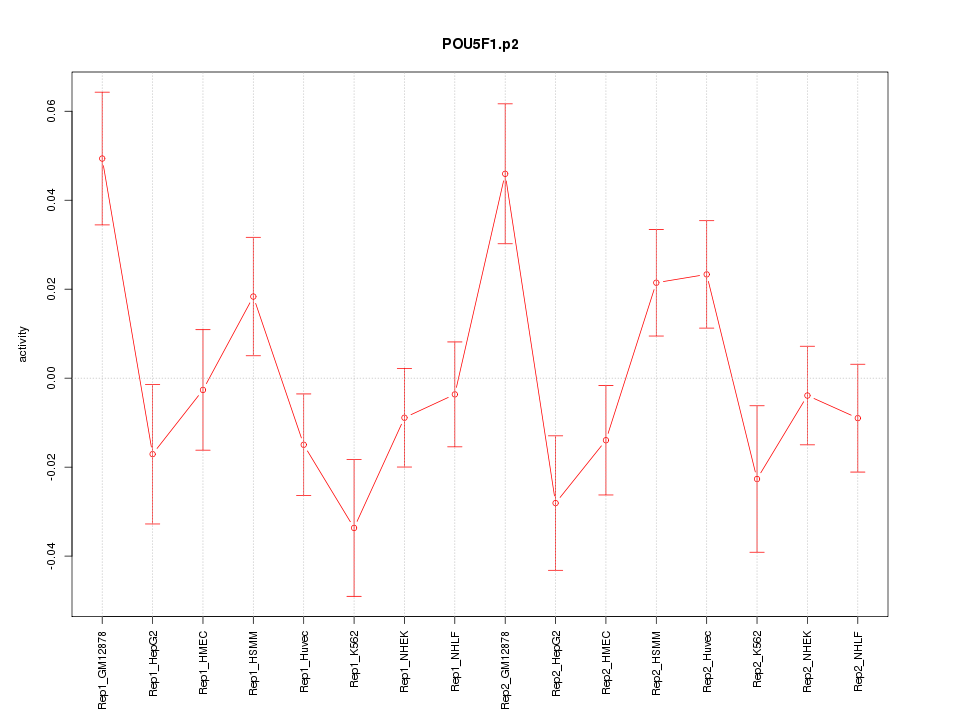
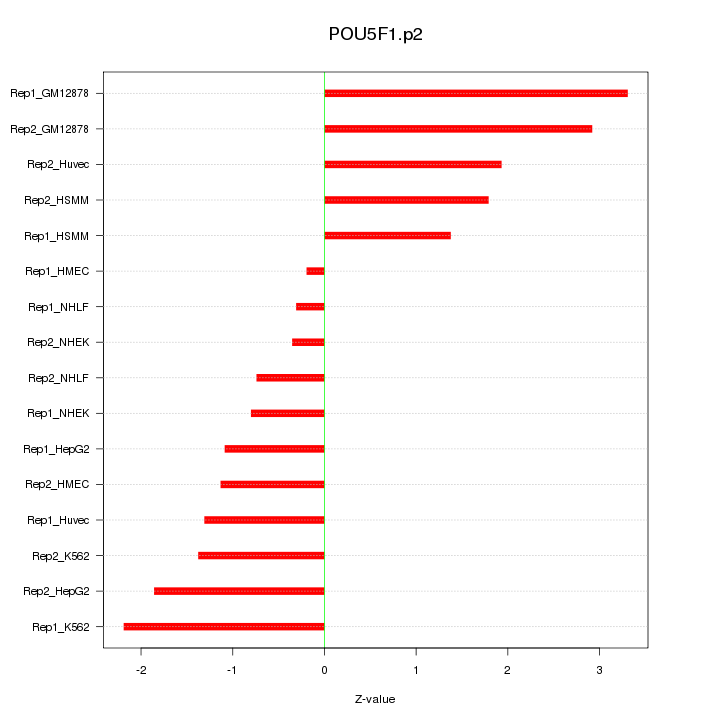
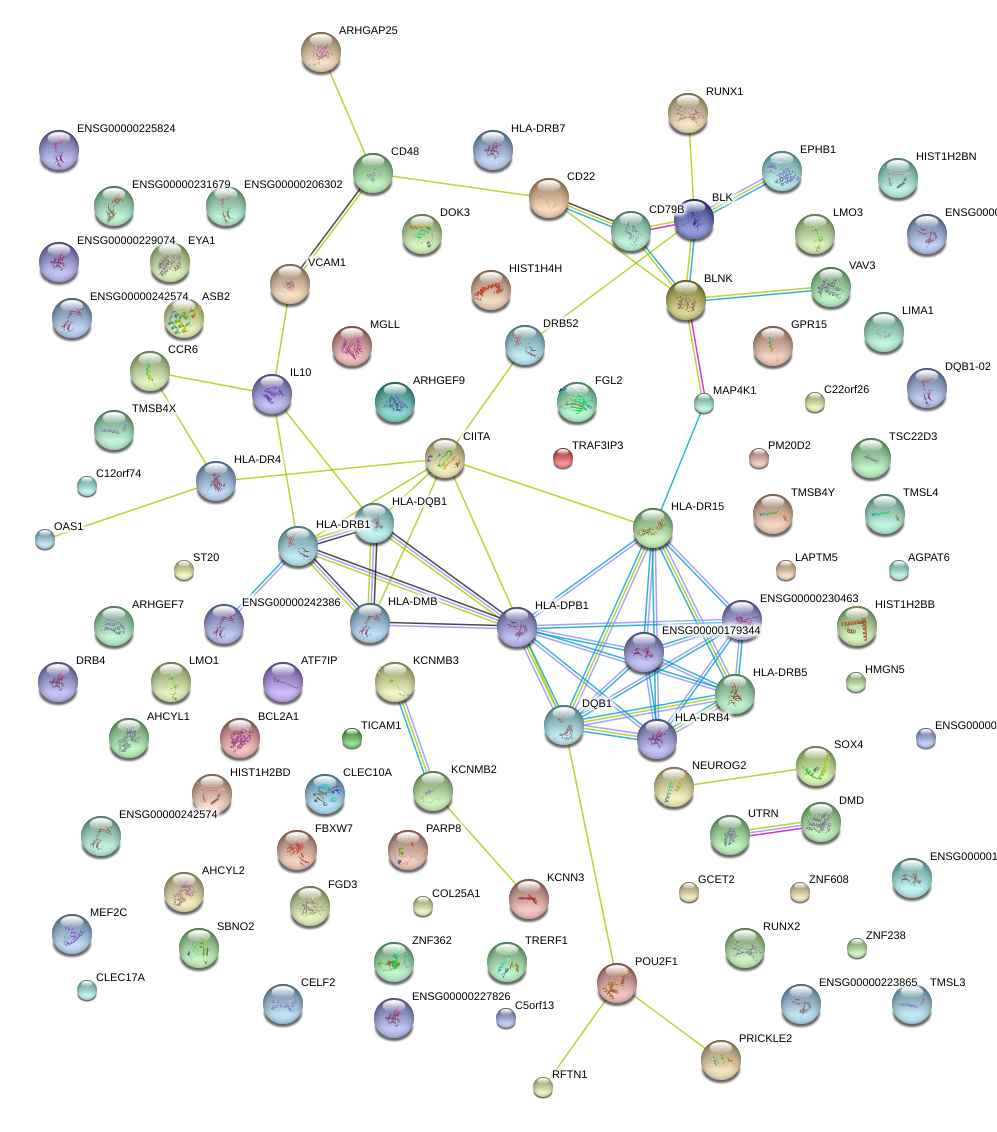
|
chr7_-_76667045
|
3.765
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr1_-_153098811
|
3.327
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr10_-_98021144
|
3.170
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr5_-_124108672
|
2.848
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr10_+_11246934
|
2.433
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr5_-_88155360
|
2.413
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_-_91979113
|
2.352
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr22_-_44828636
|
2.178
|
NM_018280
|
C22orf26
|
chromosome 22 open reading frame 26
|
|
chr19_-_43800374
|
2.152
|
NM_001042600
NM_007181
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
|
chr1_+_165565006
|
2.019
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr5_-_111119846
|
1.961
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr12_-_16649211
|
1.904
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr1_-_197173159
|
1.880
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr12_+_14409875
|
1.827
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr3_+_179736917
|
1.809
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr14_-_93512818
|
1.793
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr6_-_42527760
|
1.769
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr3_+_99733432
|
1.737
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr1_+_165564904
|
1.737
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr15_-_78050515
|
1.729
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr6_-_26151863
|
1.692
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chrX_-_62891066
|
1.686
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr1_+_242281175
|
1.669
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr17_-_59363352
|
1.653
|
NM_000626
NM_001039933
NM_021602
|
CD79B
|
CD79b molecule, immunoglobulin-associated beta
|
|
chr2_+_68815416
|
1.651
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr3_-_64186151
|
1.651
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr1_+_33494745
|
1.639
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr5_+_140871107
|
1.580
|
|
|
|
|
chr3_-_129023850
|
1.578
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr5_-_88215034
|
1.528
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_+_242281199
|
1.499
|
|
ZNF238
|
zinc finger protein 238
|
|
chr19_+_14554895
|
1.488
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr1_-_205012348
|
1.486
|
NM_000572
|
IL10
|
interleukin 10
|
|
chr14_-_93512647
|
1.461
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr6_+_21701942
|
1.450
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr12_+_111829121
|
1.420
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr12_-_16650697
|
1.405
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr6_-_33016561
|
1.394
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr2_+_88966170
