|
chr7_-_22363036
|
8.870
|
NM_012294
|
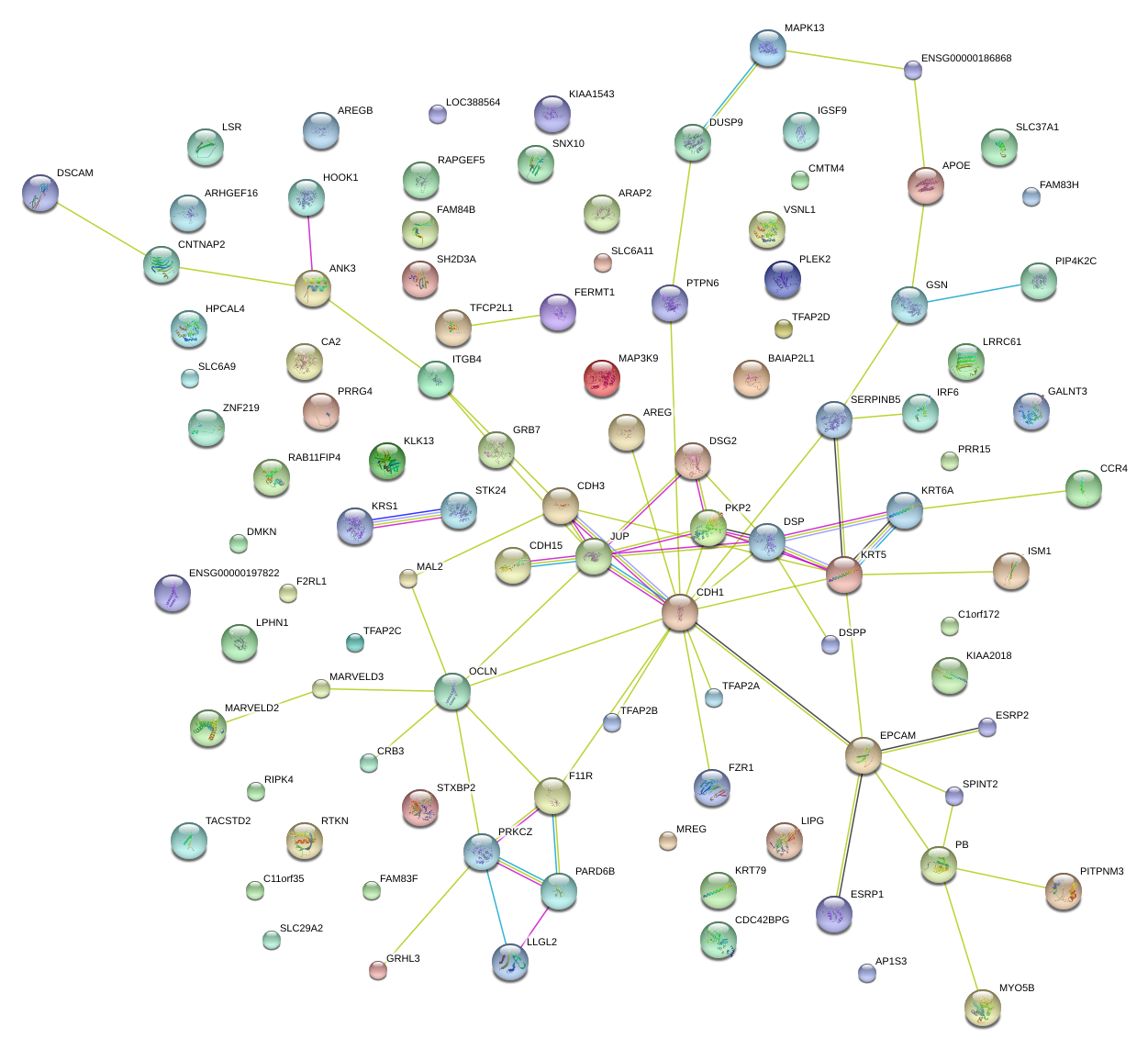
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr19_+_6415259
|
8.786
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr19_+_40431359
|
8.259
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr19_+_40431618
|
8.197
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr1_-_27159462
|
7.743
|
NM_152365
|
C1orf172
|
chromosome 1 open reading frame 172
|
|
chr8_+_120289735
|
7.661
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr22_+_38720881
|
7.568
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr4_-_35922288
|
7.358
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr19_+_43447262
|
6.873
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_40431122
|
5.969
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr8_-_144887876
|
5.887
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr19_+_40431678
|
5.768
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr19_+_43447078
|
5.661
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr18_+_27332024
|
5.040
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr1_-_208046011
|
5.033
|
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr19_+_43446937
|
4.957
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_7566698
|
4.859
|
NM_001080429
NM_020902
|
KIAA1543
|
KIAA1543
|
|
chr7_+_26298018
|
4.853
|
NM_013322
|
SNX10
|
sorting nexin 10
|
|
chr17_+_26839143
|
4.548
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr11_+_32808045
|
4.384
|
NM_024081
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane)
|
|
chr6_-_10527772
|
4.329
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr1_+_1971762
|
4.228
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr16_-_66827449
|
4.198
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr19_-_60587414
|
4.161
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr3_+_10832867
|
4.026
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr11_-_64368616
|
4.005
|
NM_017525
|
CDC42BPG
|
CDC42 binding protein kinase gamma (DMPK-like)
|
|
chr2_-_166358951
|
3.973
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr18_-_45975163
|
3.965
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr2_-_166359016
|
3.903
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr2_-_166358792
|
3.895
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr1_+_60053050
|
3.859
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr5_+_68746694
|
3.836
|
NM_001038603
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr8_+_95722448
|
3.747
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr21_-_42060266
|
3.693
|
NM_020639
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr2_+_17585462
|
3.678
|
|
VSNL1
|
visinin-like 1
|
|
chr2_+_17585287
|
3.639
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr7_+_26298136
|
3.559
|
|
SNX10
|
sorting nexin 10
|
|
chr17_-_37196463
|
3.530
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr11_-_65895640
|
3.514
|
NM_001532
|
SLC29A2
|
solute carrier family 29 (nucleoside transporters), member 2
|
|
chr21_-_42060334
|
3.437
|
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr1_-_159257595
|
3.377
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr16_+_67328693
|
3.371
|
NM_004360
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr1_-_44269719
|
3.357
|
NM_001024845
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr16_+_67328715
|
3.351
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr21_-_41140908
|
3.295
|
NM_001389
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr20_+_48781457
|
3.272
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr1_-_159257567
|
3.244
|
|
F11R
|
F11 receptor
|
|
chr1_+_24518398
|
3.174
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr1_-_159257486
|
3.172
|
|
F11R
|
F11 receptor
|
|
chr17_+_35147738
