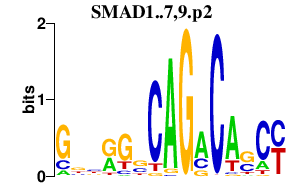
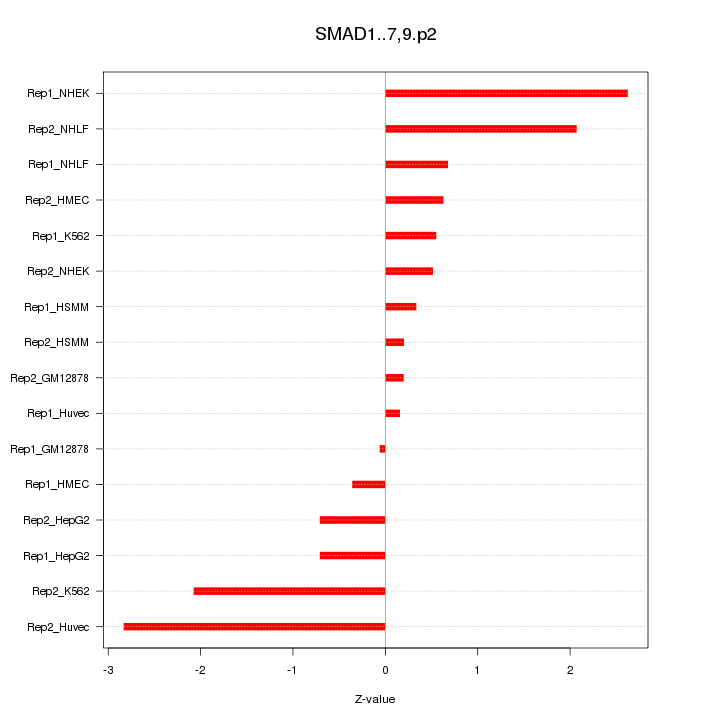
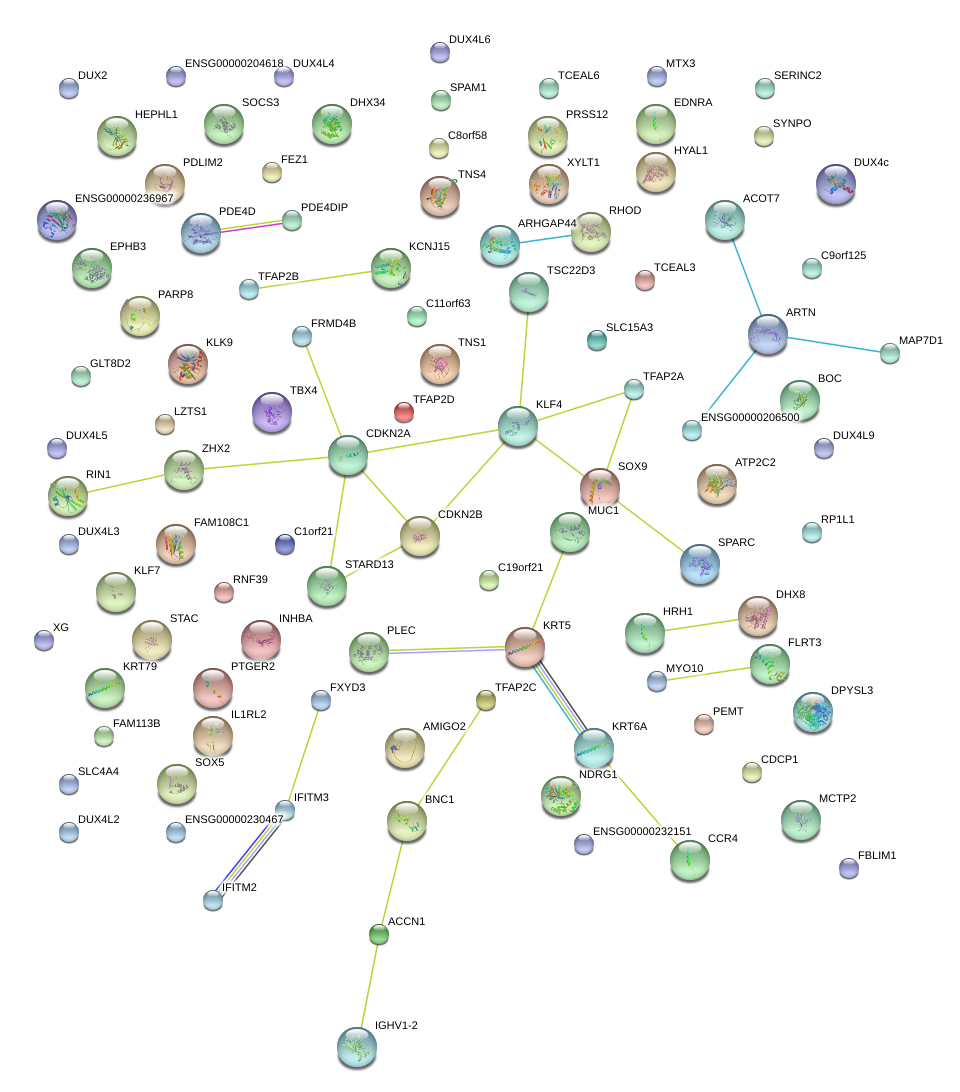
|
chr3_+_11171213
|
1.478
|
NM_001098212
|
HRH1
|
histamine receptor H1
|
|
chr15_-_81744469
|
1.396
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr11_-_65860450
|
1.322
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr5_-_151046638
|
1.292
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr5_-_151046680
|
1.217
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr1_-_6368324
|
1.082
|
NM_181864
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr9_-_109291130
|
1.039
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_+_15145001
|
1.027
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr8_+_22493928
|
0.979
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr2_+_102169864
|
0.890
|
NM_003854
|
IL1RL2
|
interleukin 1 receptor-like 2
|
|
chrX_+_102749489
|
0.874
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr10_+_22765996
|
0.868
|
|
LOC100289455
|
hypothetical LOC100289455
|
|
chr9_-_109291575
|
0.815
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr14_+_51850754
|
0.811
|
NM_000956
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa
|
|
chr13_-_32822759
|
0.798
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr11_-_310925
|
0.739
|
|
IFITM3
|
interferon induced transmembrane protein 3 (1-8U)
|
|
chr6_-_30151526
|
0.732
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr19_+_40299058
|
0.728
|
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr11_+_66580884
|
0.707
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr3_+_114414064
|
0.680
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr15_+_92575767
|
0.680
|
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr1_+_31659246
|
0.677
|
NM_001199039
NM_018565
|
SERINC2
|
serine incorporator 2
|
|
chr3_+_185762233
|
0.674
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chr8_+_123863169
|
0.644
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr4_+_191242540
|
0.628
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr11_-_310775
|
0.624
|
NM_021034
|
IFITM3
|
interferon induced transmembrane protein 3 (1-8U)
|
|
chr21_+_38550739
|
0.613
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr12_-_45759691
|
0.611
|
NM_001143668
NM_181847
|
AMIGO2
|
adhesion molecule with Ig-like domain 2
|
|
chr8_+_123863075
|
0.602
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr6_-_10520264
|
0.595
|
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr5_-_79322549
|
0.593
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chr17_-_35911340
|
0.593
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr17_-_73867748
|
0.592
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr16_-_17472238
|
0.591
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr11_-_124871258
|
0.586
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr5_-_58370693
|
0.571
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_+_191229360
|
0.570
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr19_-_56204615
|
0.562
|
NM_012315
|
KLK9
|
kallikrein-related peptidase 9
|
|
chr4_+_72271218
|
0.547
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr17_+_12633504
|
0.540
|
NM_014859
|
ARHGAP44
|
Rho GTPase activating protein 44
|
|
chr5_-_16989290
|
0.536
|
|
MYO10
|
myosin X
|
|
chr5_+_149960820
|
0.532
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr17_+_67628749
|
0.530
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr12_+_56406296
|
0.526
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr8_-_10550026
|
0.525
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr17_-_67628523
|
0.524
|
|
FLJ37644
|
hypothetical LOC400618
|
|
chr1_+_182622850
|
0.524
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr2_-_218575922
|
0.520
|
|
TNS1
|
tensin 1
|
|
chrX_-_106846253
|
0.516
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr10_+_135343649
|
0.516
