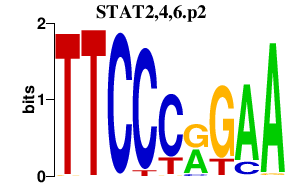
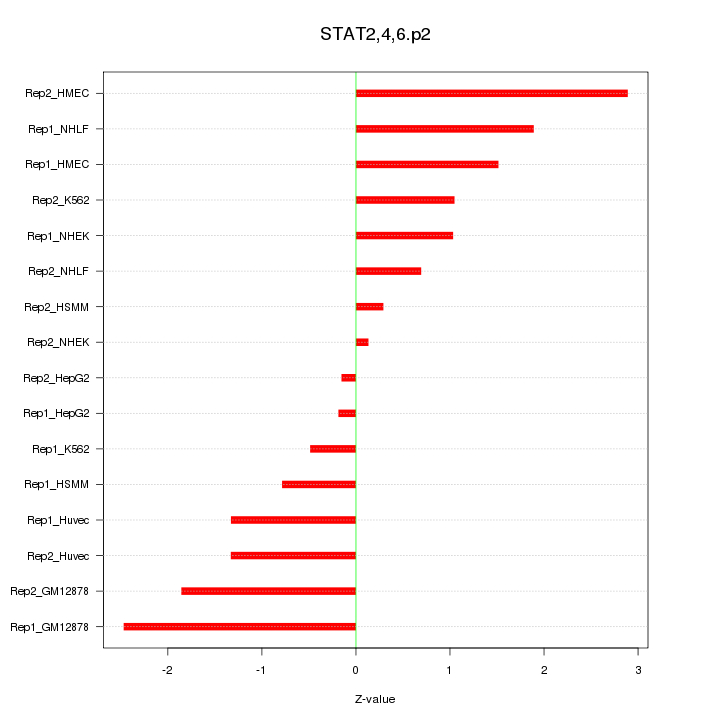
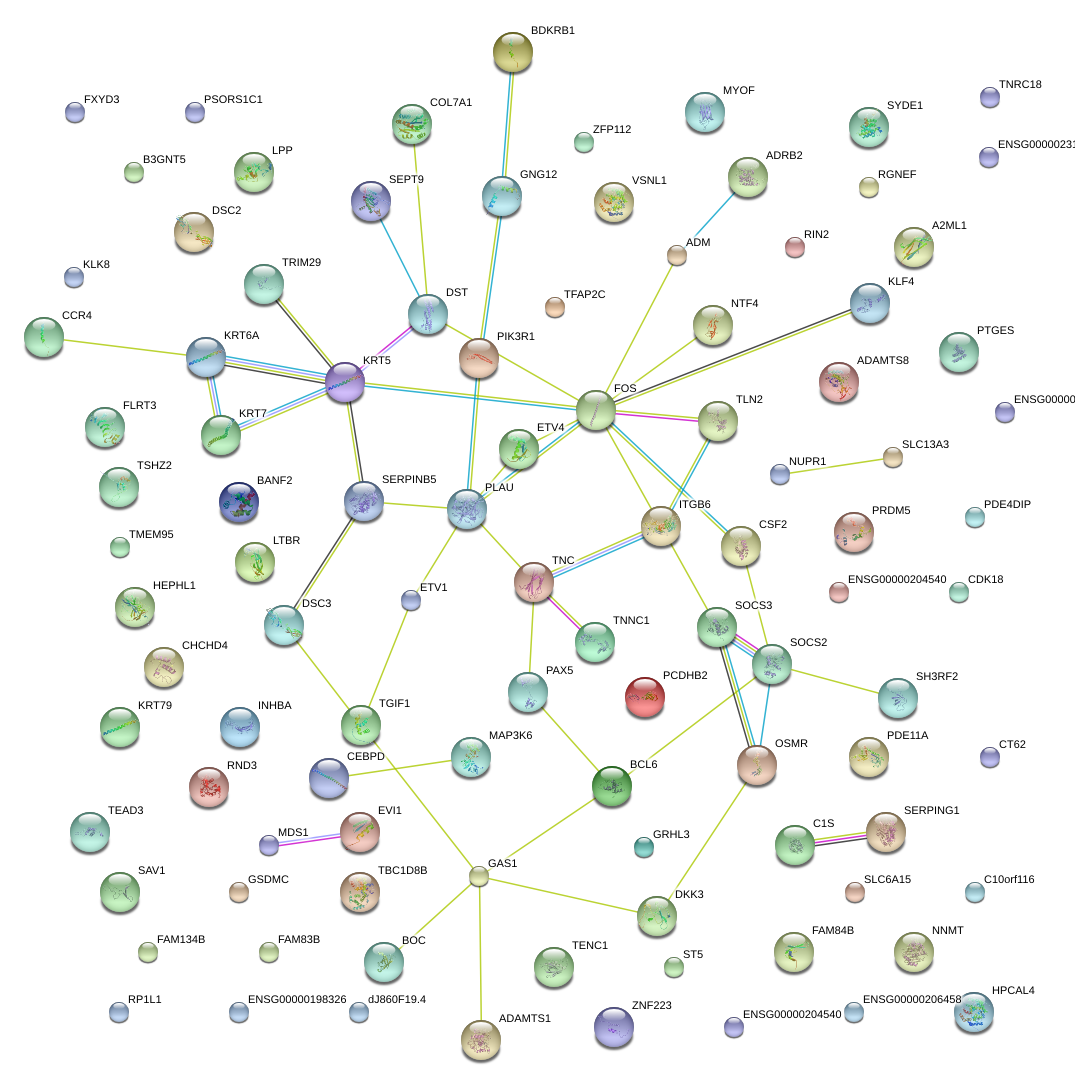
|
chr12_-_51200347
|
3.065
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr3_-_188945921
|
2.554
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr18_+_59295123
|
2.268
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr19_-_49552638
|
2.040
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr3_-_188946168
|
1.929
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr9_-_116920232
|
1.824
|
NM_002160
|
TNC
|
tenascin C
|
|
chr10_+_75343352
|
1.376
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr5_+_38882017
|
1.370
|
|
OSMR
|
oncostatin M receptor
|
|
chr1_-_68071635
|
1.366
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr2_-_178645718
|
1.309
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr9_-_88751923
|
1.248
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr19_-_54258935
|
1.221
|
NM_006179
|
NTF4
|
neurotrophin 4
|
|
chr7_-_5429702
|
1.205
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr3_+_189353502
|
1.198
|
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr12_-_92488767
|
1.151
|
|
|
|
|
chr12_+_92488932
|
1.136
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chrX_+_105932536
|
1.067
|
NM_017752
NM_198881
|
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain)
|
|
chr1_-_27565923
|
1.050
|
NM_004672
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6
|
|
chr5_+_145296313
|
1.041
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr6_+_54819527
|
1.028
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr12_+_51730101
|
1.012
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr5_+_38881719
|
0.991
|
NM_003999
|
OSMR
|
oncostatin M receptor
|
|
chr20_+_54637714
|
0.981
|
NM_003222
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
|
|
chr1_+_203740391
|
0.959
|
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr5_-_38881636
|
0.947
|
|
|
|
|
chr19_+_15079141
|
0.944
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr7_-_13992351
|
0.938
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr6_-_35572694
|
0.899
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr2_-_151052391
|
0.892
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr2_-_160764819
|
0.887
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr19_+_49248003
|
0.879
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chr9_-_131555111
|
0.873
|
NM_004878
|
PTGES
|
prostaglandin E synthase
|
|
chr19_+_40299058
|
0.858
|
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr8_-_130868315
|
0.857
|
NM_031415
|
GSDMC
|
gasdermin C
|
|
chr20_+_19903778
|
0.856
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr10_+_88718167
|
0.849
|
NM_006829
|
C10orf116
|
chromosome 10 open reading frame 116
|
|
chr3_-_48607596
|
0.843
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr11_-_11986704
|
0.839
|
NM_001018057
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr11_-_119504726
|
0.814
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr20_-_14266200
|
0.805
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr11_-_119513429
|
0.803
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr11_-_8788765
|
0.802
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr8_-_127639892
|
0.801
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr9_-_109291130
|
0.796
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr10_-_95231933
|
0.795
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr1_-_20682541
|
0.791
|
|
|
|
|
chr12_-_83830670
|
0.772
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr5_+_38881716
|
0.770
|
NM_001168355
|
OSMR
|
oncostatin M receptor
|
|
chr2_-_160765008
|
0.764
|
|
ITGB6
|
integrin, beta 6
|
|
chr2_+_17585287
|
0.756
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr12_-_83830710
|
0.747
