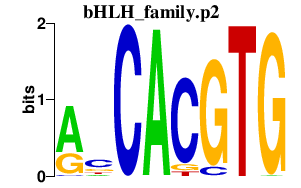
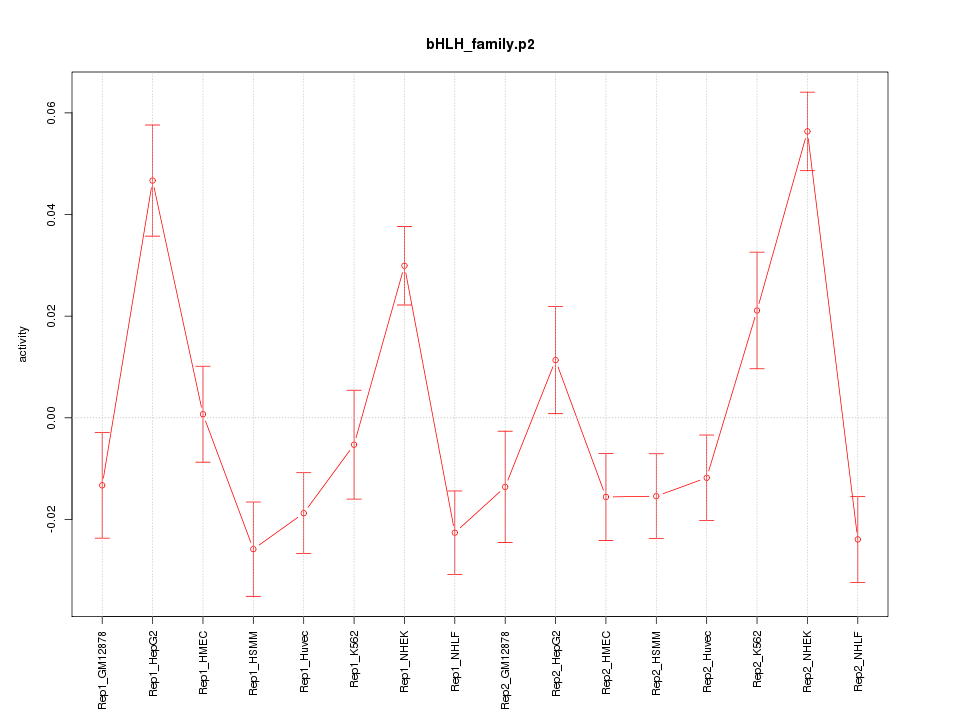
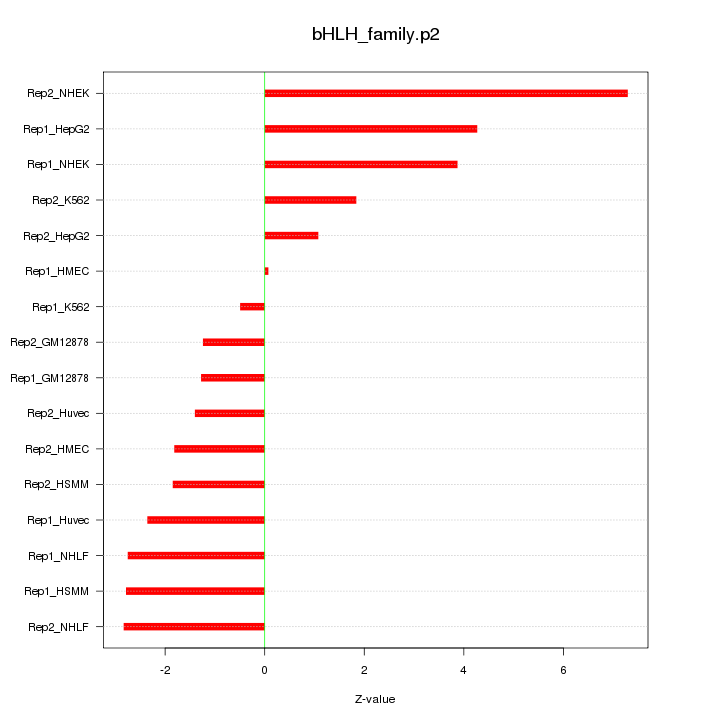
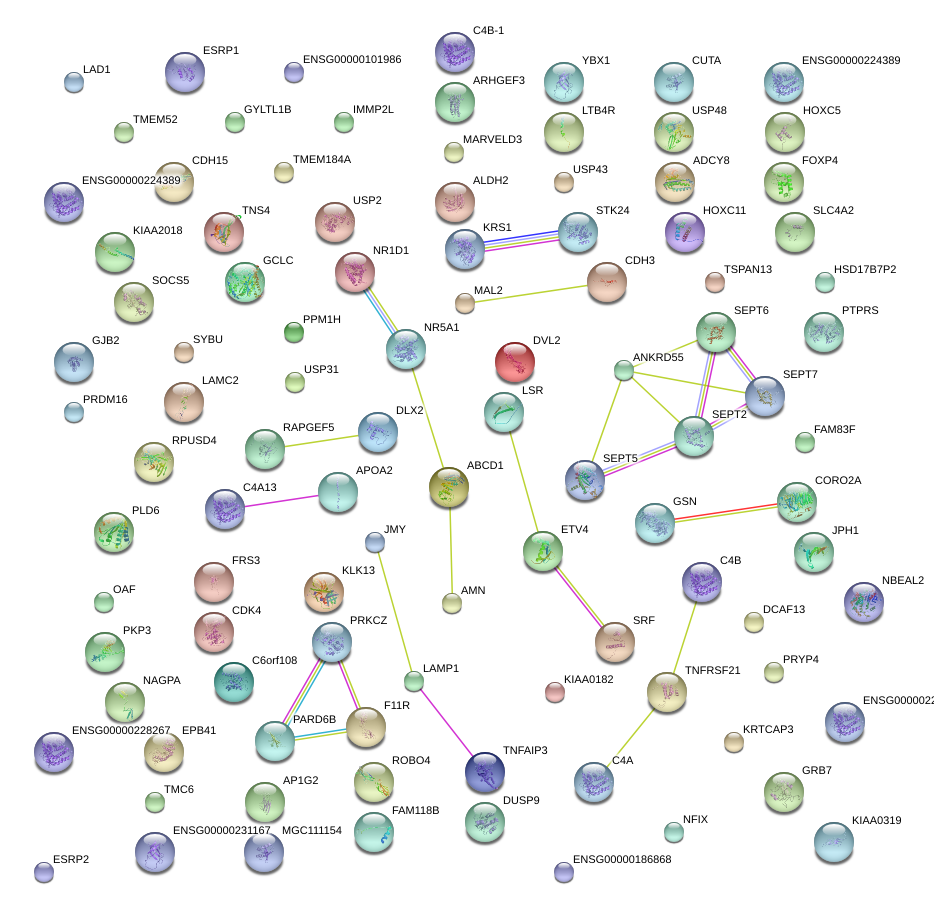
|
chr8_+_120289735
|
5.532
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr16_-_66827449
|
4.608
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr22_+_38720881
|
4.540
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr8_+_95722448
|
3.392
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr8_-_17702991
|
3.117
|
|
|
|
|
chr16_+_67236699
|
3.115
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr17_-_73636362
|
3.106
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr12_-_56432292
|
2.989
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr17_+_9489578
|
2.942
|
NM_153210
|
USP43
|
ubiquitin specific peptidase 43
|
|
chr16_-_23068010
|
2.815
|
NM_020718
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr8_+_95722575
|
2.734
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr2_-_172675873
|
2.694
|
|
DLX2
|
distal-less homeobox 2
|
|
chr8_+_95722516
|
2.690
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr12_-_56432377
|
2.643
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_56432375
|
2.604
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_56432385
|
2.600
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr11_+_384199
|
2.564
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr5_+_78567604
|
2.551
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr11_+_384238
|
2.360
|
|
PKP3
|
plakophilin 3
|
|
chr11_+_45900731
|
2.301
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr17_+_35148084
|
2.295
|
NM_001030002
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr5_+_78568094
|
2.275
|
|
|
|
|
chr2_-_172675480
|
2.257
|
NM_004405
|
DLX2
|
distal-less homeobox 2
|
|
chr19_+_40431122
|
2.216
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr19_-_5291700
|
2.177
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chrX_+_152562013
|
2.175
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr11_-_118757477
|
2.123
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr17_+_35147697
|
2.117
|
NM_005310
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr6_+_138230045
|
2.098
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr17_+_35147738
|
2.094
|
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr17_-_35510074
|
2.056
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr2_+_27518736
|
1.962
|
NM_001168364
NM_173853
|
KRTCAP3
|
keratinocyte associated protein 3
|
|
chr17_-_35510431
|
1.898
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr1_-_159257486
|
1.896
|
|
F11R
|
F11 receptor
|
|
chr1_-_159257567
|
1.892
|
|
F11R
|
F11 receptor
|
|
chr16_+_70217564
|
1.864
|
NM_001017967
NM_052858
|
MARVELD3
|
MARVEL domain containing 3
|
|
chr8_-_75396016
|
1.826
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr7_-_1562272
|
1.814
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr16_-_5023914
|
1.801
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr16_-_5023934
|
1.791
|
NM_016256
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr19_+_40431359
|
1.756
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr2_-_172572993
|
1.739
|
|
|
|
|
chr7_-_22363036
|
1.711
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr16_+_67236730
|
1.680
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr3_-_57088332