|
3.151
|
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr5_+_76150637
|
3.125
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr17_+_71229191
|
3.076
|
|
ITGB4
|
integrin, beta 4
|
|
chr5_+_76150588
|
3.069
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr1_-_44269615
|
3.063
|
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr16_+_67236730
|
3.024
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr17_+_71229151
|
3.018
|
|
ITGB4
|
integrin, beta 4
|
|
chr17_+_71229110
|
2.994
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr6_+_7486891
|
2.988
|
|
DSP
|
desmoplakin
|
|
chr16_+_67328805
|
2.977
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr1_+_24518501
|
2.973
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr6_+_7486836
|
2.961
|
|
DSP
|
desmoplakin
|
|
chr8_-_127639647
|
2.882
|
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr6_+_7486862
|
2.837
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr19_+_7608010
|
2.822
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr2_+_47449790
|
2.822
|
NM_002354
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr16_+_70217564
|
2.759
|
NM_001017967
NM_052858
|
MARVELD3
|
MARVEL domain containing 3
|
|
chr17_+_71033339
|
2.756
|
NM_001015002
NM_001031803
NM_004524
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr12_-_32940931
|
2.754
|
NM_001005242
NM_004572
|
PKP2
|
plakophilin 2
|
|
chr19_+_7608015
|
2.716
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr2_-_224410444
|
2.714
|
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr19_+_7607976
|
2.687
|
NM_001127396
NM_006949
|
STXBP2
|
syntaxin binding protein 2
|
|
chrX_+_152562013
|
2.677
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr7_+_149651202
|
2.632
|
NM_001142928
NM_023942
|
LRRC61
|
leucine rich repeat containing 61
|
|
chr19_-_7607852
|
2.615
|
|
|
|
|
chr19_+_7608020
|
2.581
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr14_-_66948523
|
2.546
|
NM_016445
|
PLEK2
|
pleckstrin 2
|
|
chr7_+_29570024
|
2.541
|
|
PRR15
|
proline rich 15
|
|
chr2_+_47449951
|
2.533
|
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr17_+_35147697
|
2.519
|
NM_005310
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr14_-_66948612
|
2.505
|
|
PLEK2
|
pleckstrin 2
|
|
chr8_+_144888275
|
2.500
|
|
LOC100128338
|
hypothetical LOC100128338
|
|
chr17_-_6400470
|
2.480
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr10_-_62163083
|
2.479
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr16_+_67328737
|
2.433
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr2_-_224410544
|
2.428
|
NM_001039569
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr16_-_65287967
|
2.368
|
NM_178818
NM_181521
|
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4
|
|
chr1_+_36544549
|
2.363
|
NM_001162530
|
SH3D21
|
SH3 domain containing 21
|
|
chr19_-_14177980
|
2.337
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr18_+_59295123
|
2.329
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr12_+_6926014
|
2.279
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr8_-_17702991
|
2.278
|
|
|
|
|
chr11_-_550706
|
2.269
|
NM_173573
|
C11orf35
|
chromosome 11 open reading frame 35
|
|
chr7_-_97868101
|
2.231
|
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr20_-_6051562
|
2.217
|
|
FERMT1
|
fermitin family member 1
|
|
chr5_+_68824143
|
2.196
|
|
OCLN
|
occludin
|
|
chr2_-_216586488
|
2.171
|
NM_018000
|
MREG
|
melanoregulin
|
|
chr1_-_158182006
|
2.163
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chr14_-_70345485
|
2.162
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr21_+_42807052
|
2.155
|
|
SLC37A1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1
|
|
chr20_+_13150417
|
2.145
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr14_-_20636337
|
2.142
|
|
ZNF219
|
zinc finger protein 219