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr12_-_83830700
|
0.746
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr18_-_26876622
|
0.733
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr1_+_24518501
|
0.720
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr20_+_51023159
|
0.714
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr3_+_114414064
|
0.711
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr7_-_41709191
|
0.695
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr12_+_51729239
|
0.683
|
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr8_-_10550026
|
0.682
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr19_-_56196611
|
0.681
|
NM_007196
NM_144505
NM_144506
NM_144507
|
KLK8
|
kallikrein-related peptidase 8
|
|
chr20_+_2743613
|
0.680
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr17_+_72827227
|
0.671
|
|
SEPT9
|
septin 9
|
|
chr16_-_28457728
|
0.666
|
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr1_-_27565412
|
0.665
|
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6
|
|
chr17_-_68100537
|
0.659
|
|
LOC100499467
|
hypothetical LOC100499467
|
|
chr1_+_68070573
|
0.655
|
|
|
|
|
chr5_-_16561936
|
0.650
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr5_+_131437383
|
0.649
|
NM_000758
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage)
|
|
chr19_+_40298571
|
0.642
|
NM_001136007
NM_001136008
NM_001136009
NM_001136010
NM_001136011
NM_001136012
NM_005971
NM_021910
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr11_+_113672378
|
0.637
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr14_+_95792299
|
0.637
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr5_+_67558217
|
0.633
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr5_+_73145098
|
0.632
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr17_+_7199178
|
0.631
|
NM_198154
|
TMEM95
|
transmembrane protein 95
|
|
chr1_+_24518398
|
0.622
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr17_-_38979178
|
0.617
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr9_+_676640
|
0.609
|
|
|
|
|
chr1_-_143706300
|
0.609
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr17_-_73867748
|
0.608
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr16_-_28457825
|
0.602
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr3_+_189354167
|
0.599
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr15_+_60838053
|
0.596
|
|
TLN2
|
talin 2
|
|
chr6_-_56615544
|
0.595
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr11_+_113672407
|
0.595
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr8_-_127639647
|
0.594
|
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr12_+_6363482
|
0.591
|
NM_002342
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3)
|
|
chr3_-_170346760
|
0.591
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr12_+_50913220
|
0.585
|
NM_005556
|
KRT7
|
keratin 7
|
|
chr11_+_10283171
|
0.579
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr6_+_31190505
|
0.579
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr20_+_51022352
|
0.570
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr17_+_72827185
|
0.569
|
NM_006640
|
SEPT9
|
septin 9
|
|
chr11_+_93394025
|
0.568
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr14_-_50204612
|
0.567
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr11_+_57122153
|
0.563
|
NM_001032295
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr20_+_17622319
|
0.561
|
NM_001159495
|
BANF2
|
barrier to autointegration factor 2
|
|
chr14_+_74815233
|
0.553
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr5_+_140454382
|
0.546
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr3_+_184453547
|
0.545
|
NM_032047
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
|
|
chr21_-_27139563
|
0.541
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr9_-_109291575
|
0.540
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr20_-_44746529