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr4_+_191239247
|
0.513
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chrX_+_2680092
|
0.513
|
NM_001141919
NM_001141920
NM_175569
|
XGPY2
XG
|
Xg pseudogene, Y-linked 2
Xg blood group
|
|
chr1_-_197173159
|
0.513
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr21_+_38550533
|
0.510
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr8_-_134378630
|
0.508
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr12_-_102968044
|
0.507
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr5_+_49998677
|
0.505
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr1_+_15964045
|
0.496
|
NM_001024216
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr15_+_78775008
|
0.493
|
|
FAM108C1
|
family with sequence similarity 108, member C1
|
|
chr10_+_135333667
|
0.489
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_148621694
|
0.487
|
|
EDNRA
|
endothelin receptor type A
|
|
chr1_-_143750988
|
0.482
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr3_-_50324815
|
0.478
|
NM_153281
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr7_-_41709191
|
0.470
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr3_-_69674412
|
0.469
|
|
FRMD4B
|
FERM domain containing 4B
|
|
chr3_-_45162901
|
0.458
|
NM_022842
NM_178181
|
CDCP1
|
CUB domain containing protein 1
|
|
chr16_+_82959626
|
0.457
|
NM_014861
|
ATP2C2
|
ATPase, Ca++ transporting, type 2C, member 2
|
|
chr20_-_14266200
|
0.455
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr9_-_103289154
|
0.453
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr3_+_36396948
|
0.453
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr12_-_24606616
|
0.450
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr4_-_119493322
|
0.447
|
NM_003619
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin)
|
|
chr11_+_122258600
|
0.446
|
NM_024806
NM_199124
|
C11orf63
|
chromosome 11 open reading frame 63
|
|
chr8_+_123863224
|
0.443
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr19_+_702108
|
0.442
|
NM_173481
|
C19orf21
|
chromosome 19 open reading frame 21
|
|
chr1_+_182622751
|
0.440
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr17_-_17421050
|
0.435
|
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr9_-_21965131
|
0.435
|
NM_000077
NM_001195132
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr3_-_45162847
|
0.432
|
|
CDCP1
|
CUB domain containing protein 1
|
|
chr2_-_207738858
|
0.430
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr12_-_51200347
|
0.425
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr1_+_182622987
|
0.419
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr11_-_60475558
|
0.414
|
NM_016582
|
SLC15A3
|
solute carrier family 15, member 3
|
|
chr1_+_44171578
|
0.412
|
NM_001136215
NM_057090
NM_057091
|
ARTN
|
artemin
|
|
chr17_+_56884560
|
0.410
|
|
TBX4
|
T-box 4
|
|
chr1_+_36409277
|
0.406
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr12_+_45759634
|
0.405
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr10_+_135330357
|
0.404
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_148621567
|
0.403
|
|
EDNRA
|
endothelin receptor type A
|
|
chr5_-_146813343
|
0.403
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr11_+_93394025
|
0.400
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr8_-_145085616
|
0.394
|
NM_201384
|
PLEC
|
plectin
|
|
chr2_+_17923404
|
0.393
|
NM_002252
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3
|
|
chr1_-_26553168
|
0.393
|
NM_001039775
|
AIM1L
|
absent in melanoma 1-like
|
|
chrX_+_110226030
|
0.390
|
NM_002578
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr18_+_40513889
|
0.390
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr12_-_102967828
|
0.390
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr15_+_97010219
|
0.388
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr12_+_2032964
|
0.388
|
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr9_-_21964825
|
0.387
|
NM_058197
