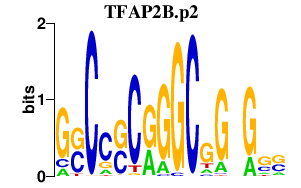
|
chr18_-_5533967
|
6.173
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr19_+_39664706
|
4.813
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr2_-_127580975
|
4.465
|
NM_004305
NM_139343
NM_139344
NM_139345
NM_139346
NM_139347
NM_139348
NM_139349
NM_139350
NM_139351
|
BIN1
|
bridging integrator 1
|
|
chr7_-_139987044
|
4.424
|
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr20_+_56701224
|
4.122
|
NM_024663
|
NPEPL1
|
aminopeptidase-like 1
|
|
chr5_+_10617431
|
3.846
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr11_+_93917214
|
3.474
|
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chrX_+_9943793
|
3.458
|
NM_015691
|
WWC3
|
WWC family member 3
|
|
chr2_-_160972606
|
3.419
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr3_-_130807865
|
3.396
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr16_-_79395379
|
3.382
|
NM_152342
|
CDYL2
|
chromodomain protein, Y-like 2
|
|
chr19_-_1518875
|
3.288
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr7_+_1536847
|
3.264
|
NM_002360
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr9_-_38059082
|
3.067
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr17_+_78630793
|
3.039
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr22_-_48607449
|
2.991
|
|
BRD1
|
bromodomain containing 1
|
|
chr14_+_105012106
|
2.905
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr17_-_74432800
|
2.799
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr1_+_148388870
|
2.767
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr19_+_16296632
|
2.702
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr13_+_110565587
|
2.661
|
NM_001113511
NM_001113512
NM_145735
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr1_+_148389039
|
2.643
|
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr1_-_95165089
|
2.600
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr11_+_1367665
|
2.589
|
NM_003957
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr4_+_89147821
|
2.588
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr2_+_241586927
|
2.584
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr17_-_53387493
|
2.566
|
|
CUEDC1
|
CUE domain containing 1
|
|
chr20_+_8060762
|
2.565
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr9_+_127549437
|
2.549
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr14_-_29466562
|
2.544
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr13_-_27967215
|
2.515
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr16_+_88169612
|
2.469
|
NM_014427
NM_153636
|
CPNE7
|
copine VII
|
|
chr20_-_4177519
|
2.463
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr12_+_26003218
|
2.444
|
NM_001164748
NM_007211
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr19_+_18391423
|
2.440
|
|
|
|
|
chr17_-_74433048
|
2.409
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr4_+_14614462
|
2.400
|
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr4_-_7991992
|
2.395
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr1_+_148388719
|
2.385
|
NM_016274
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr17_+_56831951
|
2.360
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr19_+_16296734
|
2.352
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr11_-_110087982
|
2.326
|
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr22_-_27405708
|
2.319
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr6_+_1335067
|
2.307
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr22_-_41446740
|
2.303
|
NM_017436
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr15_-_53667800
|
2.303
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr1_+_199884152
|
2.272
|
|
NAV1
|
neuron navigator 1
|
|
chr19_+_1358664
|
2.266
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr1_+_9275542
|
2.241
|
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr1_-_1465602
|
2.224
|
NM_001114748
|
C1orf70
|
chromosome 1 open reading frame 70
|
|
chr10_-_725522
|
2.203
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr1_-_29321599
|
2.180
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chr17_-_74432671
|
2.175
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr14_+_99329198
|
2.174
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr12_-_105165805
|
2.159
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr8_+_17058170
|
2.128
|
NM_016353
|
ZDHHC2
|
zinc finger, DHHC-type containing 2
|
|
chr12_+_63958949
|
2.123
|
NM_001193461
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr7_+_55922623
|
2.119
|
|
ZNF713
|
zinc finger protein 713
|
|
chr4_+_151218862
|
2.116
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr4_+_6835363
|
2.099
|
|
KIAA0232
|
KIAA0232
|
|
chr10_+_123913094
|
2.097
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr5_+_14196287
|
2.094
|
NM_007118
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr1_+_179148935
|
2.094
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr11_-_110088330
|
2.094
|
NM_020809
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr2_+_109112428
|
2.093
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr5_+_98132898
|
2.087
