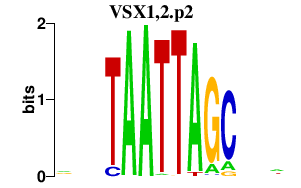
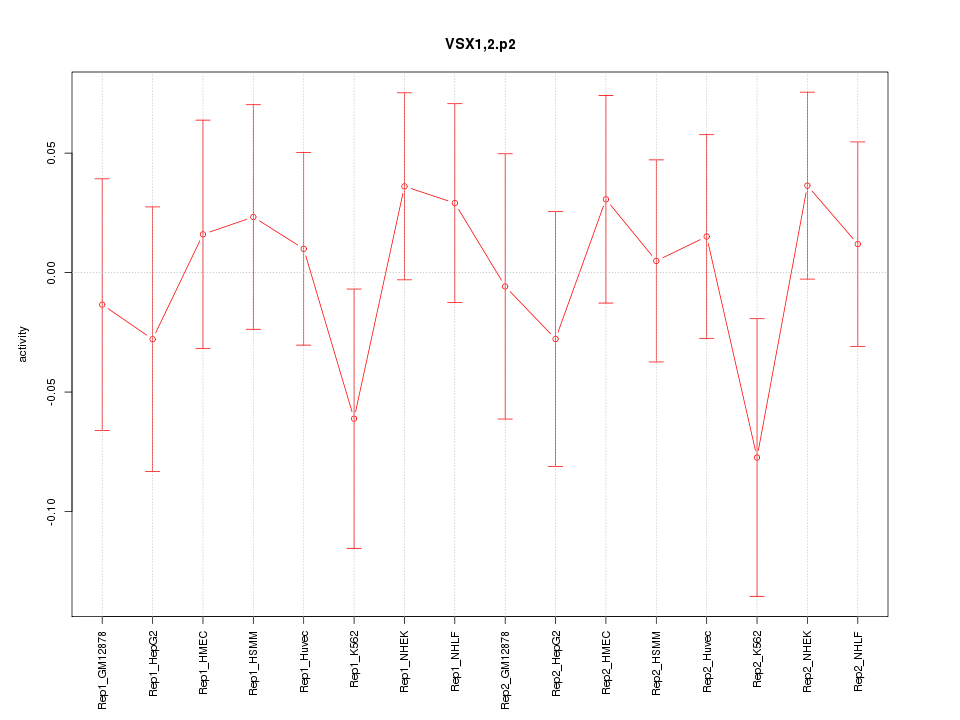
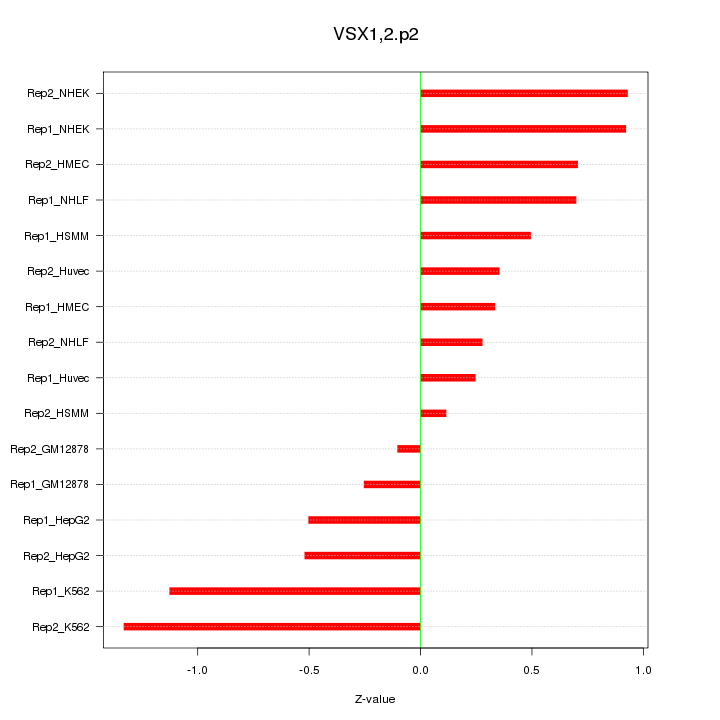
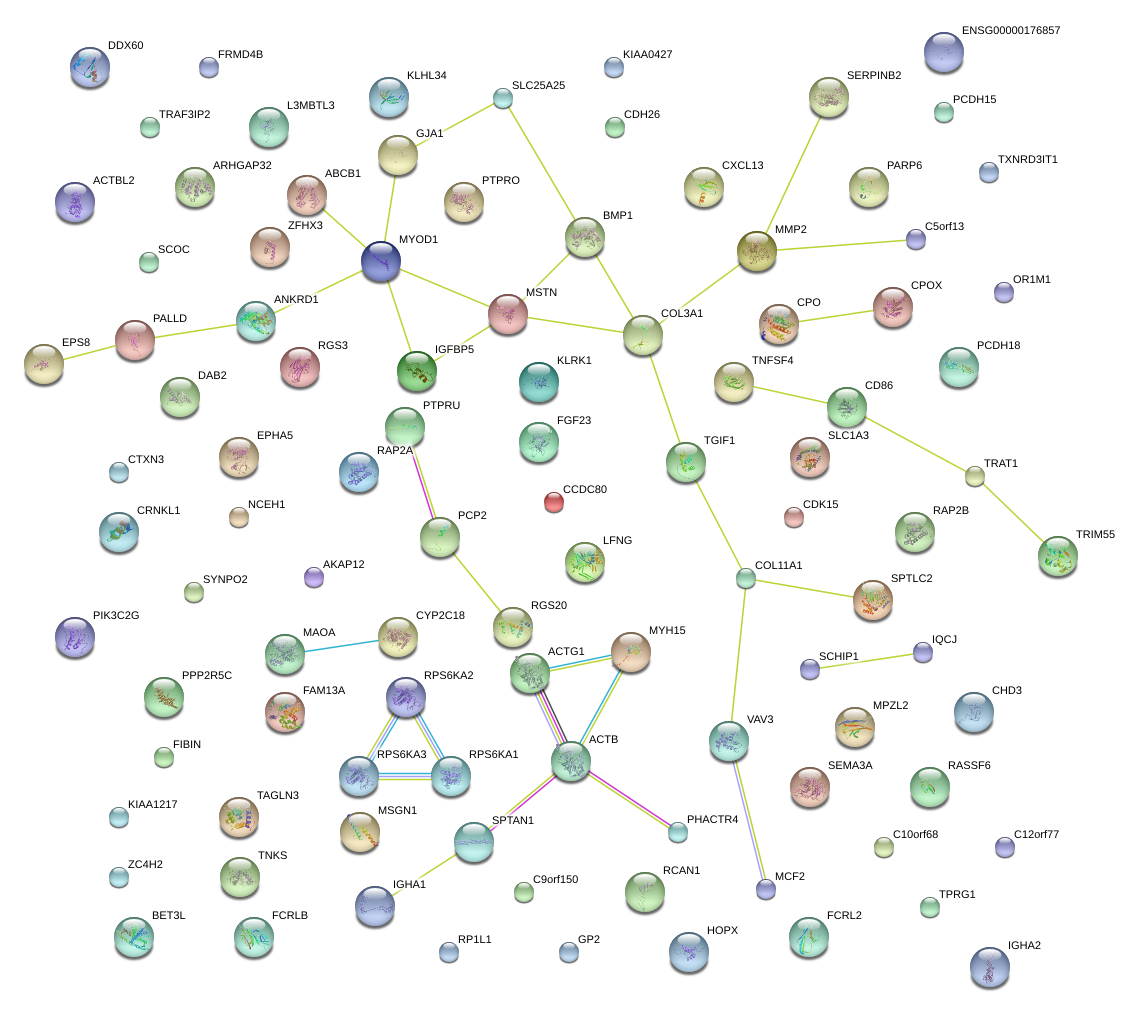
|
chr3_+_190147633
|
1.816
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr6_-_166960496
|
1.804
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr9_+_12764988
|
1.609
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chrX_+_43400385
|
1.579
|
|
MAOA
|
monoamine oxidase A
|
|
chrX_+_43400475
|
1.531
|
|
MAOA
|
monoamine oxidase A
|
|
chr21_-_34820916
|
1.529
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chrX_+_43400350
|
1.490
|
NM_000240
|
MAOA
|
monoamine oxidase A
|
|
chr8_-_10550026
|
1.236
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr6_-_166960690
|
1.225
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr2_+_17861266
|
1.184
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr4_-_169476391
|
1.178
|
NM_017631
|
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60
|
|
chr5_+_127012611
|
1.029
|
NM_001048252
|
CTXN3
|
cortexin 3
|
|
chr11_-_117640407
|
0.950
|
NM_005797
NM_144765
|
MPZL2
|
myelin protein zero-like 2
|
|
chr4_-_138673014
|
0.932
|
|
PCDH18
|
protocadherin 18
|
|
chr6_+_130381408
|
0.919
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr6_-_166960649
|
0.874
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr4_-_138673078
|
0.814
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr16_+_54072969
|
0.708
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr16_-_71650034
|
0.685
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr4_+_169654742
|
0.662
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr11_-_128399218
|
0.644
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr10_+_24795446
|
0.607
|
|
KIAA1217
|
KIAA1217
|
|
chr10_-_56230953
|
0.606
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr18_+_44319365
|
0.552
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr4_+_120029426
|
0.516
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr7_+_2524022
|
0.506
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr7_-_83662152
|
0.493
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr6_-_112033752
|
0.462
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr2_-_217268492
|
0.446
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr11_-_128567302
|
0.414
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr19_-_41514390
|
0.405
|
|
|
|
|
chr6_+_121798443
|
0.389
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr3_+_113200696
