
|
0.520
|
NM_005259
|
MSTN
|
myostatin
|
|
chr6_+_136214495
|
0.518
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr2_-_202271598
|
0.516
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr4_-_152368492
|
0.511
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr15_+_47502666
|
0.509
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr8_+_79591074
|
0.508
|
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr10_+_31650069
|
0.502
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr19_+_12036513
|
0.495
|
NM_001136501
|
ZNF844
|
zinc finger protein 844
|
|
chr7_+_28692122
|
0.491
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr1_-_110735158
|
0.484
|
NM_004696
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr7_+_28305464
|
0.480
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr3_-_52269467
|
0.478
|
|
WDR82
|
WD repeat domain 82
|
|
chr12_-_90029672
|
0.476
|
NM_002345
|
LUM
|
lumican
|
|
chr8_-_72431274
|
0.461
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr7_-_76667045
|
0.458
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr20_+_11846536
|
0.456
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr2_+_12775814
|
0.451
|
|
|
|
|
chr12_+_74071116
|
0.444
|
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chr7_-_137337354
|
0.443
|
NM_194071
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr1_+_242691295
|
0.440
|
NM_001130957
NM_173807
|
C1orf101
|
chromosome 1 open reading frame 101
|
|
chr3_-_150903749
|
0.436
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr8_+_79590890
|
0.431
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr12_+_118100977
|
0.430
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr2_+_66519949
|
0.423
|
|
MEIS1
|
Meis homeobox 1
|
|
chr10_+_86174665
|
0.422
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr2_+_33213111
|
0.409
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr6_-_56824371
|
0.402
|
|
DST
|
dystonin
|
|
chr8_+_42671718
|
0.399
|
NM_000749
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3
|
|
chr1_-_152394215
|
0.393
|
NM_001159484
NM_207308
|
NUP210L
|
nucleoporin 210kDa-like
|
|
chr3_-_170348215
|
0.390
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr21_+_29439768
|
0.385
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr2_-_188127430
|
0.378
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr14_-_60185659
|
0.376
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr2_+_108751033
|
0.375
|
|
RANBP2
|
RAN binding protein 2
|
|
chr8_+_133060353
|
0.370
|
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr8_-_133841988
|
0.369
|
NM_001145153
NM_144649
|
TMEM71
|
transmembrane protein 71
|
|
chr6_+_88026338
|
0.369
|
|
ZNF292
|
zinc finger protein 292
|
|
chr6_+_26071255
|
0.367
|
|
TRIM38
|
tripartite motif containing 38
|
|
chr8_-_72437020
|
0.360
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr8_-_37916803
|
0.356
|
NM_152413
|
GOT1L1
|
glutamic-oxaloacetic transaminase 1-like 1
|
|
chrX_+_47326746
|
0.355
|
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr1_+_15145001
|
0.352
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr1_-_219982049
|
0.350
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr11_+_12265022
|
0.346
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr4_+_120167721
|
0.342
|
|
SYNPO2
|
synaptopodin 2
|
|
chr1_-_219982081
|
0.339
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_-_195818927
|
0.337
|
|
DENND1B
|
DENN/MADD domain containing 1B
|
|
chr15_-_74941339
|
0.336
|
NM_001145923
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr5_-_64813459
|
0.336
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr1_-_149115667
|
0.334
|
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr11_-_7684516
|
0.327
|
NM_198185
|
OVCH2
|
ovochymase 2 (gene/pseudogene)
|
|
chr7_-_13992351
|
0.327
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr1_-_219982016
|
0.327
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr12_+_9871343
|
0.325
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr11_+_65026382
|
0.321
|
|
|
|
|
chr2_-_188127294
|
0.321
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr7_+_72034735
|
0.316
|
|
POM121
|
POM121 membrane glycoprotein
|
|
chr1_+_154120170
|
0.313
|
|
SYT11
|
synaptotagmin XI
|
|
chr2_-_237987554
|
0.307
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr9_-_72673753
|
0.304
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr13_-_98757634
|
0.303
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr1_+_12149662
|
0.303
|
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr10_-_69661860
|
0.300
|
NM_145178
|
ATOH7
|
atonal homolog 7 (Drosophila)
|
|
chr6_+_26548678
|
0.298
|
NM_006994
NM_197974
|
BTN3A3
|
butyrophilin, subfamily 3, member A3
|
|
chr3_+_195688974
|
0.295
|
|
FLJ34208
|
hypothetical LOC401106
|
|
chr3_-_116272911
|
0.295
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chrX_+_47326594
|
0.294
|
NM_003254
|
TIMP1
|
TIMP metallopeptidase inhibitor 1
|
|
chr2_-_144994038
|
0.294
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_237987545
|
0.290
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_217268492
|
0.287
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr2_-_162808123
|
0.287
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr6_+_26070957
|
0.287
|
NM_006355
|
TRIM38
|
tripartite motif containing 38
|
|
chr13_-_46368968
|
0.285
|
NM_000621
NM_001165947
|
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A
|
|
chr2_-_237987579
|
0.282
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_237987553
|
0.281
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr6_-_80303822
|
0.280
|
NM_001122769
NM_181714
|
LCA5
|
Leber congenital amaurosis 5
|
|
chr1_-_199613427
|
0.274
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr11_+_93916653
|
0.273
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr18_-_12646730
|
0.272
|
NM_001128627
|
SPIRE1
|
spire homolog 1 (Drosophila)