|
0.540
|
NM_001011554
NM_001193340
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr12_+_50913274
|
0.535
|
|
KRT7
|
keratin 7
|
|
chr4_-_122063118
|
0.535
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr3_-_14141371
|
0.533
|
NM_001098502
NM_144636
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr18_+_3437607
|
0.532
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr12_+_8866416
|
0.526
|
NM_144670
|
A2ML1
|
alpha-2-macroglobulin-like 1
|
|
chr3_-_14141247
|
0.524
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr12_+_7038240
|
0.523
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|
|
chr5_+_148186348
|
0.508
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr8_-_48813236
|
0.507
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr15_-_69194854
|
0.503
|
NM_001102658
|
CT62
|
cancer/testis antigen 62
|
|
chrX_+_73557809
|
0.498
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr4_-_89963177
|
0.497
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr7_+_134114957
|
0.495
|
|
CALD1
|
caldesmon 1
|
|
chr16_+_88515917
|
0.491
|
NM_001197181
|
TUBB3
|
tubulin, beta 3
|
|
chr4_+_128773529
|
0.489
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr19_+_702108
|
0.488
|
NM_173481
|
C19orf21
|
chromosome 19 open reading frame 21
|
|
chr3_+_184453907
|
0.487
|
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
|
|
chr2_-_189752575
|
0.486
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr1_-_143706265
|
0.483
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr11_+_131286513
|
0.481
|
|
NTM
|
neurotrimin
|
|
chr8_-_48813256
|
0.481
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr1_-_225572419
|
0.481
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr19_+_49023283
|
0.479
|
NM_181845
|
ZNF283
|
zinc finger protein 283
|
|
chr3_+_114192489
|
0.477
|
NM_014170
NM_138485
|
GTPBP8
|
GTP-binding protein 8 (putative)
|
|
chr11_+_129823667
|
0.477
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr22_+_29848923
|
0.475
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr3_-_152517204
|
0.473
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr7_+_134114912
|
0.472
|
|
CALD1
|
caldesmon 1
|
|
chr5_-_138238877
|
0.471
|
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr3_-_14141229
|
0.469
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr6_-_42218692
|
0.468
|
NM_001164446
|
C6orf132
|
chromosome 6 open reading frame 132
|
|
chr18_-_7107757
|
0.467
|
NM_005559
|
LAMA1
|
laminin, alpha 1
|
|
chr3_+_11153778
|
0.466
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr2_+_235525355
|
0.465
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr11_+_7574992
|
0.460
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr9_-_106730157
|
0.458
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr2_+_159533329
|
0.458
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr19_-_56214765
|
0.457
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr16_+_68516401
|
0.455
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr12_-_6356783
|
0.447
|
NM_001159575
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr9_-_106730256
|
0.446
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr3_-_14141293
|
0.446
|
|
CHCHD4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr5_-_138238891
|
0.440
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr1_-_143706196
|
0.438
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr14_-_50204771
|
0.438
|
NM_021818
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr10_+_74540215
|
0.437
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chr17_-_77884899
|
0.432
|
|
SECTM1
|
secreted and transmembrane 1
|
|
chr19_-_49644470
|
0.431
|
NM_014518
|
ZNF229
|
zinc finger protein 229
|
|
chr2_+_165858586
|
0.430
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr12_+_8958397
|
0.429
|
NM_004426
|
PHC1