|
0.378
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr19_+_9064854
|
0.335
|
NM_001004456
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1
|
|
chr5_+_36642424
|
0.320
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr18_+_59705918
|
0.315
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr3_-_127810087
|
0.314
|
NM_001039783
|
TXNRD3IT1
|
thioredoxin reductase 3 intronic transcript 1
|
|
chr10_-_92671005
|
0.312
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr12_+_18305740
|
0.288
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr9_+_115396020
|
0.283
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_-_171441234
|
0.279
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr8_+_67201684
|
0.259
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr11_+_17697685
|
0.236
|
NM_002478
|
MYOD1
|
myogenic differentiation 1
|
|
chr14_-_77152862
|
0.228
|
NM_004863
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2
|
|
chr3_-_173911467
|
0.226
|
NM_001146276
NM_001146277
NM_001146278
NM_020792
|
NCEH1
|
neutral cholesterol ester hydrolase 1
|
|
chr18_+_3437607
|
0.218
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr16_-_20246309
|
0.215
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chrX_-_21586337
|
0.201
|
NM_153270
|
KLHL34
|
kelch-like 34 (Drosophila)
|
|
chr12_-_15706340
|
0.198
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr10_+_96433240
|
0.195
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr3_+_123256902
|
0.180
|
NM_175862
|
CD86
|
CD86 molecule
|
|
chr6_+_151688327
|
0.178
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr1_+_28637247
|
0.175
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr1_-_103269356
|
0.173
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr2_+_189547358
|
0.171
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr11_-_117640078
|
0.170
|
|
|
|
|
chrX_-_64112993
|
0.168
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr20_+_58004811
|
0.152
|
NM_021810
|
CDH26
|
cadherin 26
|
|
chrX_+_55951280
|
0.144
|
|
|
|
|
chr1_-_108032645
|
0.144
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr2_+_202379442
|
0.139
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr4_-_57242492
|
0.137
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr11_-_117640166
|
0.137
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr6_-_112034166
|
0.131
|
NM_001164281
NM_147686
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr4_-_66218777
|