|
1.671
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr14_+_102458745
|
1.667
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr9_-_126309510
|
1.654
|
NM_004959
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1
|
|
chr17_-_38979178
|
1.651
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr17_-_73636386
|
1.645
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr2_+_172573049
|
1.633
|
NM_199227
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial)
|
|
chr6_-_47385205
|
1.616
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr1_-_159459999
|
1.576
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr7_+_16759845
|
1.561
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr19_+_40431618
|
1.554
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr1_-_159257595
|
1.552
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr6_-_33493792
|
1.548
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr6_-_43305156
|
1.544
|
NM_006443
NM_199184
|
C6orf108
|
chromosome 6 open reading frame 108
|
|
chr14_+_23855291
|
1.542
|
|
LTB4R
|
leukotriene B4 receptor
|
|
chr11_+_119586879
|
1.496
|
NM_178507
|
OAF
|
OAF homolog (Drosophila)
|
|
chr1_-_199635057
|
1.494
|
|
LAD1
|
ladinin 1
|
|
chr1_-_1840564
|
1.489
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chrX_+_130985316
|
1.476
|
NM_001042452
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr16_+_5023672
|
1.452
|
|
LOC100507589
|
hypothetical LOC100507589
|
|
chr1_-_199635240
|
1.450
|
NM_005558
|
LAD1
|
ladinin 1
|
|
chr17_-_17050326
|
1.438
|
|
PLD6
|
phospholipase D family, member 6
|
|
chr17_+_77478858
|
1.438
|
|
LOC92659
|
hypothetical LOC92659
|
|
chr1_+_42920946
|
1.436
|
|
YBX1
|
Y box binding protein 1
|
|
chr16_+_84204424
|
1.436
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr1_+_181422016
|
1.432
|
|
LAMC2
|
laminin, gamma 2
|
|
chr16_-_5023562
|
1.417
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr2_+_241903395
|
1.415
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr7_-_110989582
|
1.413
|
NM_032549
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae)
|
|
chr6_-_47385313
|
1.400
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr6_+_41622267
|
1.399
|
|
FOXP4
|
forkhead box P4
|
|
chr17_-_35911340
|
1.399
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr2_+_46779553
|
1.382
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr6_-_53517397
|
1.380
|
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr1_-_199635339
|
1.380
|
|
LAD1
|
ladinin 1
|
|
chr9_-_99994704
|
1.379
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr17_-_7078446
|
1.373
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr19_+_12966970
|
1.358
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr12_+_52713098
|
1.351
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr17_-_7078304
|
1.347
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr11_-_118757574
|
1.338
|
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr17_-_7078399
|
1.333
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr1_+_29113589
|
1.319
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr11_-_125586733
|
1.316
|
NM_001144827
NM_032795
|
RPUSD4
|
RNA pseudouridylate synthase domain containing 4
|
|
chr1_+_1971762
|
1.312
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr17_-_17050355
|
1.296
|
NM_178836
|
PLD6
|
phospholipase D family, member 6