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr2_-_239987629
|
2.078
|
|
HDAC4
|
histone deacetylase 4
|
|
chr17_-_19711580
|
2.042
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr18_-_24011349
|
2.037
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr9_-_38059207
|
2.035
|
NM_003028
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr7_-_50828608
|
2.033
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr2_-_133144094
|
1.999
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr4_-_177950658
|
1.979
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr1_+_22909916
|
1.969
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr16_+_88517187
|
1.967
|
NM_006086
|
TUBB3
|
tubulin, beta 3
|
|
chr10_+_123912930
|
1.962
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr5_+_34692239
|
1.961
|
|
RAI14
|
retinoic acid induced 14
|
|
chr13_+_97593411
|
1.961
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr1_-_95165037
|
1.956
|
|
CNN3
|
calponin 3, acidic
|
|
chr15_-_62854749
|
1.947
|
NM_194272
|
RBPMS2
|
RNA binding protein with multiple splicing 2
|
|
chr10_+_11100111
|
1.938
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_2233504
|
1.938
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr1_+_199884072
|
1.935
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr14_+_23908141
|
1.935
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr17_+_56832492
|
1.933
|
|
TBX2
|
T-box 2
|
|
chr6_-_117193505
|
1.909
|
NM_001085480
|
FAM162B
|
family with sequence similarity 162, member B
|
|
chr12_+_50271283
|
1.907
|
NM_014191
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr14_+_23907065
|
1.862
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr17_+_17525571
|
1.858
|
|
|
|
|
chr7_-_150576634
|
1.851
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr7_-_150576660
|
1.851
|
NM_001003801
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr21_+_45649429
|
1.835
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr6_+_72055229
|
1.820
|
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr13_+_30378311
|
1.819
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr19_+_18391211
|
1.807
|
NM_001009998
NM_032627
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr4_-_1232882
|
1.778
|
NM_001012614
NM_001328
|
CTBP1
|
C-terminal binding protein 1
|
|
chr21_+_45649719
|
1.775
|
|
|
|
|
chr7_+_90063543
|
1.771
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr12_+_47498778
|
1.762
|
NM_000725
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr10_+_11100075
|
1.750
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr9_+_138680064
|
1.730
|
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr17_+_72880967
|
1.727
|
|
SEPT9
|
septin 9
|
|
chr2_-_239987557
|
1.717
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr17_+_1906262
|
1.716
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr1_-_85703321
|
1.712
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr19_-_1188840
|
1.708
|
NM_152769
|
C19orf26
|
chromosome 19 open reading frame 26
|
|
chr7_+_18093068
|
1.707
|
|
HDAC9
|
histone deacetylase 9
|
|
chr3_-_126257425
|
1.707
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr19_+_1358531
|
1.702
|
NM_018959
NM_170711
|
DAZAP1
|
DAZ associated protein 1
|
|
chr17_-_8006979
|
1.702
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr6_-_112682038
|
1.701
|
|
LAMA4
|
laminin, alpha 4
|
|
chr4_+_72271218
|
1.697
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr22_-_45512708
|
1.692
|
|
CERK
|
ceramide kinase
|
|
chr11_+_12355600
|
1.692
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr4_+_1842947
|
1.690
|
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr11_-_77806500
|
1.680
|
NM_080491
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr19_-_8581559
|
1.679
|
NM_030957
|
ADAMTS10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10
|
|
chr5_+_34692352
|
1.677
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr6_+_3696224
|
1.676
|
|
|
|
|
chr11_-_77806413
|
1.673
|
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr14_+_58174489
|
1.672
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr8_-_93184629
|
1.671
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr6_-_169865990
|
1.664
|
|
PHF10
|
PHD finger protein 10
|
|
chr14_+_54102518
|
1.650
|
|
LOC644925
|
hypothetical LOC644925
|
|
chr2_-_110769670
|
1.650
|
NM_001164463
NM_005054
|
RGPD8
RGPD5
|
RANBP2-like and GRIP domain containing 8
RANBP2-like and GRIP domain containing 5
|
|
chr6_-_3697244
|
1.646
|
NM_183373
|
C6orf145
|
chromosome 6 open reading frame 145
|
|
chr2_-_197883739
|
1.637
|
NM_001195144
NM_153697
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr22_-_49041786
|
1.635
|
|
MAPK12
|
mitogen-activated protein kinase 12
|
|
chr16_+_11669736
|
1.635
|
NM_003498
|
SNN
|
stannin
|
|
chr13_+_109757593
|
1.634
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr4_-_2233606
|
1.634
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr6_+_72055197
|
1.631
|
NM_024576
|
OGFRL1
|
opioid growth factor receptor-like 1
|
|
chr12_+_120132319
|
1.621
|
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4
|
|
chr10_-_30388439
|
1.601