|
1.389
|
|
|
|
|
chr5_-_176868483
|
1.388
|
|
DOK3
|
docking protein 3
|
|
chr8_+_11388918
|
1.294
|
NM_001715
|
BLK
|
B lymphoid tyrosine kinase
|
|
chr16_+_10878539
|
1.292
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chrX_-_106846684
|
1.283
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr12_-_48902692
|
1.248
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr1_-_158948176
|
1.248
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr1_-_108308997
|
1.228
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr7_+_128795280
|
1.191
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chrX_+_12903145
|
1.178
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr6_-_32665409
|
1.172
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr8_-_72431274
|
1.172
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr12_+_91620749
|
1.171
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr3_-_16530132
|
1.171
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chrX_-_106846924
|
1.144
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_100957883
|
1.144
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr1_-_31003181
|
1.138
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr21_-_35343065
|
1.129
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_208008517
|
1.120
|
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr1_-_31003260
|
1.119
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr19_-_1083109
|
1.111
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr10_+_11247059
|
1.108
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr3_+_135996990
|
1.104
|
|
EPHB1
|
EPH receptor B1
|
|
chr5_+_140870881
|
1.099
|
|
|
|
|
chr4_-_110443247
|
1.098
|
NM_032518
NM_198721
|
COL25A1
|
collagen, type XXV, alpha 1
|
|
chr6_+_167456230
|
1.089
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr1_-_42738535
|
1.081
|
|
TMSB4XP1
|
thymosin beta 4, X-linked pseudogene 1
|
|
chr3_+_135996734
|
1.069
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr9_+_94766379
|
1.065
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr19_+_40511909
|
1.055
|
NM_001185099
NM_001185100
NM_001185101
NM_001771
|
CD22
|
CD22 molecule
|
|
chrX_-_33267646
|
1.052
|
NM_000109
|
DMD
|
dystrophin
|
|
chr13_+_110637173
|
1.050
|
NM_001113513
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr4_-_153493133
|
1.042
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr5_+_49998677
|
1.041
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr4_-_113656762
|
1.040
|
NM_024019
|
NEUROG2
|
neurogenin 2
|
|
chrX_-_80344096
|
1.033
|
NM_030763
|
HMGN5
|
high-mobility group nucleosome binding domain 5
|
|
chr3_-_113334657
|
1.028
|
NM_001190259
NM_001190260
NM_152785
|
GCET2
|
germinal center expressed transcript 2
|
|
chr18_+_40514772
|
1.017
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr1_+_145479805
|
1.010
|
NM_004326
|
BCL9
|
B-cell CLL/lymphoma 9
|
|
chr17_-_16816113
|
1.009
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr12_-_101115677
|
0.998
