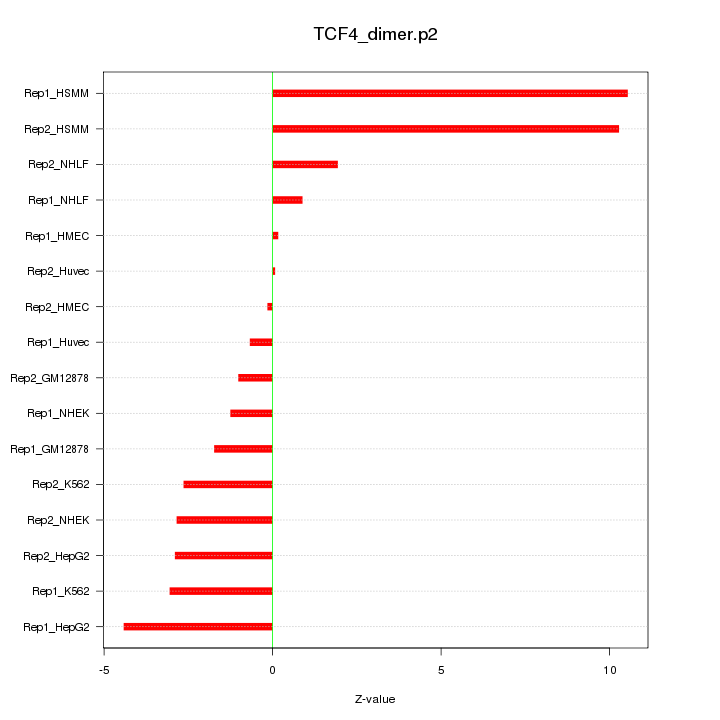
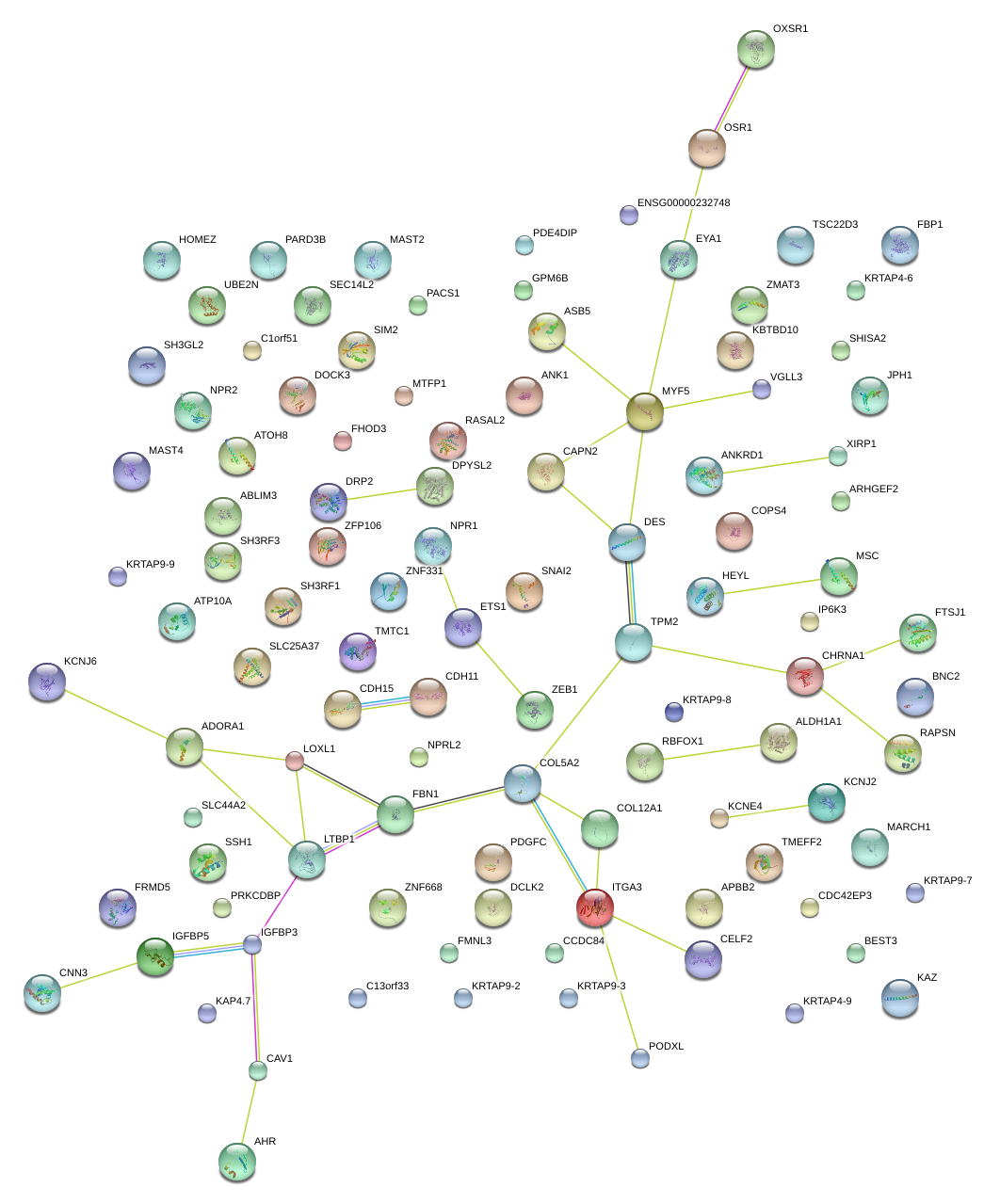
|
chr8_-_49996353
|
10.893
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr8_-_41641881
|
8.957
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr3_-_87122936
|
8.135
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr1_+_46151845
|
7.709
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr2_+_219991342
|
7.579
|
NM_001927
|
DES
|
desmin
|
|
chr11_-_47427263
|
7.530
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr4_-_177427252
|
6.988
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr1_+_201364050
|
6.916
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr1_+_181259317
|
6.873
|
|
|
|
|
chr2_-_175337386
|
6.810
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr9_-_35679799
|
6.767
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr15_+_72006015
|
6.753
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr9_-_35679902
|
6.450
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr19_+_10597167
|
6.354
|
NM_020428
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chrX_-_13866366
|
6.039
|
|
|
|
|
chr2_+_85834111
|
5.831
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr2_+_205118961
|
5.779
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr3_-_87122958
|
5.776
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr2_+_205118801
|
5.757
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr10_+_11100111
|
5.743
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_+_11100075
|
5.597
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr15_-_46725087
|
5.487
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr9_-_74757735
|
5.470
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr10_+_11099860
|
5.467
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_40911212
|
5.440
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr16_-_30983909
|
5.366
|
NM_001172669
NM_001172670
|
ZNF668
|
zinc finger protein 668
|
|
chr15_-_42274760
|
5.354
|
|
FRMD5
|
FERM domain containing 5
|
|
chr2_+_223625105
|
5.340
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr2_+_205118710
|
5.304
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr15_-_42274529
|
5.245
|
NM_032892
|
FRMD5
|
FERM domain containing 5
|
|
chr7_+_115953582
|
5.122
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr1_-_143643064
|
5.038
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr2_+_170074457
|
4.988
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr17_+_36493927
|
4.970
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr15_-_23659329
|
4.944
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr10_-_92671005
|
4.938
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr10_-_92670764
|
4.923
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr1_+_221955876
|
4.895
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr11_-_6298284
|
4.873
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr12_-_29827952
|
4.866
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chrX_-_13866451
|
4.704
|
|
GPM6B
|
glycoprotein M6B
|
|
chr1_-_39871226
|
4.700
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr12_-_107745455
|
4.595
|
NM_001161331
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr2_+_33213111
|
4.593
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr21_+_36993860
|
4.555
|
NM_005069
NM_009586
|
SIM2
|
single-minded homolog 2 (Drosophila)
|
|
chr8_-_72918697
|
4.533
|
|
MSC
|
musculin
|
|
chr7_-_45927263
|
4.437
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr6_-_75972473
|
4.372
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr13_+_30378311
|
4.368
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr2_+_56265260
|
4.343
|
|
|
|
|
chr8_-_75396016
