|
chr7_-_27108826
|
3.777
|
NM_006735
|
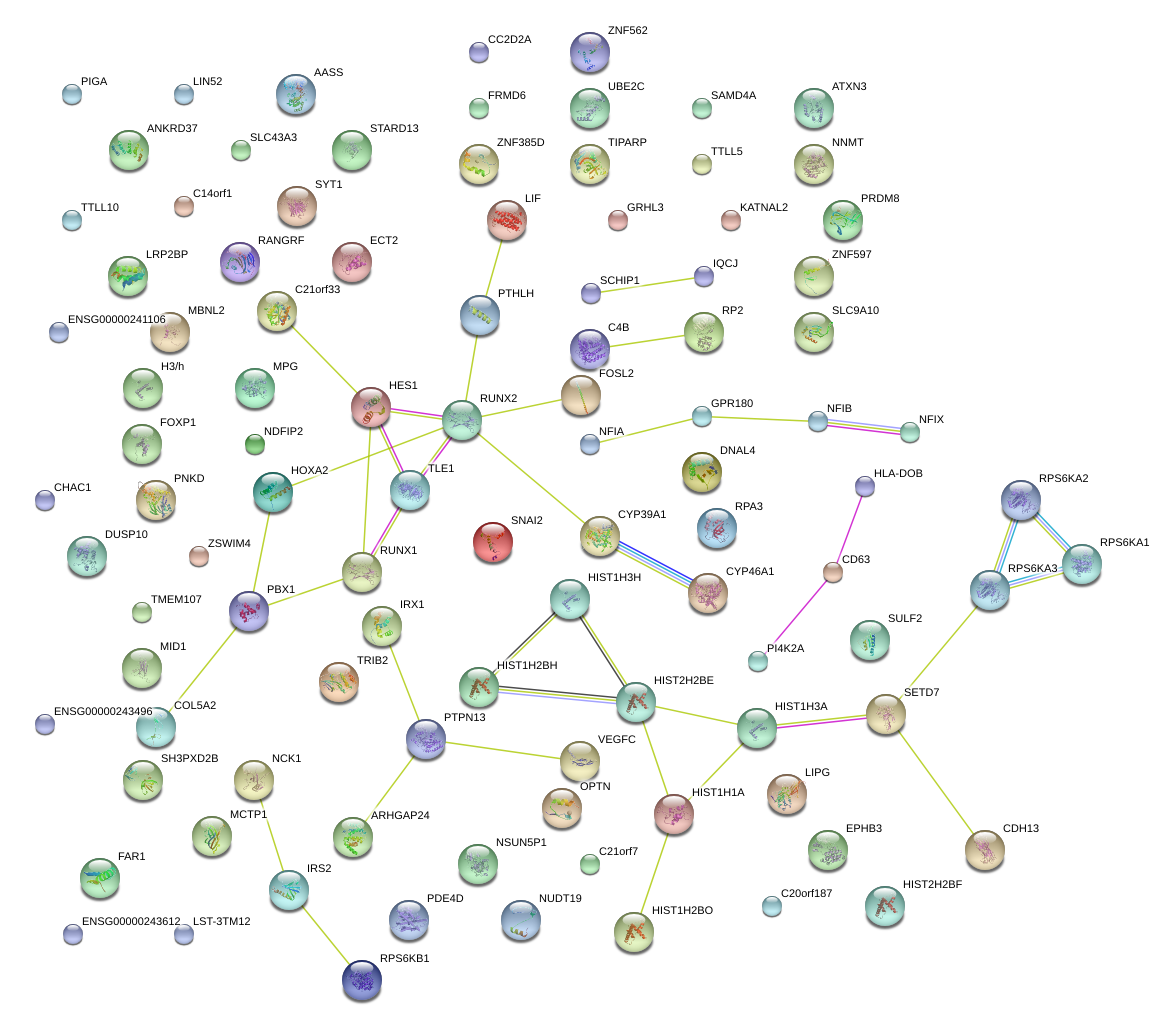
HOXA2
|
homeobox A2
|
|
chr13_-_32757835
|
3.274
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_-_189752575
|
3.045
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr5_+_3649167
|
3.041
|
NM_024337
|
IRX1
|
iroquois homeobox 1
|
|
chr18_+_42780784
|
2.983
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr1_+_24518501
|
2.921
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr1_+_24518398
|
2.609
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr3_-_113495763
|
2.487
|
NM_183061
|
SLC9A10
|
solute carrier family 9, member 10
|
|
chr12_+_77782719
|
2.455
|
|
SYT1
|
synaptotagmin I
|
|
chr6_-_166960690
|
2.360
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr11_+_113671744
|
2.351
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr6_-_166960649
|
2.257
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr14_+_51025604
|
2.088
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr5_-_58607673
|
2.086
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_28016182
|
2.063
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr21_+_29439768
|
1.891
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr3_+_157877146
|
1.853
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr6_-_166960496
|
1.834
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr4_-_177950658
|
1.798
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr1_+_162795328
|
1.774
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_+_195336625
|
1.762
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr19_+_13767207
|
1.739
|
NM_023072
|
ZSWIM4
|
zinc finger, SWIM-type containing 4
|
|
chr13_-_109236897
|
1.714
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr18_+_45342398
|
1.699
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr3_+_195336631
|
1.646
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr22_-_28972683
|
1.627
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr6_-_26125939
|
1.542
|
NM_005325
|
HIST1H1A
|
histone cluster 1, H1a
|
|
chrX_-_10548458
|
1.539
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr3_-_21767819
|
1.537
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr5_-_171814026
|
1.529
|
NM_001017995
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr14_-_75196911
|
1.491
|
|
C14orf1
|
chromosome 14 open reading frame 1
|
|
chr9_-_83494198
|
1.440
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr13_+_94052022
|
1.426
|
NM_180989
|
GPR180
|
G protein-coupled receptor 180
|
|
chr21_-_35174742
|
1.420
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr2_+_12774264
|
1.342
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr6_-_26379567
|
1.339
|
NM_003534
|
HIST1H3G
|
histone cluster 1, H3g
|
|
chr14_+_75197303
|
1.251
|
NM_015072
|
TTLL5
|
tubulin tyrosine ligase-like family, member 5
|
|
chr16_+_81218074
|
1.234
|
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr4_+_87734489
|
1.143
|
NM_006264
NM_080683
NM_080684
NM_080685
|
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase)
|
|
chr5_-_171813960
|
1.126
|
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr20_-_45848043
|
1.111
|
NM_018837
|
SULF2
|
sulfatase 2
|
|
chr14_+_75197345
|
1.103
|
|
TTLL5
|
tubulin tyrosine ligase-like family, member 5
|
|
chr14_+_75197409
|
1.094
|
|
TTLL5
|
tubulin tyrosine ligase-like family, member 5
|
|
chr14_+_54104383
|
1.094
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr1_-_147665783
|
1.065
|
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr15_+_39032927
|
1.058
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr8_-_49996353
|
1.041
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr4_+_86968069
|
1.040
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr3_+_173951163
|
1.034
|
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr4_-_186537145
|
1.025
|
NM_018409
|
LRP2BP
|
LRP2 binding protein
|
|
chr16_+_81218142
|
1.010
