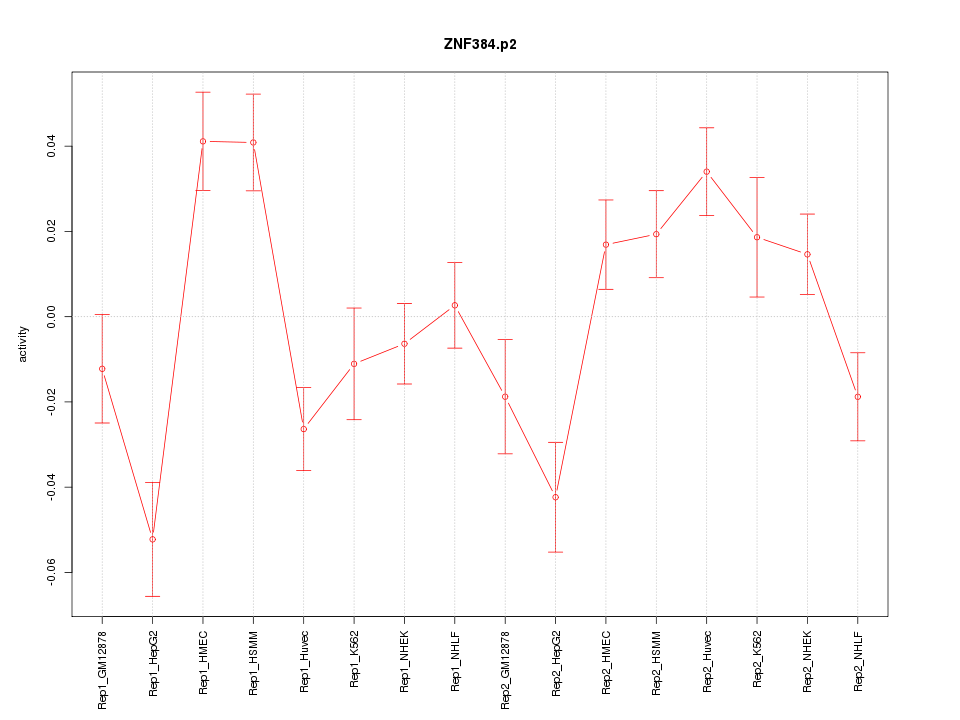
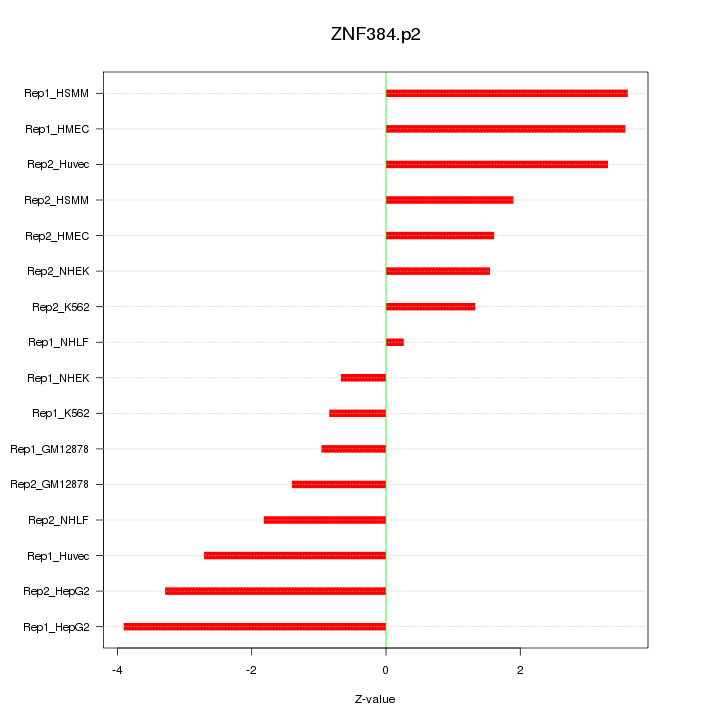
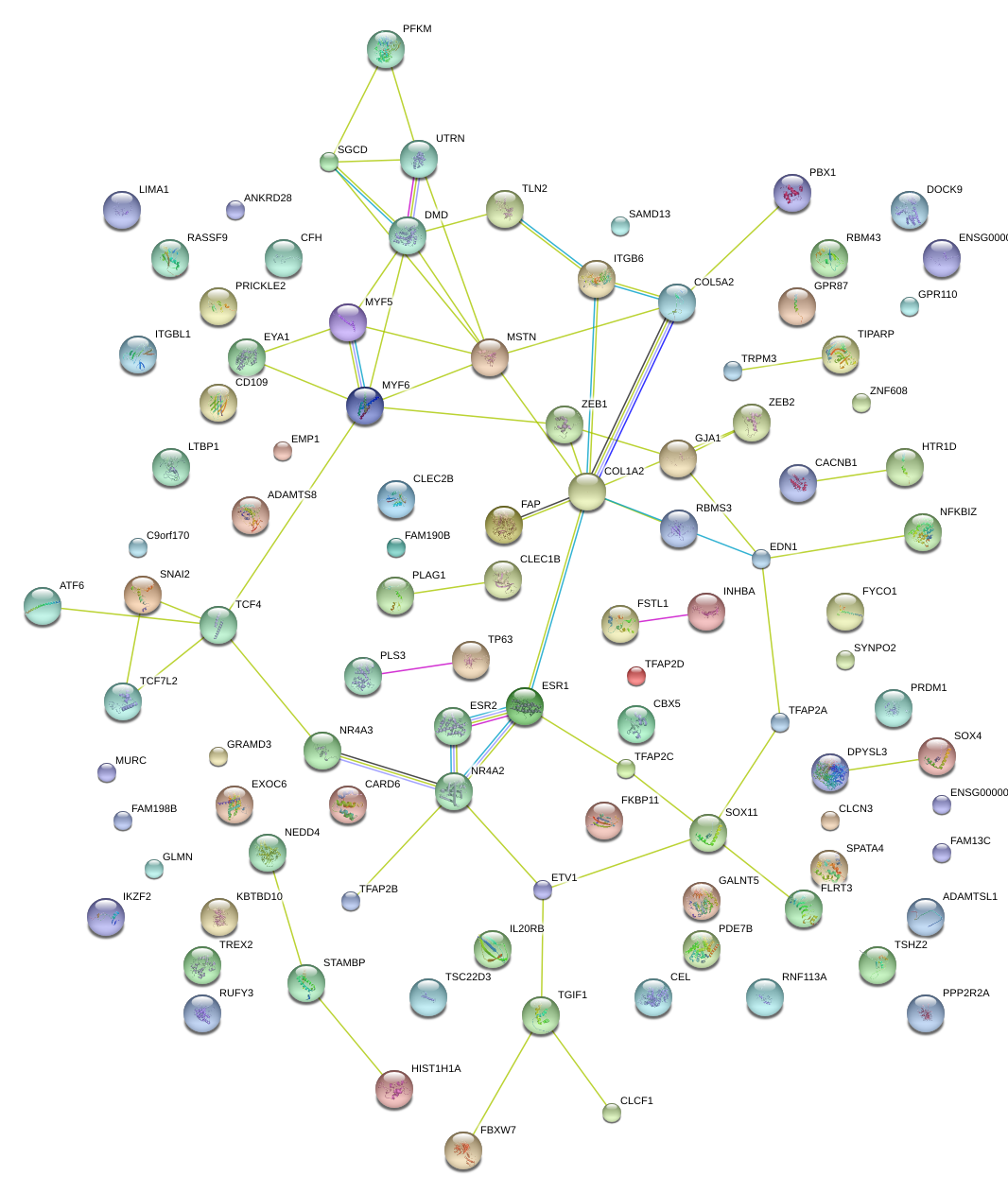
|
chr2_-_189752575
|
5.397
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr12_-_48902692
|
3.308
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr6_+_121798443
|
3.177
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr2_+_170074457
|
2.923
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr6_+_106653429
|
2.823
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr2_+_5750229
|
2.605
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr7_-_13995811
|
2.604
|
|
ETV1
|
ets variant 1
|
|
chr7_-_13996166
|
2.578
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr13_+_100902942
|
2.486
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr2_-_144994038
|
2.450
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_162808123
|
2.427
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr5_+_125786975
|
2.403
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr4_+_170817787
|
2.388
|
|
CLCN3
|
chloride channel 3
|
|
chr20_-_14266200
|
2.367
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr2_-_144994349
|
2.334
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_+_84540921
|
2.292
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr1_+_162795328
|
2.276
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_+_157877146
|
2.259
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr5_+_155686750
|
2.215
|
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr1_+_194887630
|
2.202
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr3_+_138159396
|
2.139
|
NM_144717
|
IL20RB
|
interleukin 20 receptor beta
|
|
chr12_-_9913724
|
2.122
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr10_+_94584449
|
2.073
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr7_-_41709191
|
2.067
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr3_+_29297806
|
1.994
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr12_-_52939636
|
1.965
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr11_+_65027721
|
1.954
|
|
|
|
|
chr18_+_3437607
|
1.895
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr12_-_52939584
|
1.880
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr18_-_51406841
|
1.838
|
NM_001083962
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr10_+_86174665
|
1.811
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr5_+_40877053
|
1.809
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr1_-_23393808
|
1.782
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr6_+_12398514
|
1.764
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr12_+_13240868
|
1.752
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_-_9913199
|
1.750
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr2_-_151826504
|
1.740
|
NM_198557
|
RBM43
|
RNA binding motif protein 43
|
|
chr3_+_190990142
|
1.722
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr6_+_74462654
|
1.720
|
|
CD109
|
CD109 molecule
|
|
chr2_+_33213111
|
1.707
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chrX_+_114734074
|
1.691
|
NM_001136025
NM_001172335
|
PLS3
|
plastin 3
|
|
chr4_-_153493133
|
1.683
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr3_-_152517204
|
1.651
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr5_+_125786717
|
1.636
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr4_+_71806538
|
1.634
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr2_-_156897286
|
1.633
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr2_-_213723298
|
1.608
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr1_-_197173159
|
1.606
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr12_+_13240987
|
1.603
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr4_-_159313167
|
1.571