|
4.327
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr12_+_79634820
|
4.309
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr1_-_95165089
|
4.281
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr1_-_154205675
|
4.275
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr6_-_75972286
|
4.251
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr8_-_72918938
|
4.234
|
|
MSC
|
musculin
|
|
chr15_-_40537002
|
4.203
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr5_+_148501178
|
4.146
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chrX_-_13866565
|
4.144
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr8_+_72918772
|
4.128
|
|
LOC100132891
|
hypothetical LOC100132891
|
|
chr5_+_66160359
|
4.116
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr21_-_38210531
|
4.105
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr8_+_72918889
|
4.103
|
|
|
|
|
chr15_+_72005839
|
4.100
|
NM_005576
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr15_+_72005847
|
4.055
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr11_-_127897098
|
4.052
|
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr9_-_96441409
|
4.003
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr22_+_29122932
|
3.985
|
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr6_-_33822659
|
3.940
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr8_-_72436832
|
3.919
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr17_+_45488356
|
3.877
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr16_+_87765661
|
3.860
|
NM_004933
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule)
|
|
chr1_+_176330254
|
3.778
|
|
RASAL2
|
RAS protein activator like 2
|
|
chr17_+_45488330
|
3.701
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr9_+_17568952
|
3.592
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chrX_+_48221323
|
3.572
|
NM_177434
|
FTSJ1
|
FtsJ homolog 1 (E. coli)
|
|
chr2_-_217267742
|
3.565
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr17_+_36642240
|
3.557
|
NM_031962
|
KRTAP9-9
KRTAP9-3
|
keratin associated protein 9-9
keratin associated protein 9-3
|
|
chr18_+_32131628
|
3.549
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr16_+_7322751
|
3.530
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chrX_-_106905545
|
3.524
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_148521852
|
3.510
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr19_+_58750305
|
3.503
|
NM_001079907
|
ZNF331
|
zinc finger protein 331
|
|
chr17_+_65677255
|
3.485
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr4_-_158111504
|
3.459
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr2_-_37752731
|
3.455
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr2_-_189752575
|
3.391
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr3_-_39209075
|
3.357
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr7_+_17304707
|
3.329
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr3_+_50687675
|
3.314
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr3_-_180272229
|
3.290
|
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr4_-_165523856
|
3.289
|
NM_001166373
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr10_-_31647925
|
3.266
|
|
LOC220930
|
hypothetical LOC220930
|
|
chr12_-_68379331
|
3.243
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr9_+_35782405
|
3.242
|
NM_003995
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B)
|
|
chr2_-_192767494
|
3.230
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chrX_+_100361588
|
3.179
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr10_+_31648103
|
3.179
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr16_-_63713466
|
3.178
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr7_-_130891861