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr5_+_49998528
|
0.992
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr18_-_59137488
|
0.988
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr7_+_128795178
|
0.985
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr10_+_91051685
|
0.965
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chr1_-_23758503
|
0.949
|
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr2_-_207738858
|
0.947
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr2_+_5750229
|
0.943
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr17_-_8756524
|
0.936
|
NM_014308
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5
|
|
chr2_+_68445821
|
0.923
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr7_+_18501876
|
0.919
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr3_-_52235218
|
0.918
|
NM_017442
|
TLR9
|
toll-like receptor 9
|
|
chr11_+_5610060
|
0.915
|
NM_130390
|
TRIM34
|
tripartite motif containing 34
|
|
chr11_+_46339691
|
0.913
|
NM_001105540
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr12_-_16650687
|
0.911
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_-_3049872
|
0.909
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr4_+_81325447
|
0.905
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr2_+_237143118
|
0.897
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr22_+_20715340
|
0.888
|
|
|
|
|
chr3_+_129124591
|
0.886
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr1_+_161305558
|
0.878
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_190552744
|
0.864
|
NM_001039152
|
RGS21
|
regulator of G-protein signaling 21
|
|
chr2_+_170742900
|
0.864
|
NM_001083615
NM_001171642
NM_138995
|
MYO3B
|
myosin IIIB
|
|
chr11_+_128068982
|
0.861
|
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr1_+_66570683
|
0.861
|
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr2_+_189547358
|
0.857
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chrX_-_77469635
|
0.856
|
NM_006639
|
CYSLTR1
|
cysteinyl leukotriene receptor 1
|
|
chr5_-_36337756
|
0.847
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr8_+_123863075
|
0.847
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr18_+_40513889
|
0.845
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr7_+_45033757
|
0.844
|
NM_001029835
|
CCM2
|
cerebral cavernous malformation 2
|
|
chr5_-_111120764
|
0.839
|
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chrX_+_80344253
|
0.839
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr14_-_105804211
|
0.837
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr8_+_123863169
|
0.836
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr3_-_16529996
|
0.830
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr9_-_35608300
|
0.828
|
NM_001782
|
CD72
|
CD72 molecule
|
|
chr7_-_129868059
|
0.821
|
NM_018718
|
TSGA14
|
testis specific, 14
|
|
chr10_+_68355797
|
0.818
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr12_+_111829191
|
0.817
|
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr11_+_128069203
|
0.814