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr3_-_45162783
|
0.386
|
|
|
|
|
chr13_-_39075304
|
0.382
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr4_+_191232653
|
0.381
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr1_-_10779291
|
0.372
|
NM_001079843
NM_017766
|
CASZ1
|
castor zinc finger 1
|
|
chr17_+_74531873
|
0.372
|
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr17_+_72795567
|
0.371
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr1_-_93919789
|
0.371
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr19_-_50977586
|
0.371
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr9_+_36026900
|
0.370
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr11_-_110888273
|
0.370
|
NM_017589
|
BTG4
|
B-cell translocation gene 4
|
|
chr8_-_145122884
|
0.370
|
NM_000445
|
PLEC
|
plectin
|
|
chr15_-_68971665
|
0.369
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr9_+_90339835
|
0.366
|
NM_001161625
NM_145283
|
NXNL2
|
nucleoredoxin-like 2
|
|
chr14_+_64240902
|
0.366
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr15_+_39918302
|
0.366
|
NM_001114633
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic)
|
|
chr1_-_115682374
|
0.365
|
NM_002506
|
NGF
|
nerve growth factor (beta polypeptide)
|
|
chrX_-_62891730
|
0.363
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chrX_+_149280486
|
0.363
|
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr3_-_150858274
|
0.362
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr16_+_22732982
|
0.361
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr14_-_89155078
|
0.361
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr6_+_106640881
|
0.359
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr17_-_68100537
|
0.358
|
|
LOC100499467
|
hypothetical LOC100499467
|
|
chr1_+_182622809
|
0.357
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr22_+_44828338
|
0.357
|
|
|
|
|
chr8_+_128817658
|
0.355
|
|
MYC
|
v-myc myelocytomatosis viral oncogene homolog (avian)
|
|
chr7_-_27108826
|
0.354
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr11_+_66580861
|
0.352
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr16_-_73830025
|
0.352
|
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr17_+_7199178
|
0.350
|
NM_198154
|
TMEM95
|
transmembrane protein 95
|
|
chr17_-_72093529
|
0.348
|
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2
|
|
chr6_-_32929570
|
0.347
|
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr15_-_40236061
|
0.346
|
NM_213600
|
PLA2G4F
|
phospholipase A2, group IVF
|
|
chr12_-_113328271
|
0.345
|
NM_181486
|
TBX5
|
T-box 5
|
|
chr17_-_72093739
|
0.344
|
NM_006456
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2
|
|
chr22_+_38720881
|
0.341
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr14_+_92330383
|
0.341
|
NM_005113
|
GOLGA5
|
golgin A5
|
|
chr9_-_90456824
|
0.336
|
NM_001100111
|
LOC286238
|
hypothetical protein LOC286238
|
|
chrX_+_106956115
|
0.336
|
|
MID2
|
midline 2
|
|
chr4_+_148621518
|
0.335
|
NM_001166055
NM_001957
|
EDNRA
|
endothelin receptor type A
|
|
chr16_+_55580910
|
0.335
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr8_-_70909754
|
0.335
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr7_+_1543644
|
0.335
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr1_+_199884152
|
0.332
|
|
NAV1
|
neuron navigator 1
|
|
chr12_+_55896844
|
0.332
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chrX_-_106846684
|
0.332
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr15_-_38420414
|
0.332
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr1_+_178466064
|
0.332
|
NM_033343
|
LHX4
|
LIM homeobox 4
|
|
chr17_-_72093476
|
0.331
|
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2
|
|
chr17_+_74531820
|
0.330
|
NM_030968
NM_198594
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr8_-_57395207
|
0.330
|
|
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5
|
|
chr9_-_37566248
|
0.329
|
NM_012166
|
FBXO10
|
F-box protein 10