|
3.167
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr13_-_25523164
|
3.162
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr2_+_109112428
|
3.160
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr11_+_118374071
|
3.159
|
|
CCDC84
|
coiled-coil domain containing 84
|
|
chr4_+_151218862
|
3.158
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr16_-_63713485
|
3.125
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr9_-_16717887
|
3.124
|
|
BNC2
|
basonuclin 2
|
|
chr4_+_159311353
|
3.121
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr4_-_158111916
|
3.105
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr4_-_158111995
|
3.090
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr2_-_19421852
|
3.086
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr21_+_29439768
|
3.067
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr11_+_131286513
|
3.038
|
|
NTM
|
neurotrimin
|
|
chr11_+_12088639
|
3.024
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr1_-_231498081
|
3.015
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr7_+_116099648
|
3.012
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr1_-_39877883
|
3.003
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr9_-_109291575
|
2.972
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr8_-_72431274
|
2.950
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr17_+_36636404
|
2.948
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr9_+_129893948
|
2.937
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr1_-_84237316
|
2.898
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr1_-_39877928
|
2.890
|
NM_014571
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chrX_-_106905657
|
2.883
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_-_143643324
|
2.874
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr8_-_122722674
|
2.861
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr5_-_136862130
|
2.850
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr6_+_129245978
|
2.844
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr12_-_50871953
|
2.834
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr9_-_109291130
|
2.810
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr7_+_93862137
|
2.785
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr10_+_31647425
|
2.772
|
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr15_-_72981595
|
2.770
|
|
C15orf17
|
chromosome 15 open reading frame 17
|
|
chrX_-_33267646
|
2.769
|
NM_000109
|
DMD
|
dystrophin
|
|
chr3_+_8750485
|
2.700
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr16_-_52876390
|
2.689
|
|
IRX3
|
iroquois homeobox 3
|
|
chr15_+_60838053
|
2.681
|
|
TLN2
|
talin 2
|
|
chr17_+_1905104
|
2.677
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr11_-_85108030
|
2.666
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr10_+_99248605
|
2.659
|
NM_024954
|
UBTD1
|
ubiquitin domain containing 1
|
|
chr10_-_75080784
|
2.648
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr5_+_159276317
|
2.645
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr13_-_98428244
|
2.642
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr1_-_191422346
|
2.642
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr1_-_160260210
|
2.641
|
NM_015441
|
OLFML2B
|
olfactomedin-like 2B
|
|
chr1_-_85703198
|
2.636
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr9_-_130980302
|
2.634
|
|
IER5L
|
immediate early response 5-like
|
|
chr5_-_136862317
|
2.607
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr10_-_99248355
|
2.607
|
NM_022362
|
MMS19
|
MMS19 nucleotide excision repair homolog (S. cerevisiae)
|
|
chr11_+_113436438
|
2.601
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr2_-_216944911