0.128
|
|
EPHA5
|
EPH receptor A5
|
|
chr8_+_9450832
|
0.120
|
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr11_+_26972244
|
0.113
|
|
FIBIN
|
fin bud initiation factor homolog (zebrafish)
|
|
chr3_-_113842255
|
0.107
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_+_154362726
|
0.106
|
|
RAP2B
|
RAP2B, member of RAS oncogene family
|
|
chr8_+_9450800
|
0.104
|
|
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr17_+_7732759
|
0.099
|
NM_001005273
NM_005852
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr12_+_15590552
|
0.098
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr4_+_78651929
|
0.097
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr9_+_130428519
|
0.097
|
|
SPTAN1
|
spectrin, alpha, non-erythrocytic 1 (alpha-fodrin)
|
|
chr5_-_111119846
|
0.096
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_-_190635566
|
0.095
|
NM_005259
|
MSTN
|
myostatin
|
|
chr3_-_109730858
|
0.094
|
NM_014981
|
MYH15
|
myosin, heavy chain 15
|
|
chr15_-_70350609
|
0.093
|
NM_020214
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6
|
|
chr3_+_110338250
|
0.092
|
|
FLJ22763
|
hypothetical LOC401081
|
|
chr7_-_87180499
|
0.090
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr5_-_56814310
|
0.089
|
NM_001017992
|
ACTBL2
|
actin, beta-like 2
|
|
chr8_+_54926920
|
0.086
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr12_-_4358968
|
0.083
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr4_+_141397889
|
0.083
|
NM_032547
|
SCOC
|
short coiled-coil protein
|
|
chr5_-_39430180
|
0.081
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr6_-_116973465
|
0.080
|
NM_001139444
|
BET3L
|
BET3 like (S. cerevisiae)
|
|
chr4_-_74704964
|
0.079
|
NM_177532
NM_201431
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr3_+_113200275
|
0.078
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr12_-_25041639
|
0.078
|
NM_001101339
|
C12orf77
|
chromosome 12 open reading frame 77
|
|
chr14_+_101345878
|
0.076
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr19_-_7604569
|
0.075
|
NM_174895
|
PCP2
|
Purkinje cell protein 2
|
|
chr10_+_32896656
|
0.074
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chrX_-_138617994
|
0.074
|
NM_001099855
NM_001171876
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr11_+_26972203
|
0.071
|
NM_203371
|
FIBIN
|
fin bud initiation factor homolog (zebrafish)
|
|
chr3_+_110024234
|
0.068
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr9_+_129893948
|
0.066