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr2_+_218843358
|
0.960
|
NM_001077399
NM_015488
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chrX_-_15263596
|
0.950
|
NM_002641
NM_020473
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chrX_+_46581418
|
0.931
|
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive)
|
|
chr1_-_219982081
|
0.930
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr11_+_13646727
|
0.925
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr10_+_99390459
|
0.924
|
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr3_+_185762233
|
0.918
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chrX_-_15263586
|
0.910
|
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr1_-_219982049
|
0.903
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chrX_-_15263555
|
0.887
|
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr17_+_8132539
|
0.849
|
NM_001177801
NM_001177802
NM_016492
|
RANGRF
|
RAN guanine nucleotide release factor
|
|
chr3_+_138132006
|
0.847
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr6_-_32892705
|
0.845
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr3_-_71196696
|
0.844
|
|
FOXP1
|
forkhead box P1
|
|
chrX_+_46581368
|
0.831
|
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive)
|
|
chr11_+_13646839
|
0.830
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr11_+_13646822
|
0.822
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr1_-_219982016
|
0.822
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr12_+_21059896
|
0.820
|
NM_001009562
|
LST-3TM12
|
organic anion transporter LST-3b
|
|
chr12_+_77782648
|
0.801
|
|
SYT1
|
synaptotagmin I
|
|
chr22_-_37520049
|
0.797
|
|
DNAL4
|
dynein, axonemal, light chain 4
|
|
chr6_-_46728188
|
0.792
|
NM_016593
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1
|
|
chr4_-_140696740
|
0.792
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr7_-_7646573
|
0.788
|
|
RPA3
|
replication protein A3, 14kDa
|
|
chr12_-_54409710
|
0.788
|
|
CD63
|
CD63 molecule
|
|
chr13_+_96726197
|
0.780
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr3_+_161040439
|
0.766
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr20_+_43874735
|
0.763
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr5_-_94442941
|
0.763
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chrX_+_46581290
|
0.762
|
NM_006915
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive)
|
|
chr7_+_7646840
|
0.761
|
|
LOC729852
|
hypothetical LOC729852
|
|
chr14_+_73621408
|
0.758
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr16_-_3433490
|
0.755
|
NM_152457
|
ZNF597
|
zinc finger protein 597
|
|
chr9_-_14303509
|
0.745
|
|
NFIB
|
nuclear factor I/B
|
|
chr10_+_13182061
|
0.729
|
NM_001008211
NM_001008212
NM_001008213
NM_021980
|
OPTN
|
optineurin
|
|
chr14_+_99220349
|
0.726
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr4_+_81325447
|
0.722
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr3_+_161040343
|
0.718
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr14_-_91642703
|
0.712
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr2_+_28469850
|
0.707
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr7_-_121571472
|
0.689
|
NM_005763
|
AASS
|
aminoadipate-semialdehyde synthase
|
|
chr13_+_78953258
|
0.681
|
NM_001161407
NM_019080
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr11_-_56951628
|
0.678
|
NM_017611
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr19_-_63092099
|
0.647
|
NM_001144989
|
ZNF814
|
zinc finger protein 814
|
|
chr11_+_122258600
|
0.646
|
NM_024806
NM_199124
|
C11orf63
|
chromosome 11 open reading frame 63
|
|
chr21_-_31012965
|
0.641
|
NM_001164435
|
KRTAP21-3
|
keratin associated protein 21-3
|
|
chr10_+_17310263
|
0.639
|
NM_003380
|
VIM
|
vimentin
|
|
chr7_+_106596641
|
0.637
|
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr20_+_56369012
|
0.636
|
|
|
|
|
chr1_-_149586341
|
0.634
|
NM_000449
NM_001025603