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr12_+_13240983
|
1.569
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_+_160002733
|
1.564
|
|
ATF6
|
activating transcription factor 6
|
|
chr6_+_136214495
|
1.554
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr6_-_26125939
|
1.554
|
NM_005325
|
HIST1H1A
|
histone cluster 1, H1a
|
|
chr18_+_3443771
|
1.542
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr4_-_177353707
|
1.530
|
NM_144644
|
SPATA4
|
spermatogenesis associated 4
|
|
chr2_-_190635566
|
1.494
|
NM_005259
|
MSTN
|
myostatin
|
|
chr12_+_79634820
|
1.480
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chrX_-_33139349
|
1.477
|
NM_004006
|
DMD
|
dystrophin
|
|
chr3_+_103029523
|
1.473
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr9_+_18464138
|
1.470
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr4_+_159311353
|
1.467
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chrX_-_106846253
|
1.446
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_160002693
|
1.445
|
NM_007348
|
ATF6
|
activating transcription factor 6
|
|
chr9_-_72673753
|
1.428
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr2_+_157822355
|
1.416
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr2_-_160765008
|
1.414
|
|
ITGB6
|
integrin, beta 6
|
|
chr6_+_152053323
|
1.409
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr1_+_160002746
|
1.405
|
|
ATF6
|
activating transcription factor 6
|
|
chr1_+_162795538
|
1.404
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_-_64186151
|
1.402
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr6_-_10520264
|
1.397
|
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr11_-_66898223
|
1.382
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr3_-_15814678
|
1.378
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr20_+_51023159
|
1.365
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr8_-_49996353
|
1.362
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr15_-_53996597
|
1.354
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr6_-_47117951
|
1.354
|
NM_025048
NM_153840
|
GPR110
|
G protein-coupled receptor 110
|
|
chr5_+_155686333
|
1.349
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr5_-_124108672
|
1.336
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr4_+_120168002
|
1.333
|
|
SYNPO2
|
synaptopodin 2
|
|
chr9_+_88953378
|
1.324
|
NM_001001709
|
C9orf170
|
chromosome 9 open reading frame 170
|
|
chr2_+_73911477
|
1.322
|
|
STAMBP
|
STAM binding protein
|
|
chr3_-_121652183
|
1.313
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr1_+_207668787
|
1.313
|
NM_001104548
|
LOC642587
|
NPC-A-5
|
|
chr9_+_102380156
|
1.312
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr5_-_146813343
|
1.309
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr13_-_98428244
|
1.304
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr15_+_60838053
|
1.288
|
|
TLN2
|
talin 2
|
|
chr8_-_72436519
|
1.283
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr10_-_60792225
|
1.278
|
NM_001001971
NM_001166698
NM_198215
|
FAM13C
|
family with sequence similarity 13, member C
|
|
chr6_+_74462577
|
1.269
|
|
CD109
|
CD109 molecule
|
|
chr7_+_93862137
|
1.240
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr8_+_26206648
|
1.239
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr12_+_79625538
|
1.238
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr12_+_46799279
|
1.232
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chr12_-_84754289
|
1.230
|
NM_005447
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|
|
chr11_-_56846246
|
1.228
|
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr2_+_36777336
|
1.218
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr12_-_26169074
|
1.211
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr2_+_189547358