|
|
chr2_-_74522513
|
2.133
|
|
RTKN
|
rhotekin
|
|
chr19_-_6718417
|
2.111
|
NM_005490
|
SH2D3A
|
SH2 domain containing 3A
|
|
chr7_+_147667461
|
2.105
|
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chrX_+_130985316
|
2.101
|
NM_001042452
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr2_-_74522534
|
2.096
|
|
RTKN
|
rhotekin
|
|
chr12_-_51200347
|
2.095
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr2_-_74522563
|
2.081
|
NM_001015055
|
RTKN
|
rhotekin
|
|
chr4_+_75699652
|
2.079
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr8_+_86563382
|
2.060
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr12_+_56271181
|
2.051
|
NM_001146258
NM_001146259
NM_001146260
NM_024779
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma
|
|
chr21_-_41140457
|
2.028
|
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr19_-_40693176
|
2.014
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr18_+_45342398
|
2.008
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr1_-_58815415
|
1.988
|
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr3_-_45162783
|
1.975
|
|
|
|
|
chr12_+_56271259
|
1.970
|
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma
|
|
chr6_+_36206239
|
1.946
|
NM_002754
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr2_-_121759237
|
1.945
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr1_+_3360880
|
1.945
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr7_+_29569978
|
1.941
|
|
PRR15
|
proline rich 15
|
|
chr2_-_36384
|
1.930
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr1_+_29086206
|
1.916
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr1_+_29086156
|
1.914
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr20_-_60375695
|
1.899
|
NM_005560
|
LAMA5
|
laminin, alpha 5
|
|
chr4_+_75529716
|
1.881
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr16_+_751050
|
1.866
|
NM_013404
|
MSLN
|
mesothelin
|
|
chr11_+_45900731
|
1.863
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr3_-_45162847
|
1.841
|
|
CDCP1
|
CUB domain containing protein 1
|
|
chr4_+_152549776
|
1.792
|
NM_001109977
|
FAM160A1
|
family with sequence similarity 160, member A1
|
|
chr12_+_6925997
|
1.771
|
NM_080548
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr17_+_35148084
|
1.771
|
NM_001030002
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr14_-_37134191
|
1.767
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr1_-_58815753
|
1.754
|
NM_002353
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr3_-_45162865
|
1.734
|
|
CDCP1
|
CUB domain containing protein 1
|
|
chr1_-_21821597
|
1.734
|
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr14_+_30413393
|
1.729
|
NM_001135058
NM_004086
|
COCH
|
coagulation factor C homolog, cochlin (Limulus polyphemus)
|
|
chr16_-_11588257
|
1.725
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr3_+_46996150
|
1.724
|
NM_015175
|
NBEAL2
|
neurobeachin-like 2
|
|
chr22_-_49317408
|
1.715
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr14_-_60817946
|
1.703
|
|
|
|
|
chr3_-_45162901
|
1.688
|
NM_022842
NM_178181
|
CDCP1
|
CUB domain containing protein 1
|
|
chr1_-_27212035
|
1.667
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr6_+_54819527
|
1.666
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr20_-_62081752
|
1.663
|
|
SAMD10
|
sterile alpha motif domain containing 10
|
|
chr2_+_111594921
|
1.660
|
NM_006538
NM_138621
NM_207002
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr11_+_551449
|
1.657
|
NM_001143994
NM_003475
|
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7
|
|
chr11_+_75157414
|
1.656
|
NM_032564
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr9_-_125070675
|
1.649
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr12_+_6290127
|
1.640
|
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr8_-_81246215
|
1.627