|
2.598
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr6_+_87921980
|
2.595
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr3_-_11598829
|
2.589
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr3_-_180272291
|
2.588
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr19_+_43447078
|
2.580
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr6_-_53638464
|
2.572
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr11_+_118374051
|
2.568
|
NM_198489
|
CCDC84
|
coiled-coil domain containing 84
|
|
chr16_-_63713382
|
2.567
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr1_+_238321794
|
2.561
|
NM_020066
|
FMN2
|
formin 2
|
|
chr19_-_50977485
|
2.541
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr3_+_106568243
|
2.537
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr10_-_116382089
|
2.530
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr7_-_127458231
|
2.511
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr9_+_34980266
|
2.507
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr12_-_16650687
|
2.500
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr21_+_36429079
|
2.497
|
NM_001236
|
CBR3
|
carbonyl reductase 3
|
|
chr6_-_139737042
|
2.476
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr19_+_43446937
|
2.471
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr11_-_8695974
|
2.461
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr1_-_153107084
|
2.450
|
|
|
|
|
chr8_+_37773991
|
2.447
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chrX_+_12066505
|
2.436
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr17_+_58058774
|
2.422
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr3_+_106568334
|
2.409
|
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr4_+_3341521
|
2.406
|
NM_198227
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr17_-_15495534
|
2.405
|
|
TRIM16
|
tripartite motif containing 16
|
|
chr5_-_176832476
|
2.399
|
NM_080881
|
DBN1
|
drebrin 1
|
|
chr4_-_75938710
|
2.398
|
NM_001729
|
BTC
|
betacellulin
|
|
chr9_-_130980359
|
2.391
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr2_+_238712101
|
2.375
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr5_-_58370693
|
2.375
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_16650697
|
2.374
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr9_-_88751923
|
2.370
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr15_-_53822468
|
2.332
|
NM_173814
|
PRTG
|
protogenin
|
|
chr1_+_31658549
|
2.326
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr5_+_102229612
|
2.325
|
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr17_-_36559579
|
2.322
|
NM_033188
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr5_-_151046680
|
2.322
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr9_+_71848555
|
2.318
|
|
MAMDC2
|
MAM domain containing 2
|
|
chr7_+_93861808
|
2.312
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr9_+_34980715
|
2.305
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr1_-_199613427
|
2.301
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr14_-_88087575
|
2.300
|
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr2_-_192419896
|
2.300
|
|
SDPR
|
serum deprivation response
|
|
chr13_-_35603513
|
2.289
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr8_+_32525269
|
2.280
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr9_-_116920232
|
2.262
|
NM_002160
|
TNC
|
tenascin C
|
|
chr11_-_85107569
|
2.262
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr2_-_227371698
|
2.262
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr5_+_102229425
|
2.255
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr11_-_118739838
|
2.255
|
|
USP2
|
ubiquitin specific peptidase 2
|