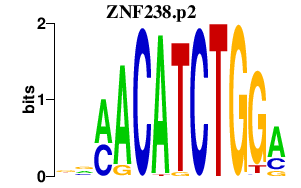
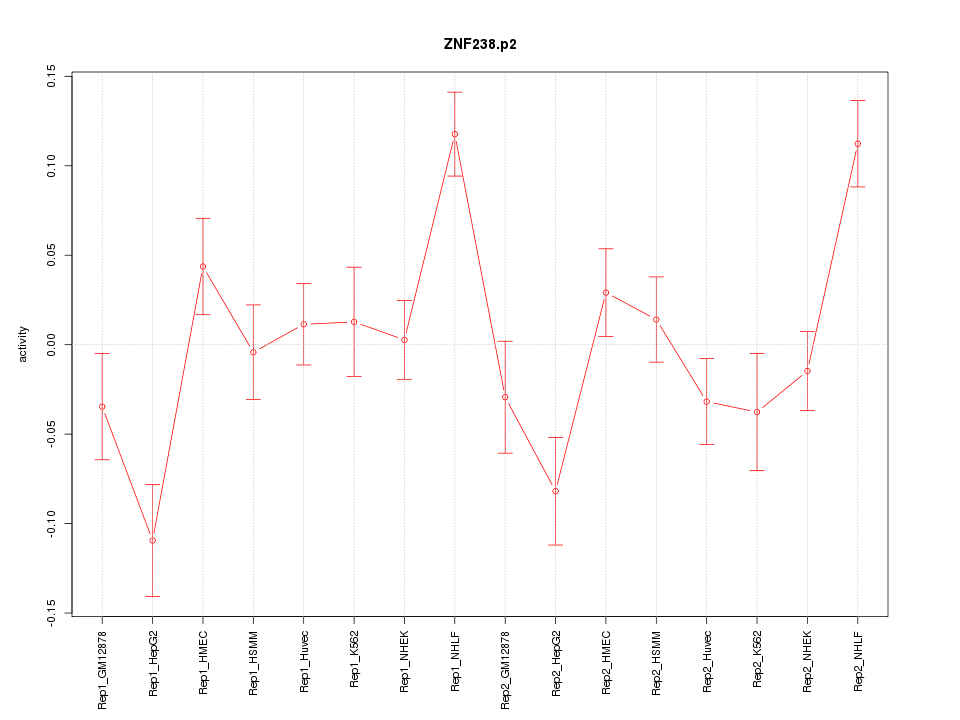
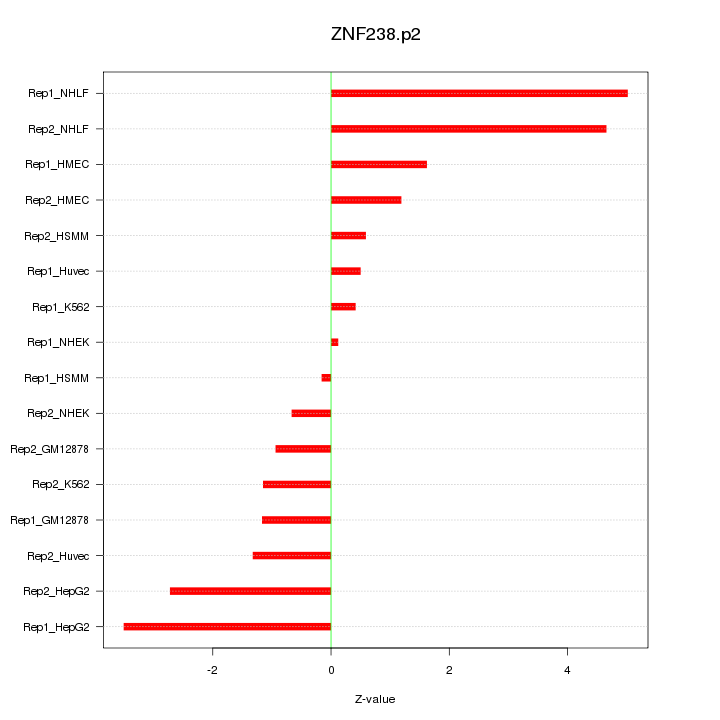
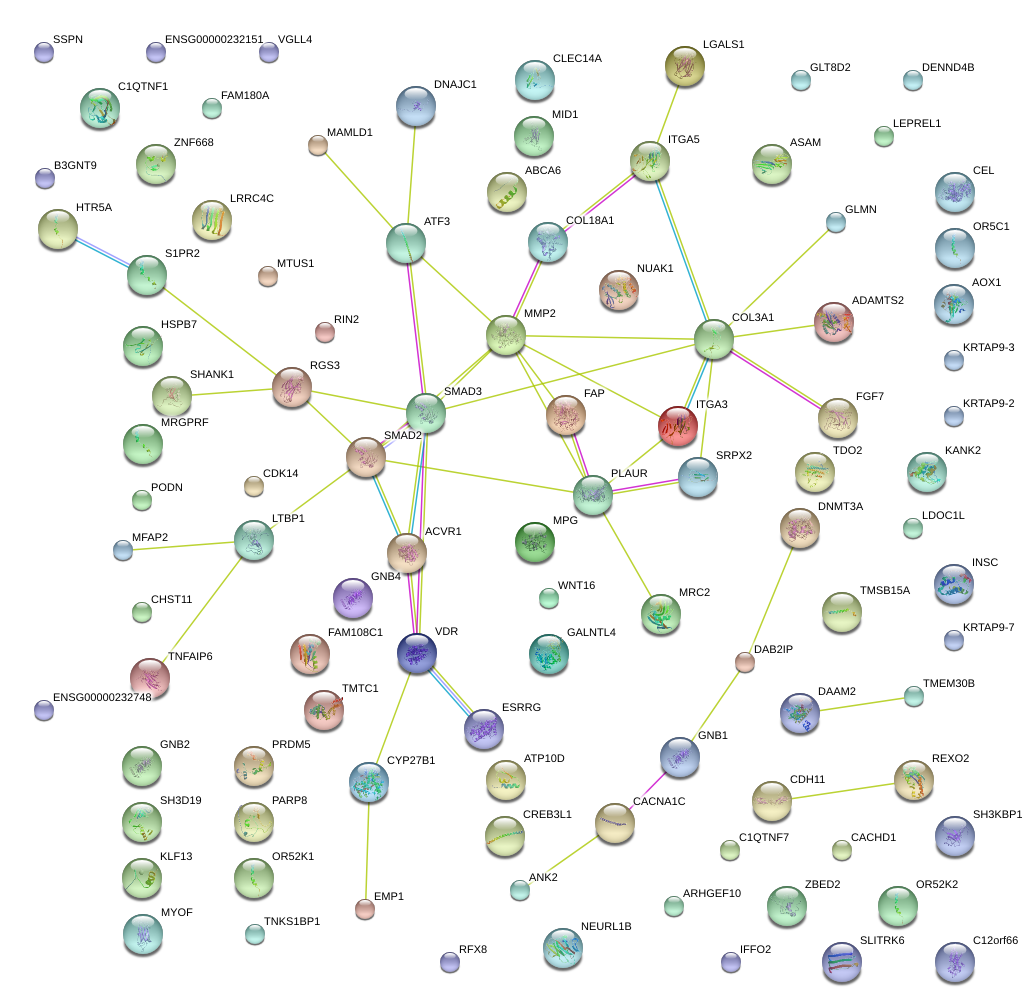
|
chr11_-_122571046
|
10.367
|
|
CLMP
|
CXADR-like membrane protein
|
|
chr16_+_54070302
|
8.251
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr2_-_162808123
|
4.436
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr20_+_19903778
|
4.124
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr11_-_68537243
|
3.798
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr10_+_99463446
|
3.340
|
NM_031484
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr10_+_99463468
|
3.176
|
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr1_+_53300410
|
3.132
|
NM_153703
|
PODN
|
podocan
|
|
chr12_+_13240987
|
3.073
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr7_+_90063543
|
3.043
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr12_+_13240983
|
3.035
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr17_+_74531820
|
2.905
|
NM_030968
NM_198594
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr12_-_29827952
|
2.790
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr5_-_178704934
|
2.688
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr5_-_178705036
|
2.640
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr2_+_151922350
|
2.528
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr7_-_135083906
|
2.428
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr8_-_17599438
|
2.371
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr17_+_58058774
|
2.353
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr12_-_102968044
|
2.327
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr19_-_48866111
|
2.282
|
NM_001005376
NM_001005377
NM_002659
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr12_-_102967828
|
2.258
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr4_+_14985272
|
2.228
|
NM_001135171
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr19_-_10202947
|
2.160
|
NM_004230
|
S1PR2
|
sphingosine-1-phosphate receptor 2
|
|
chr5_+_49998677
|
2.152
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr12_+_26239755
|
2.125
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr3_-_191321601
|
2.120
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr12_-_105056577
|
2.091
|
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr1_-_17179694
|
2.037
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr22_-_43271843
|
2.020
|
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr12_+_2032649
|
2.015
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chrX_-_19598912
|
1.942
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_-_215329569
|
1.885
|
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr8_+_1759502
|
1.880
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chrX_+_99785818
|
1.862
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr17_+_74531873
|
1.816
|
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr11_-_56846246
|
1.809
|
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr4_-_152368492
|
1.800
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr2_+_189547377
|
1.792
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr19_-_11166186
|
1.751