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr20_-_19981005
|
0.066
|
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr1_-_156013487
|
0.065
|
NM_030764
|
FCRL2
|
Fc receptor-like 2
|
|
chr2_+_207512522
|
0.060
|
NM_173077
|
CPO
|
carboxypeptidase O
|
|
chr3_-_69381995
|
0.059
|
|
FRMD4B
|
FERM domain containing 4B
|
|
chr19_+_4720144
|
0.058
|
|
C19orf30
|
chromosome 19 open reading frame 30
|
|
chr4_-_90197140
|
0.058
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr3_+_161040343
|
0.054
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr14_-_106249999
|
0.054
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chrX_+_107174855
|
0.052
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr13_+_110637173
|
0.049
|
NM_001113513
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr17_+_12510030
|
0.049
|
|
MYOCD
|
myocardin
|
|
chr3_+_99350859
|
0.049
|
NM_001005514
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14
|
|
chr12_-_55979413
|
0.049
|
|
|
|
|
chr3_-_168674502
|
0.048
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr1_-_156635879
|
0.048
|
NM_001004475
|
OR10T2
|
olfactory receptor, family 10, subfamily T, member 2
|
|
chr4_+_5577783
|
0.047
|
NM_005750
|
C4orf6
|
chromosome 4 open reading frame 6
|
|
chr12_+_15590811
|
0.046
|
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr22_+_39789945
|
0.044
|
|
|
|
|
chr12_-_14740694
|
0.043
|
NM_004963
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor)
|
|
chr13_-_33148899
|
0.042
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_-_34948758
|
0.042
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr3_-_48634192
|
0.041
|
NM_001008269
|
TMEM89
|
transmembrane protein 89
|
|
chr5_-_9683462
|
0.041
|
NM_019599
|
TAS2R1
|
taste receptor, type 2, member 1
|
|
chr17_+_18027816
|
0.040
|
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr9_+_66197108
|
0.040
|
|
|
|
|
chr2_-_101291483
|
0.038
|
NM_173647
|
RNF149
|
ring finger protein 149
|
|
chr1_-_150653351
|
0.038
|
NM_016190
|
CRNN
|
cornulin
|
|
chr15_-_40236061
|
0.036
|
NM_213600
|
PLA2G4F
|
phospholipase A2, group IVF
|
|
chr3_+_196933422
|
0.035
|
NM_152673
|
MUC20
|
mucin 20, cell surface associated
|
|
chr8_-_134156437
|
0.033
|
|
SLA
|
Src-like-adaptor
|
|
chr9_-_21067804
|
0.033
|
NM_002176
|
IFNB1
|
interferon, beta 1, fibroblast
|
|
chr20_+_30161173
|
0.030
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr10_+_83627422
|
0.030
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr11_+_65740789
|
0.030
|