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr17_-_39094787
|
0.810
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr11_+_128069183
|
0.801
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr3_+_29297806
|
0.799
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr11_+_128069165
|
0.799
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chrX_-_77801480
|
0.798
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr6_-_56824672
|
0.797
|
|
DST
|
dystonin
|
|
chr11_-_26700121
|
0.797
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr6_+_26381122
|
0.796
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr13_-_98757634
|
0.794
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr10_+_11247243
|
0.787
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_38460983
|
0.775
|
NM_001017388
NM_001195106
NM_001195107
NM_001195108
NM_030956
|
TLR10
|
toll-like receptor 10
|
|
chr4_+_113958652
|
0.773
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr22_+_21370394
|
0.772
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr2_+_189547377
|
0.771
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr9_+_18464138
|
0.769
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr21_+_25933938
|
0.766
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr8_+_123863224
|
0.766
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr2_+_108751033
|
0.762
|
|
RANBP2
|
RAN binding protein 2
|
|
chr5_-_111120817
|
0.755
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr4_-_164754106
|
0.754
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr8_+_70567634
|
0.752
|
|
SULF1
|
sulfatase 1
|
|
chr1_+_190811479
|
0.750
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr8_+_86287161
|
0.746
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr21_+_25933916
|
0.743
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr2_-_225519974
|
0.740
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr10_+_63331401
|
0.737
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr6_-_32892705
|
0.733
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr6_-_25044156
|
0.730
|
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chrX_-_135690699
|
0.728
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr1_+_179719338
|
0.727
|
NM_000721
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr1_+_66570363
|
0.726
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr22_-_34886727
|
0.724
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chr17_+_64922420
|
0.720
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chrX_-_33139349
|
0.718
|
NM_004006
|
DMD
|
dystrophin
|
|
chrX_+_80344215
|
0.718
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr9_+_112470871
|
0.718
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr6_-_149847809
|
0.717
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chrX_+_154264931
|
0.716
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr19_-_53444733
|
0.713
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr3_-_58175437
|
0.711