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr6_+_39868770
|
1.742
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr4_+_114433551
|
1.741
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr17_+_45488330
|
1.737
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr3_-_11585264
|
1.735
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chrX_+_99786044
|
1.730
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr17_+_45488356
|
1.703
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr2_+_33213111
|
1.694
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr1_-_152185764
|
1.677
|
NM_014856
|
DENND4B
|
DENN/MADD domain containing 4B
|
|
chr5_+_49998528
|
1.658
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr1_+_64709063
|
1.607
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr10_-_95231933
|
1.606
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr2_+_189547358
|
1.568
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189547323
|
1.553
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr9_+_123369219
|
1.523
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chrX_-_101658297
|
1.478
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chr11_-_40272239
|
1.472
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr12_+_26239535
|
1.460
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr2_-_101457596
|
1.447
|
NM_001145664
|
RFX8
|
regulatory factor X, 8
|
|
chr5_+_172000793
|
1.360
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr15_+_65205107
|
1.338
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr4_-_122063118
|
1.285
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr10_-_22332600
|
1.251
|
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr15_+_29406335
|
1.239
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr1_+_210848592
|
1.233
|
NM_001040619
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr16_-_65742323
|
1.232
|
NM_033309
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9
|
|
chr16_-_30983909
|
1.216
|
NM_001172669
NM_001172670
|
ZNF668
|
zinc finger protein 668
|
|
chr2_-_158439868
|
1.209
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr2_+_201182187
|
1.197
|
|
AOX1
|
aldehyde oxidase 1
|
|
chrX_+_149280486
|
1.189
|
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr16_-_63713466
|
1.162
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr15_+_78775008
|
1.162
|
|
FAM108C1
|
family with sequence similarity 108, member C1
|
|
chr1_-_146398501
|
1.141
|
|
LOC100286793
FLJ39739
|
hypothetical LOC100286793
hypothetical FLJ39739
|
|
chr3_-_112796855
|
1.136
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr14_-_60818263
|
1.131
|
NM_001017970
|
TMEM30B
|
transmembrane protein 30B
|
|
chr1_+_210848726
|
1.130
|
|
ATF3
|
activating transcription factor 3
|
|
chr1_+_210848763
|
1.123
|
|
ATF3
|
activating transcription factor 3
|
|
chr9_+_124590970
|
1.113
|
NM_001001923
|
OR5C1
|
olfactory receptor, family 5, subfamily C, member 1
|
|
chr4_+_157044296
|
1.091
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr11_-_11600118
|
1.087
|
NM_198516
|
GALNTL4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4
|
|
chr12_-_46585032
|
1.084
|
NM_000376
NM_001017535