|
polyhomeotic homolog 1 (Drosophila)
|
|
chr16_+_4485859
|
0.428
|
NM_001127205
|
HMOX2
|
heme oxygenase (decycling) 2
|
|
chr21_-_27139143
|
0.428
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr10_+_114699953
|
0.427
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr3_-_170347019
|
0.424
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr12_-_3472245
|
0.423
|
|
LOC100129223
|
hypothetical LOC100129223
|
|
chr1_-_199635240
|
0.423
|
NM_005558
|
LAD1
|
ladinin 1
|
|
chr5_-_138238953
|
0.422
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr9_-_109291866
|
0.421
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr15_-_65334022
|
0.420
|
NM_024666
|
AAGAB
|
alpha- and gamma-adaptin binding protein
|
|
chr3_+_137224266
|
0.417
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr9_+_129900637
|
0.416
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr20_-_48741481
|
0.416
|
|
|
|
|
chr17_+_71893014
|
0.412
|
|
SPHK1
|
sphingosine kinase 1
|
|
chr3_+_114192467
|
0.408
|
|
GTPBP8
|
GTP-binding protein 8 (putative)
|
|
chr14_-_50204791
|
0.407
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr20_-_42249146
|
0.405
|
|
JPH2
|
junctophilin 2
|
|
chr5_+_148631636
|
0.405
|
|
AFAP1L1
|
actin filament associated protein 1-like 1
|
|
chr1_+_170077234
|
0.404
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr5_+_148631588
|
0.403
|
NM_001146337
NM_152406
|
AFAP1L1
|
actin filament associated protein 1-like 1
|
|
chr5_-_102926349
|
0.402
|
NM_031438
|
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12
|
|
chr16_-_73842884
|
0.400
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr19_+_45973139
|
0.400
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr17_+_71892271
|
0.398
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr1_+_167603817
|
0.397
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr3_+_199171467
|
0.397
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chrX_-_134013644
|
0.397
|
NM_001078172
NM_001134321
|
FAM127B
|
family with sequence similarity 127, member B
|
|
chr3_-_139315011
|
0.396
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr6_-_29081015
|
0.396
|
NM_001010877
|
ZNF311
|
zinc finger protein 311
|
|
chr1_+_36409277
|
0.396
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr15_+_65334169
|
0.395
|
NM_001031715
NM_022784
|
IQCH
|
IQ motif containing H
|
|
chr1_+_35020425
|
0.395
|
NM_001005752
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr15_-_88256922
|
0.393
|
NM_001199058
NM_182616
|
C15orf38-AP3S2
C15orf38
|
C15orf38-AP3S2 readthrough
chromosome 15 open reading frame 38
|
|
chr7_+_89870647
|
0.392
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr9_-_129729518
|
0.390
|
NM_173492
|
PIP5KL1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1
|
|
chr22_+_27609889
|
0.388
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr17_-_77884926
|
0.387
|
|
SECTM1
|
secreted and transmembrane 1
|
|
chr1_-_116184846
|
0.387
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr4_+_77575276
|
0.385
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr10_+_71482362
|
0.384
|
NM_018649
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr1_-_151788341
|
0.383
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr11_-_11987115
|
0.382
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr17_+_71892781
|
0.381
|
NM_001142602
|
SPHK1
|
sphingosine kinase 1
|
|
chr15_+_68971835
|
0.380
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chrX_-_10761729
|
0.380
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr7_-_13997574
|
0.379
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr3_-_11585264
|
0.378
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr20_+_43470042
|
0.377
|
NM_001048225
NM_001048226
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr1_+_167604056
|
0.377
|
|
BLZF1
|
basic leucine zipper nuclear factor 1
|