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr12_-_14982708
|
0.633
|
NM_152321
|
ERP27
|
endoplasmic reticulum protein 27
|
|
chr16_+_3433606
|
0.633
|
NM_001083601
|
NAT15
|
N-acetyltransferase 15 (GCN5-related, putative)
|
|
chr6_+_26153590
|
0.621
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr2_+_191454157
|
0.617
|
|
GLS
|
glutaminase
|
|
chr1_-_205272619
|
0.614
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr11_+_13646806
|
0.606
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr4_-_118226119
|
0.604
|
NM_152402
|
TRAM1L1
|
translocation associated membrane protein 1-like 1
|
|
chr3_-_191322919
|
0.602
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr17_+_70495318
|
0.596
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr2_-_162808123
|
0.594
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr6_+_26381122
|
0.589
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr6_+_26333361
|
0.587
|
NM_003532
|
HIST1H3E
|
histone cluster 1, H3e
|
|
chr17_+_72380323
|
0.577
|
NM_198955
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr5_-_171643247
|
0.563
|
|
UBTD2
|
ubiquitin domain containing 2
|
|
chr14_-_90596665
|
0.555
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr14_-_91642641
|
0.553
|
|
ATXN3
ATXN8
|
ataxin 3
ataxin 8
|
|
chr8_-_108579270
|
0.550
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr22_-_37520092
|
0.549
|
NM_005740
|
DNAL4
|
dynein, axonemal, light chain 4
|
|
chr19_-_63301386
|
0.547
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr22_-_41741073
|
0.533
|
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chrX_-_107568211
|
0.531
|
NM_033641
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr22_-_41741100
|
0.521
|
NM_001184970
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr22_+_28164571
|
0.515
|
NM_021026
|
RFPL1
|
ret finger protein-like 1
|
|
chr4_-_48477038
|
0.514
|
NM_015030
|
FRYL
|
FRY-like
|
|
chr1_+_85300580
|
0.509
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr8_+_107807415
|
0.508
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr20_+_43874654
|
0.505
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr1_-_149586298
|
0.503
|
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr4_-_48477021
|
0.499
|
|
FRYL
|
FRY-like
|
|
chr3_+_130641840
|
0.498
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr7_+_29569913
|
0.489
|
NM_175887
|
PRR15
|
proline rich 15
|
|
chr12_+_52689062
|
0.487
|
NM_022658
|
HOXC8
|
homeobox C8
|
|
chr12_+_27568311
|
0.486
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr19_-_44573468
|
0.485
|
NM_019088
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae)
|
|
chr9_+_112470871
|
0.483
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr1_-_159257486
|
0.479
|
|
F11R
|
F11 receptor
|
|
chr19_+_16157198
|
0.472
|
NM_014077
|
FAM32A
|
family with sequence similarity 32, member A
|
|
chr14_-_73620912
|
0.470
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr9_+_129604962
|
0.463
|
NM_004957
|
FPGS
|
folylpolyglutamate synthase
|
|
chr2_+_191454036
|
0.460
|
|
GLS
|
glutaminase
|
|
chr8_-_25371708
|
0.454
|
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr14_-_22722680
|
0.452
|
NM_012244
|
SLC7A8
|
solute carrier family 7 (amino acid transporter, L-type), member 8
|
|
chr12_-_51173238
|
0.451
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr4_+_77575276
|
0.451
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr12_+_28234657
|
0.449
|
|
|
|
|
chr3_-_150578023
|
0.449
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr19_+_55221023
|
0.448
|
NM_001006656
NM_015428
|
ZNF473
|
zinc finger protein 473
|
|
chr17_+_17816851
|
0.442
|
NM_001130090
NM_001130091
NM_001130092
NM_031294
|
LRRC48
|
leucine rich repeat containing 48
|
|
chr8_-_25371663
|
0.439
|
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr20_+_11846536
|
0.438
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr12_-_51132129
|
0.437
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr10_+_99390431