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr14_-_37795001
|
1.079
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr4_+_47181987
|
1.078
|
NM_020453
|
ATP10D
|
ATPase, class V, type 10D
|
|
chr1_-_19155360
|
1.077
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr1_-_16217025
|
1.074
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr17_+_58058783
|
1.026
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr11_+_4466684
|
1.018
|
NM_001005171
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1
|
|
chr13_-_85271329
|
1.010
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr11_+_46256161
|
1.010
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr16_-_63713485
|
1.007
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr1_-_143232275
|
0.980
|
|
LOC728855
|
hypothetical LOC728855
|
|
chr11_+_113815317
|
0.976
|
NM_015523
|
REXO2
|
REX2, RNA exonuclease 2 homolog (S. cerevisiae)
|
|
chr9_+_115396020
|
0.966
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr15_+_47502666
|
0.964
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr2_-_25320582
|
0.961
|
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr7_+_154493478
|
0.944
|
NM_024012
|
HTR5A
|
5-hydroxytryptamine (serotonin) receptor 5A
|
|
chr10_-_22332646
|
0.923
|
NM_022365
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr21_+_45699830
|
0.923
|
NM_030582
NM_130444
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr12_-_62902279
|
0.913
|
NM_152440
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chr10_-_22332368
|
0.900
|
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr7_+_120756325
|
0.894
|
NM_057168
|
WNT16
|
wingless-type MMTV integration site family, member 16
|
|
chr11_+_15093059
|
0.894
|
NM_001042536
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chrX_-_10548458
|
0.889
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr12_+_103374771
|
0.880
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr12_+_103375116
|
0.866
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr1_-_143232344
|
0.851
|
|
LOC728855
|
hypothetical LOC728855
|
|
chr12_+_103374880
|
0.837
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr19_-_55912006
|
0.837
|
NM_016148
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1
|
|
chr7_+_100113664
|
0.832
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr17_+_36636404
|
0.828
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr6_+_39868116
|
0.825
|
|
|
|
|
chr6_+_39868126
|
0.823
|
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr12_+_103374892
|
0.820
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr12_+_103374914
|
0.788
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr17_-_64649608
|
0.786
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr22_+_36401558
|
0.785
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr12_-_53099269
|
0.774
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr14_-_37795290
|
0.769
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr3_-_64186151
|
0.757
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr17_-_31219943
|
0.756
|
NM_152781
|
C17orf66
|
chromosome 17 open reading frame 66
|
|
chr7_+_5609416
|
0.745
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chrX_-_34585287
|
0.745