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr11_+_75157450
|
1.626
|
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr17_-_35911340
|
1.626
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr6_+_150505849
|
1.618
|
NM_030949
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C
|
|
chr8_+_102574012
|
1.607
|
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr14_-_37134050
|
1.601
|
|
FOXA1
|
forkhead box A1
|
|
chr8_-_110726063
|
1.600
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr19_-_51909416
|
1.598
|
NM_001079882
|
PRKD2
|
protein kinase D2
|
|
chr3_+_188131209
|
1.578
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr17_+_72789086
|
1.568
|
NM_001113491
|
SEPT9
|
septin 9
|
|
chr14_-_37133927
|
1.563
|
|
FOXA1
|
forkhead box A1
|
|
chr22_-_20314274
|
1.554
|
NM_001017964
|
YDJC
|
YdjC homolog (bacterial)
|
|
chr1_-_27211913
|
1.553
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr12_+_7174230
|
1.545
|
NM_014718
|
CLSTN3
|
calsyntenin 3
|
|
chr19_+_43486640
|
1.537
|
NM_033520
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr8_-_110726201
|
1.536
|
NM_001099753
NM_001099754
NM_001099755
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr16_+_67236699
|
1.508
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr14_-_60817663
|
1.507
|
|
TMEM30B
|
transmembrane protein 30B
|
|
chr5_-_168660293
|
1.502
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr14_-_60817709
|
1.502
|
|
TMEM30B
|
transmembrane protein 30B
|
|
chr11_+_550868
|
1.494
|
NM_001143993
|
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7
|
|
chr4_-_82612049
|
1.490
|
NM_152545
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chr1_+_64983312
|
1.485
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr13_-_27092538
|
1.479
|
|
LNX2
|
ligand of numb-protein X 2
|
|
chr1_+_181422016
|
1.464
|
|
LAMC2
|
laminin, gamma 2
|
|
chr1_+_203740391
|
1.454
|
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr6_+_30959839
|
1.450
|
NM_001954
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr18_+_58143499
|
1.449
|
NM_003839
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator
|
|
chr22_-_30072217
|
1.442
|
NM_014323
NM_032050
NM_032051
NM_032052
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr4_-_82611995
|
1.425
|
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chr2_-_99124466
|
1.424
|
NM_182911
|
TSGA10
|
testis specific, 10
|
|
chr6_+_44203335
|
1.424
|
NM_018426
|
TMEM63B
|
transmembrane protein 63B
|
|
chr14_-_60817621
|
1.421
|
|
|
|
|
chr8_-_95720870
|
1.417
|
|
LOC100288748
|
hypothetical LOC100288748
|
|
chr8_+_95722516
|
1.410
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr1_+_232107170
|
1.406
|
NM_173508
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chr8_+_95722575
|
1.404
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr19_+_60283571
|
1.402
|
NM_017729
|
EPS8L1
|
EPS8-like 1
|
|
chr15_-_20637731
|
1.398
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr19_+_45811856
|
1.393
|
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr5_-_60175798
|
1.387
|
NM_001104558
NM_024930
|
ELOVL7
|
ELOVL family member 7, elongation of long chain fatty acids (yeast)
|
|
chr1_+_181421984
|
1.386
|
|
LAMC2
|
laminin, gamma 2
|
|
chr10_-_21502993
|
1.385
|
|
NEBL
|
nebulette
|
|
chr6_-_47385205
|
1.381
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr7_+_149651481
|
1.379
|
|
LRRC61
|
leucine rich repeat containing 61
|
|
chr17_-_40866062
|
1.372
|
NM_174919
|
SH3D20
|
SH3 domain containing 20
|
|
chr19_-_48835783
|
1.367
|
NM_145296
|
CADM4
|
cell adhesion molecule 4
|
|
chr7_-_149651594
|
1.361
|
NM_001164458
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast)
|
|
chr1_+_32979930
|
1.360
|
NM_020888
|
KIAA1522
|
KIAA1522
|
|
chr11_-_114880275
|
1.359
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|