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr2_-_218739891
|
0.028
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr1_-_45862225
|
0.026
|
NM_001114938
NM_001190182
|
CCDC17
|
coiled-coil domain containing 17
|
|
chr11_+_89403921
|
0.026
|
NM_001195234
|
TRIM49
TRIM49L2
|
tripartite motif containing 49
tripartite motif containing 49-like 2
|
|
chr22_+_42726423
|
0.026
|
NM_001003828
|
PARVB
|
parvin, beta
|
|
chr8_-_87824996
|
0.024
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr12_+_100394787
|
0.023
|
NM_152323
|
SPIC
|
Spi-C transcription factor (Spi-1/PU.1 related)
|
|
chr2_+_28892366
|
0.023
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr6_+_55300225
|
0.022
|
NM_207410
|
GFRAL
|
GDNF family receptor alpha like
|
|
chr16_+_4322225
|
0.020
|
NM_032575
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr20_-_49852353
|
0.019
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr12_-_52939636
|
0.019
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr16_+_6763810
|
0.016
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr1_+_111571803
|
0.014
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr4_-_122357104
|
0.010
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr11_-_113971681
|
0.007
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr5_+_175041069
|
0.006
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr5_-_24535386
|
0.006
|
NM_001190450
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr6_+_155626838
|
0.006
|
NM_001001346
|
CLDN20
|
claudin 20
|
|
chr1_-_77457664
|
0.006
|
NM_005482
|
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K
|
|
chr14_-_59264044
|
0.006
|
NM_206857
|
RTN1
|
reticulon 1
|
|
chr3_+_116824840
|
0.006
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr10_+_5126613
|
0.005
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr10_+_44726635
|
0.003
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr15_-_64644758
|
0.003
|
NM_207338
|
LCTL
|
lactase-like
|
|
chr11_-_89181390
|
0.003
|
NM_020358
|
TRIM49
|
tripartite motif containing 49
|
|
chr16_+_47965308
|
0.003
|
NM_144602
|
C16orf78
|
chromosome 16 open reading frame 78
|
|
chr3_-_158695832
|
0.001
|
NM_001167917
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr11_+_71578606
|
0.001
|
NM_000802
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr7_-_63660889
|
0.001
|
NM_001130022
NM_178558
|
ZNF680
|
zinc finger protein 680
|
|
chr1_+_175406685
|
0.001
|
|
FAM5B
|
family with sequence similarity 5, member B
|