|
1.195
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr10_+_88720477
|
1.190
|
NM_133447
|
AGAP11
|
ankyrin repeat and GTPase domain Arf GTPase activating protein 11
|
|
chr7_+_18502342
|
1.180
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr7_-_16471817
|
1.177
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr6_-_161614962
|
1.177
|
NM_020133
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
|
|
chr4_-_159312973
|
1.170
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr7_+_120416103
|
1.168
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr4_+_187227302
|
1.163
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr6_+_106640903
|
1.162
|
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr21_-_35343065
|
1.152
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_35020425
|
1.139
|
NM_001005752
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr8_+_95722448
|
1.135
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr11_-_71458739
|
1.132
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr3_-_55489973
|
1.127
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr5_-_16989371
|
1.127
|
NM_012334
|
MYO10
|
myosin X
|
|
chr16_+_12902955
|
1.123
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr3_+_108442205
|
1.120
|
|
LOC344595
|
hypothetical LOC344595
|
|
chr18_-_63333487
|
1.119
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr5_-_16989290
|
1.115
|
|
MYO10
|
myosin X
|
|
chr12_+_74160779
|
1.114
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr7_+_18515440
|
1.103
|
|
HDAC9
|
histone deacetylase 9
|
|
chr13_+_48582389
|
1.095
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr7_+_93388951
|
1.092
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr1_+_157246305
|
1.087
|
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr2_+_189547323
|
1.087
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr3_-_55496370
|
1.074
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr5_+_176494690
|
1.070
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr4_+_77575276
|
1.065
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr12_+_14409875
|
1.060
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr5_+_66160359
|
1.059
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr1_+_20788027
|
1.051
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr12_+_11694172
|
1.049
|
|
ETV6
|
ets variant 6
|
|
chr3_-_191322919
|
1.047
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr1_+_22107137
|
1.042
|
|
RPL21
|
ribosomal protein L21
|
|
chr1_+_216586199
|
1.041
|
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr9_+_5500544
|
1.038
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr14_+_95792299
|
1.020
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr2_-_188127430
|
1.012
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr9_+_102231616
|
1.011
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr3_-_55496607
|
1.010
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr7_+_120415958
|
1.003
|
NM_024913
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr8_+_95722575
|
0.996
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr9_-_87904306
|
0.995
|
NM_016548
|
GOLM1
|
golgi membrane protein 1
|
|
chr3_-_116272911
|
0.995
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_-_139654807
|
0.975
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr1_+_100957883
|
0.974
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr9_-_87904280
|
0.974
|
|
GOLM1
|
golgi membrane protein 1
|
|
chr5_-_58607673
|
0.973
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr9_+_74956601
|
0.971
|
|
ANXA1
|
annexin A1
|
|
chr5_+_137253023
|
0.970
|
NM_014386
|
PKD2L2
|
polycystic kidney disease 2-like 2
|
|
chr11_+_93394025
|
0.969