|
|
chr8_-_120033240
|
0.329
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr7_-_141293185
|
0.329
|
NM_013252
|
CLEC5A
|
C-type lectin domain family 5, member A
|
|
chr6_+_139055244
|
0.329
|
|
FLJ46906
|
hypothetical LOC441172
|
|
chr2_+_30307900
|
0.329
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr19_-_60573552
|
0.328
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr17_-_7996409
|
0.327
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chrX_-_62891667
|
0.325
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr11_+_129823667
|
0.323
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr15_+_75074793
|
0.322
|
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chrX_-_62891066
|
0.322
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr10_+_112247614
|
0.321
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr8_-_72918697
|
0.320
|
|
MSC
|
musculin
|
|
chr15_+_75074483
|
0.317
|
NM_003978
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr3_+_16901455
|
0.316
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr10_-_98021144
|
0.316
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr15_-_58706934
|
0.313
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr16_+_2071631
|
0.313
|
|
TSC2
|
tuberous sclerosis 2
|
|
chr4_+_191226073
|
0.310
|
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
|
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
|
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
|
|
chr4_-_8211335
|
0.310
|
NM_001130083
NM_001130084
NM_001130085
NM_001130086
NM_001130087
NM_001130088
NM_032432
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr19_-_50977592
|
0.309
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr14_+_64240996
|
0.309
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr20_+_48241334
|
0.308
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr4_-_1192749
|
0.305
|
NM_001199021
|
LOC100130872
SPON2
|
hypothetical LOC100130872
spondin 2, extracellular matrix protein
|
|
chr11_-_119104463
|
0.305
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr6_-_128883125
|
0.303
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr1_-_158091760
|
0.303
|
NM_001134233
|
C1orf204
|
chromosome 1 open reading frame 204
|
|
chr5_+_72452131
|
0.302
|
NM_001161342
NM_173490
|
TMEM171
|
transmembrane protein 171
|
|
chr3_+_89239363
|
0.301
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr21_+_33560540
|
0.301
|
NM_000628
|
IL10RB
|
interleukin 10 receptor, beta
|
|
chr10_-_73203202
|
0.299
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr18_-_45630154
|
0.298
|
|
MYO5B
|
myosin VB
|
|
chr14_-_49543987
|
0.297
|
NM_001012706
|
C14orf182
|
chromosome 14 open reading frame 182
|
|
chr1_+_204049053
|
0.296
|
|
|
|
|
chr6_-_32929390
|
0.295
|
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr10_+_88718167
|
0.293
|
NM_006829
|
C10orf116
|
chromosome 10 open reading frame 116
|
|
chr2_+_17585462
|
0.292
|
|
VSNL1
|
visinin-like 1
|
|
chr14_+_20608179
|
0.292
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr7_+_75977680
|
0.291
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr9_-_112840801
|
0.290
|
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr1_-_32036858
|
0.290
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr6_-_25019173
|
0.288
|
NM_014722
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr22_-_41583248
|
0.288
|
NM_001142293
NM_014570
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr3_+_11153778
|
0.288
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr14_+_63750611
|
0.287
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chrX_+_48915127
|
0.287
|
NM_002668
|
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched)
|
|
chr22_+_29848923
|
0.287
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr1_-_39180036
|
0.287
|
NM_017821
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila)
|
|
chr22_+_31527678
|
0.287
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|