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr15_-_50608296
|
0.709
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr11_+_128069340
|
0.703
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr6_-_149847718
|
0.698
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr1_-_42156957
|
0.690
|
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr22_+_30072739
|
0.686
|
|
FLJ20464
|
hypothetical protein FLJ20464
|
|
chr6_-_56824371
|
0.684
|
|
DST
|
dystonin
|
|
chr1_-_28376041
|
0.683
|
NM_000952
|
PTAFR
|
platelet-activating factor receptor
|
|
chr1_-_111544603
|
0.681
|
|
|
|
|
chr22_-_36212160
|
0.679
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr3_+_142587973
|
0.677
|
|
|
|
|
chr5_-_54317102
|
0.675
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr1_-_171442937
|
0.674
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chrX_-_135690770
|
0.673
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr16_+_31274020
|
0.670
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr14_-_105623358
|
0.669
|
|
IGHA1
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr5_-_83052606
|
0.668
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr8_+_70567581
|
0.653
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr5_+_49997489
|
0.648
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr7_+_73826244
|
0.648
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr2_-_213723298
|
0.648
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr11_+_65027721
|
0.647
|
|
|
|
|
chr1_-_42157077
|
0.647
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr7_+_99654806
|
0.644
|
NM_024070
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing
|
|
chr14_-_106241445
|
0.644
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr7_+_149842850
|
0.644
|
NM_153236
|
GIMAP7
|
GTPase, IMAP family member 7
|
|
chr3_-_102194938
|
0.643
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr7_-_44088561
|
0.640
|
|
POLM
|
polymerase (DNA directed), mu
|
|
chr5_-_140875591
|
0.634
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr6_-_170442090
|
0.631
|
|
|
|
|
chr16_+_31274030
|
0.626
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr8_-_119703297
|
0.623
|
|
SAMD12
|
sterile alpha motif domain containing 12
|
|
chr2_+_189547323
|
0.620
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr11_-_47356479
|
0.619
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr15_-_40537002
|
0.617
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr15_-_38387357
|
0.617
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr6_+_44295219
|
0.617
|
NM_001078174
NM_004955
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr5_+_155686333
|
0.616
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr12_-_8656705
|
0.614
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr7_-_27186400
|
0.613
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr5_+_132037271
|
0.608
|
NM_000589
NM_172348
|
IL4
|
interleukin 4
|
|
chr19_+_57531309
|
0.602
|
NM_001161425
NM_001161426
NM_001161427
|
ZNF610
|
zinc finger protein 610
|