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr15_-_53822468
|
1.598
|
NM_173814
|
PRTG
|
protogenin
|
|
chr3_+_160964574
|
1.594
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr15_-_23659329
|
1.593
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr8_+_107351519
|
1.587
|
NM_001198533
NM_018002
|
LOC346887
OXR1
|
similar to solute carrier family 16 (monocarboxylic acid transporters), member 14
oxidation resistance 1
|
|
chr4_-_1232742
|
1.579
|
|
CTBP1
|
C-terminal binding protein 1
|
|
chr15_+_29406335
|
1.576
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr22_-_48607199
|
1.575
|
|
BRD1
|
bromodomain containing 1
|
|
chr17_+_75808685
|
1.563
|
NM_001166347
NM_001166348
NM_001166349
NM_173626
|
SLC26A11
|
solute carrier family 26, member 11
|
|
chr18_+_12648191
|
1.561
|
|
|
|
|
chr1_-_85703198
|
1.560
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr6_-_132764257
|
1.551
|
NM_015529
|
MOXD1
|
monooxygenase, DBH-like 1
|
|
chr10_+_122206455
|
1.550
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr3_+_61522922
|
1.548
|
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr4_+_3264509
|
1.547
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr5_-_176833197
|
1.543
|
|
DBN1
|
drebrin 1
|
|
chr20_-_38751289
|
1.540
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr4_+_6835388
|
1.536
|
|
KIAA0232
|
KIAA0232
|
|
chr5_-_178704934
|
1.536
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr13_+_97593469
|
1.530
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr14_+_104785898
|
1.529
|
NM_033271
|
BTBD6
|
BTB (POZ) domain containing 6
|
|
chr21_-_43671355
|
1.525
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr15_+_45797977
|
1.523
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr2_+_176702667
|
1.518
|
NM_019558
|
HOXD8
|
homeobox D8
|
|
chr18_+_18969724
|
1.516
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr9_-_129701644
|
1.504
|
NM_013443
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr9_+_136673534
|
1.499
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr9_-_25668230
|
1.492
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr14_+_76297885
|
1.490
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr11_+_93916653
|
1.488
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr12_+_63958689
|
1.482
|
NM_001031679
NM_001193460
NM_198080
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr11_+_7997366
|
1.473
|
|
TUB
|
tubby homolog (mouse)
|
|
chr8_+_30361485
|
1.467
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr2_+_42128521
|
1.464
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr10_+_104394185
|
1.463
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr6_-_169866011
|
1.463
|
NM_018288
NM_133325
|
PHF10
|
PHD finger protein 10
|
|
chr1_+_181259317
|
1.461
|
|
|
|
|
chr22_-_41447214
|
1.461
|
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr1_-_40009513
|
1.459
|
NM_022120
|
OXCT2
|
3-oxoacid CoA transferase 2
|
|
chr13_-_112392051
|
1.452
|
|
LOC440149
|
hypothetical LOC440149
|
|
chr20_-_3944035
|
1.451
|
NM_007219
NM_001134337
NM_001134338
|
RNF24
|
ring finger protein 24
|
|
chr5_-_158459006
|
1.450
|
|
EBF1
|
early B-cell factor 1
|
|
chr19_+_1358750
|
1.449
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr17_-_40263124
|
1.449
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr2_+_205118801
|
1.447
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr20_-_46877811
|
1.445
|
NM_020820
|
PREX1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1
|
|
chr11_-_110088097
|
1.445
|
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr4_-_141896920
|
1.445
|
NM_015130
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain)
|
|
chr19_+_50196526
|
1.444
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr8_+_38763911
|
1.440
|
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr16_-_1404635
|
1.437
|
NM_001037125
NM_001193388
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr11_+_2355119
|
1.434
|
NM_004356
|
CD81
|
CD81 molecule
|
|
chr19_-_4016467
|
1.434
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr17_-_1029758
|
1.423
|
NM_021962
|
ABR
|
active BCR-related gene
|
|
chr15_-_68933481
|
1.420
|
NM_018357
NM_197958
|
LARP6
|
La ribonucleoprotein domain family, member 6
|
|
chr19_+_1358604
|
1.419
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr22_+_38075703
|
1.416
|
NM_004711
NM_145731
|
SYNGR1
|
synaptogyrin 1
|
|
chr16_+_68233
|
1.416
|
|
MPG
|
N-methylpurine-DNA glycosylase
|
|
chr13_+_97593711
|
1.413
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr1_-_54644269
|
1.410
|
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr2_+_149895274
|
1.398
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr22_+_36286350
|
1.391
|
NM_152243
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1
|
|
chr14_+_76297536
|
1.386
|
|
|
|
|
chr10_+_128583984
|
1.384
|
NM_001380
|
DOCK1
|
dedicator of cytokinesis 1
|
|
chr10_+_11099860
|
1.381
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_140696740
|
1.380
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr2_-_144991386
|
1.375
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|