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr19_+_44595589
|
0.743
|
NM_022835
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2
|
|
chr2_+_234302511
|
0.740
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr3_-_115585178
|
0.731
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr2_+_12774264
|
0.725
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr12_+_111047742
|
0.725
|
|
TRAFD1
|
TRAF-type zinc finger domain containing 1
|
|
chr11_+_117762432
|
0.721
|
|
UBE4A
|
ubiquitination factor E4A (UFD2 homolog, yeast)
|
|
chr9_-_35860316
|
0.717
|
NM_001004487
|
OR13J1
|
olfactory receptor, family 13, subfamily J, member 1
|
|
chr3_-_161650219
|
0.715
|
|
TRIM59
|
tripartite motif containing 59
|
|
chr1_-_33585848
|
0.713
|
|
|
|
|
chrX_+_100220491
|
0.704
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr6_-_150253729
|
0.694
|
NM_139165
|
RAET1E
|
retinoic acid early transcript 1E
|
|
chr12_-_108736983
|
0.691
|
NM_001177431
NM_001177428
NM_001177433
NM_147204
|
TRPV4
|
transient receptor potential cation channel, subfamily V, member 4
|
|
chr1_-_151784889
|
0.684
|
NM_002961
NM_019554
|
S100A4
|
S100 calcium binding protein A4
|
|
chr9_+_71848555
|
0.683
|
|
MAMDC2
|
MAM domain containing 2
|
|
chr6_-_30631481
|
0.677
|
|
GNL1
|
guanine nucleotide binding protein-like 1
|
|
chr19_-_47155340
|
0.677
|
NM_006423
|
RABAC1
|
Rab acceptor 1 (prenylated)
|
|
chr17_+_36642240
|
0.675
|
NM_031962
|
KRTAP9-9
KRTAP9-3
|
keratin associated protein 9-9
keratin associated protein 9-3
|
|
chr11_-_5099422
|
0.672
|
NM_001005222
|
OR52A4
|
olfactory receptor, family 52, subfamily A, member 4
|
|
chr11_+_74950766
|
0.668
|
NM_001235
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr1_+_152644471
|
0.662
|
|
IL6R
|
interleukin 6 receptor
|
|
chr21_+_46226072
|
0.661
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr1_-_154913712
|
0.658
|
NM_006617
|
NES
|
nestin
|
|
chr7_-_124192916
|
0.658
|
NM_005302
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr11_+_46256130
|
0.644
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr1_-_155336223
|
0.643
|
NM_001004341
|
ETV3L
|
ets variant 3-like
|
|
chr17_+_36647795
|
0.639
|
NM_031963
|
KRTAP9-8
|
keratin associated protein 9-8
|
|
chr5_+_140772691
|
0.638
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr17_-_51164443
|
0.635
|
NM_001099640
|
TMEM100
|
transmembrane protein 100
|
|
chr3_-_191321367
|
0.632
|
|
LEPREL1
|
leprecan-like 1
|
|
chr17_-_24491521
|
0.623
|
|
MYO18A
|
myosin XVIIIA
|
|
chr5_+_40715799
|
0.620
|
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr15_-_76890626
|
0.620
|
NM_014272
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7
|
|
chr18_+_7557313
|
0.618
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr9_+_71848316
|
0.616
|
NM_153267
|
MAMDC2
|
MAM domain containing 2
|
|
chr12_+_111047676
|
0.608
|
NM_001143906
NM_006700
|
TRAFD1
|
TRAF-type zinc finger domain containing 1
|
|
chr1_-_142535878
|
0.608
|
|
LOC100286793
FLJ39739
|
hypothetical LOC100286793
hypothetical FLJ39739
|
|
chr12_+_62524778
|
0.602
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr15_+_54998144
|
0.599
|
|
TCF12
|
transcription factor 12
|
|
chr16_+_28798735
|
0.596
|