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr13_-_79810972
|
0.969
|
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr9_+_74956494
|
0.969
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr14_-_49932366
|
0.961
|
NM_004196
|
CDKL1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase)
|
|
chr4_-_147079018
|
0.961
|
NM_178835
|
ZNF827
|
zinc finger protein 827
|
|
chr6_-_134680888
|
0.961
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr15_+_90198567
|
0.959
|
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1
|
|
chr7_+_22733626
|
0.959
|
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr12_-_68369322
|
0.951
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr1_+_46151845
|
0.946
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr22_-_34566276
|
0.942
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr1_+_239762179
|
0.936
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr4_-_153820535
|
0.930
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chrX_+_100361588
|
0.928
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr16_+_7322751
|
0.928
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr1_-_103346639
|
0.927
|
NM_001190709
NM_001854
NM_080629
NM_080630
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr7_+_838663
|
0.922
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chrX_+_12904207
|
0.922
|
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr2_+_36777406
|
0.910
|
|
VIT
|
vitrin
|
|
chr17_-_43977273
|
0.907
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr14_-_98807307
|
0.905
|
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr9_+_35528628
|
0.902
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr12_+_4253143
|
0.898
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr18_-_44189625
|
0.898
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr3_-_150533948
|
0.897
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr12_+_18305740
|
0.896
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr10_+_47216925
|
0.877
|
NM_001630
|
ANXA8L2
ANXA8
|
annexin A8-like 2
annexin A8
|
|
chr3_-_102194938
|
0.876
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr6_+_74462630
|
0.874
|
|
CD109
|
CD109 molecule
|
|
chr11_+_10433241
|
0.873
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr20_-_17487480
|
0.872
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr16_+_3295433
|
0.871
|
NM_153028
|
ZNF75A
|
zinc finger protein 75a
|
|
chr7_-_83662152
|
0.870
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr5_-_39460687
|
0.870
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr4_-_147079410
|
0.867
|
|
|
|
|
chr7_+_120416680
|
0.864
|
NM_001105533
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr13_-_44812246
|
0.862
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr3_-_115960807
|
0.860
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_+_49155798
|
0.860
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr13_-_43908985
|
0.860
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr7_-_13997574
|
0.853
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr2_+_189547377
|
0.851
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr5_+_36644187
|
0.846
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr3_+_138132006
|
0.843
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr22_-_34566210
|
0.839
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr4_-_159311959
|
0.837
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr21_-_38792173
|
0.836
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr1_-_68288899
|
0.835
|
NM_004675
|
DIRAS3
|
DIRAS family, GTP-binding RAS-like 3
|
|
chr3_+_11153778
|
0.833
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr9_-_36266930
|
0.829
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|