|
0.436
|
NM_018425
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr6_-_27222556
|
0.435
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr2_-_108972100
|
0.435
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr3_+_130641639
|
0.433
|
NM_018262
NM_052985
NM_052989
NM_052990
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr5_-_145503190
|
0.432
|
|
LARS
|
leucyl-tRNA synthetase
|
|
chr10_+_63092724
|
0.432
|
NM_173554
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr19_+_14501413
|
0.432
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr14_+_23008723
|
0.430
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr10_-_105835525
|
0.426
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr15_-_84139021
|
0.425
|
|
KLHL25
|
kelch-like 25 (Drosophila)
|
|
chr11_+_6904167
|
0.422
|
NM_013250
|
ZNF215
|
zinc finger protein 215
|
|
chr5_+_86712376
|
0.421
|
|
|
|
|
chr2_+_28469864
|
0.420
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr5_-_112658200
|
0.414
|
|
MCC
|
mutated in colorectal cancers
|
|
chr12_+_64982542
|
0.411
|
NM_033647
|
HELB
|
helicase (DNA) B
|
|
chr8_+_69027277
|
0.410
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr13_+_78953367
|
0.410
|
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr17_+_71174993
|
0.410
|
NM_013260
|
SAP30BP
|
SAP30 binding protein
|
|
chr19_-_55220495
|
0.407
|
NM_001025778
NM_016440
|
VRK3
|
vaccinia related kinase 3
|
|
chr6_+_46728590
|
0.405
|
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr4_+_8935987
|
0.403
|
NM_001105662
|
USP17L2
USP17L6P
USP17
|
ubiquitin specific peptidase 17-like 2
ubiquitin specific peptidase 17-like 6 (pseudogene)
ubiquitin specific peptidase 17
|
|
chr19_+_54067450
|
0.401
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr19_+_54067491
|
0.400
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr19_+_54067489
|
0.399
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr20_-_56859352
|
0.398
|
|
GNAS-AS1
|
GNAS antisense RNA 1 (non-protein coding)
|
|
chr8_-_23768242
|
0.397
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr3_-_32519362
|
0.396
|
NM_017801
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6
|
|
chr1_+_216585298
|
0.390
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr10_+_63331401
|
0.390
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr19_-_55220439
|
0.388
|
|
VRK3
|
vaccinia related kinase 3
|
|
chrX_-_23671317
|
0.388
|
NM_001033583
NM_001037171
|
ACOT9
|
acyl-CoA thioesterase 9
|
|
chr18_+_37789187
|
0.387
|
NM_002647
|
PIK3C3
|
phosphoinositide-3-kinase, class 3
|
|
chrX_-_119550402
|
0.386
|
|
CUL4B
|
cullin 4B
|
|
chr6_+_151603201
|
0.384
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr3_-_52906608
|
0.384
|
|
TMEM110-MUSTN1
|
TMEM110-MUSTN1 readthrough
|
|
chr3_-_15814678
|
0.383
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr13_+_38159172
|
0.382
|
NM_207361
|
FREM2
|
FRAS1 related extracellular matrix protein 2
|
|
chr14_+_23008742
|
0.381
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr4_+_8940733
|
0.379
|
NM_001105662
|
USP17L2
USP17L6P
USP17
|
ubiquitin specific peptidase 17-like 2
ubiquitin specific peptidase 17-like 6 (pseudogene)
ubiquitin specific peptidase 17
|
|
chr19_-_63301418
|
0.377
|
NM_001145543
NM_001145544
NM_023926
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr3_-_130641539
|
0.376
|
NM_003925
|
MBD4
|
methyl-CpG binding domain protein 4
|
|
chrX_-_92815222
|
0.376
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr3_-_52906381
|
0.374
|
|
TMEM110
|
transmembrane protein 110
|
|
chr6_+_151603346
|
0.374
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr19_+_44573858
|
0.374
|
|
MED29
|
mediator complex subunit 29
|
|
chr12_+_77782579
|
0.372
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr20_-_13567529
|
0.369
|
NM_017714
|
TASP1
|
taspase, threonine aspartase, 1
|
|
chr18_+_30427279
|
0.368
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|