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr5_+_178419938
|
0.595
|
NM_014594
|
ZNF354C
|
zinc finger protein 354C
|
|
chr15_-_81743198
|
0.591
|
|
BNC1
|
basonuclin 1
|
|
chr11_-_33678861
|
0.589
|
NM_001166692
|
C11orf91
|
chromosome 11 open reading frame 91
|
|
chr2_-_1727324
|
0.583
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr12_+_39372456
|
0.582
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr11_+_5798119
|
0.575
|
NM_001005174
|
OR52N2
|
olfactory receptor, family 52, subfamily N, member 2
|
|
chr9_-_90456824
|
0.572
|
NM_001100111
|
LOC286238
|
hypothetical protein LOC286238
|
|
chr12_-_6313219
|
0.572
|
|
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A
|
|
chrX_+_48127911
|
0.564
|
NM_001034832
NM_001040612
NM_005636
NM_175729
|
SSX4B
SSX4
|
synovial sarcoma, X breakpoint 4B
synovial sarcoma, X breakpoint 4
|
|
chr13_+_51933228
|
0.556
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr2_-_1727220
|
0.555
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr18_+_3402071
|
0.542
|
NM_174886
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr12_-_53099203
|
0.539
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr15_+_54998181
|
0.539
|
|
TCF12
|
transcription factor 12
|
|
chr22_-_40941254
|
0.536
|
NM_005650
NM_181492
|
TCF20
|
transcription factor 20 (AR1)
|
|
chr1_+_156701895
|
0.536
|
NM_001004473
|
OR10K1
|
olfactory receptor, family 10, subfamily K, member 1
|
|
chr12_-_53099277
|
0.531
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr11_-_4827013
|
0.530
|
NM_001004758
|
OR51S1
|
olfactory receptor, family 51, subfamily S, member 1
|
|
chr11_-_5037432
|
0.525
|
NM_001005164
|
OR52E2
|
olfactory receptor, family 52, subfamily E, member 2
|
|
chr2_-_1727247
|
0.524
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr7_+_93862137
|
0.524
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr1_-_143232464
|
0.523
|
|
|
|
|
chr15_+_54998115
|
0.521
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr16_+_65868913
|
0.521
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr5_-_41906459
|
0.519
|
NM_000436
|
OXCT1
|
3-oxoacid CoA transferase 1
|
|
chr12_+_76749199
|
0.518
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr1_-_142536082
|
0.514
|
|
|
|
|
chr3_+_30623376
|
0.511
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr2_-_1727292
|
0.511
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr5_+_40715778
|
0.511
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr7_+_116099648
|
0.510
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr5_+_14493905
|
0.509
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr11_+_10283171
|
0.505
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr12_-_54408391
|
0.498
|
|
CD63
|
CD63 molecule
|
|
chr1_-_153444304
|
0.496
|
NM_007112
|
THBS3
|
thrombospondin 3
|
|
chr11_-_1563056
|
0.494
|
NM_001005922
|
KRTAP5-1
|
keratin associated protein 5-1
|
|
chr1_-_160260210
|
0.494
|
NM_015441
|
OLFML2B
|
olfactomedin-like 2B
|
|
chr10_-_105427898
|
0.490
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr1_-_199742871
|
0.487
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr11_+_76171780
|
0.486
|
NM_015516
|
TSKU
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis)
|