|
|
chr11_-_56951628
|
0.271
|
NM_017611
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr21_-_34820916
|
0.270
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr12_-_52281050
|
0.268
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr1_+_108904191
|
0.267
|
|
FAM102B
|
family with sequence similarity 102, member B
|
|
chr15_+_87540238
|
0.267
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr3_-_113495763
|
0.267
|
NM_183061
|
SLC9A10
|
solute carrier family 9, member 10
|
|
chrX_+_55951280
|
0.266
|
|
|
|
|
chr2_-_178681311
|
0.264
|
NM_001077197
|
PDE11A
|
phosphodiesterase 11A
|
|
chr12_-_51887266
|
0.263
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr12_+_46799279
|
0.263
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chr1_+_12149637
|
0.262
|
NM_001066
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr2_-_65447267
|
0.259
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr8_-_23768242
|
0.257
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr2_-_227952249
|
0.255
|
NM_024795
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr4_+_120167770
|
0.253
|
|
SYNPO2
|
synaptopodin 2
|
|
chr20_-_45360903
|
0.251
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr3_-_39209075
|
0.251
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr6_+_116956868
|
0.246
|
NM_153036
|
FAM26D
|
family with sequence similarity 26, member D
|
|
chr6_-_56824672
|
0.245
|
|
DST
|
dystonin
|
|
chr19_-_53912025
|
0.245
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr6_+_96132064
|
0.245
|
NM_024641
|
MANEA
|
mannosidase, endo-alpha
|
|
chr13_+_22653059
|
0.244
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr1_-_89261021
|
0.244
|
NM_018284
|
GBP3
|
guanylate binding protein 3
|
|
chr1_+_242582559
|
0.243
|
NM_001012970
|
C1orf100
|
chromosome 1 open reading frame 100
|
|
chr2_-_144711375
|
0.241
|
NM_024659
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr5_-_88235624
|
0.241
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_-_149115686
|
0.241
|
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr5_+_35892747
|
0.239
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr6_-_50039776
|
0.239
|
NM_001037499
|
DEFB114
|
defensin, beta 114
|
|
chr12_-_92489652
|
0.235
|
|
LOC144481
|
hypothetical protein LOC144481
|
|
chr5_+_149977398
|
0.234
|
NM_001166209
|
SYNPO
|
synaptopodin
|
|
chr5_+_140719773
|
0.233
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr11_+_100063061
|
0.228
|
NM_152432
|
ARHGAP42
|
Rho GTPase activating protein 42
|
|
chr1_-_149115771
|
0.227
|
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr14_-_79748273
|
0.225
|
NM_000793
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chrX_+_80344253
|
0.224
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr1_+_12325632
|
0.224
|
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae)
|
|
chr3_-_171070314
|
0.223
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr15_-_87257553
|
0.221
|
|
MFGE8
|
milk fat globule-EGF factor 8 protein
|
|
chr17_-_36475652
|
0.220
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr4_-_87593306
|
0.218
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr21_-_35174742
|
0.218
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_-_199613406
|
0.218
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chrX_+_51503219
|
0.216
|
NM_018094
|
GSPT2
|
G1 to S phase transition 2
|
|
chr11_-_126375895
|
0.215
|
NM_001161707
NM_032531
|
KIRREL3
|
kin of IRRE like 3 (Drosophila)
|
|
chr4_+_166097548
|
0.215
|
NM_153027
|
C4orf39
|
chromosome 4 open reading frame 39
|
|
chr1_+_170656450
|
0.211
|
NM_139240
|
C1orf105
|
chromosome 1 open reading frame 105
|
|
chr13_-_20533629
|
0.209
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr10_+_28918672
|
0.208
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr1_-_20374273
|
0.208
|
NM_001105572
|
PLA2G2C
|
phospholipase A2, group IIC
|
|
chr14_-_94669547
|
0.206
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr2_-_198248828
|
0.205
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr3_+_51550635
|
0.200
|
NM_015106
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae)
|
|
chr14_+_87921818
|
0.199
|
|
SPATA7
|
spermatogenesis associated 7
|
|
chr11_+_59027624
|
0.197
|
NM_001004706
|
OR4D11
|
olfactory receptor, family 4, subfamily D, member 11
|
|
chr13_-_20533703
|
0.196
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr4_+_54790190
|
0.193
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr14_+_100365162
|
0.193
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr2_+_87536174
|
0.193
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr13_-_98708549
|
0.193
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr3_-_106903734
|
0.193
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr1_+_242281199
|
0.193
|
|
ZNF238
|
zinc finger protein 238
|
|
chr1_+_156229686
|
0.191
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr11_-_73371945
|
0.188
|
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier)
|
|
chrX_-_19898302
|
0.187
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr1_+_15128882
|
0.186
|
NM_001018001
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr6_+_147568795
|
0.183
|
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr6_-_66346417
|
0.183
|
NM_198283
|
EYS
|
eyes shut homolog (Drosophila)
|
|
chr5_+_95092384
|
0.182
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chrX_+_80344215
|
0.182
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr12_+_10348311
|
0.180
|
NM_001114396
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr1_+_85300580
|
0.180
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr4_+_187227302
|
0.180
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr4_+_40953652
|
0.179
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr6_+_39868116
|
0.178
|
|
|
|
|
chr5_+_31229549
|
0.178
|
NM_004932
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|