|
|
chr13_-_19665036
|
1.295
|
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr12_-_61614930
|
1.287
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr8_-_110726063
|
1.281
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr6_+_32090516
|
1.275
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr14_-_23106372
|
1.268
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr20_+_48781457
|
1.257
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr12_+_52653176
|
1.256
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr13_+_112999538
|
1.250
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr12_-_61615124
|
1.244
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr3_+_46996150
|
1.243
|
NM_015175
|
NBEAL2
|
neurobeachin-like 2
|
|
chr7_+_150390566
|
1.236
|
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr11_+_125586828
|
1.229
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr7_-_23476497
|
1.224
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr6_-_33493898
|
1.213
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr1_-_12599934
|
1.211
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr6_-_10527772
|
1.210
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr19_+_7608010
|
1.196
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr13_+_112999590
|
1.186
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr2_+_17585287
|
1.169
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr2_-_220116508
|
1.168
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr6_-_138862271
|
1.161
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr2_-_171999473
|
1.159
|
NM_024770
|
METTL8
|
methyltransferase like 8
|
|
chr14_-_54438913
|
1.158
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr8_-_144887876
|
1.156
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr2_+_218992053
|
1.155
|
NM_007127
|
VIL1
|
villin 1
|
|
chr2_+_17585462
|
1.154
|
|
VSNL1
|
visinin-like 1
|
|
chr1_-_21868380
|
1.153
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr1_-_158182006
|
1.151
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chr6_-_53517466
|
1.146
|
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr12_-_83830700
|
1.143
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr1_+_65385799
|
1.142
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr5_+_3649167
|
1.141
|
NM_024337
|
IRX1
|
iroquois homeobox 1
|
|
chr19_+_7608015
|
1.138
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr17_-_34084682
|
1.137
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr6_+_41622111
|
1.136
|
NM_001012426
NM_001012427
NM_138457
|
FOXP4
|
forkhead box P4
|
|
chr12_+_52618815
|
1.136
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr20_+_43953606
|
1.135
|
|
CTSA
|
cathepsin A
|
|
chr6_+_160310118
|
1.122
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr16_+_29726195
|
1.112
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr6_+_105511615
|
1.109
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr12_-_83830710
|
1.109
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr19_-_38485161
|
1.107
|
|
|
|
|
chr17_-_68769613
|
1.104
|
NM_001129885
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like
|
|
chr1_-_11042555
|
1.097
|
|
SRM
|
spermidine synthase
|
|
chr3_-_49916304
|
1.095
|
NM_002447
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase)
|
|
chr19_+_7607976
|
1.094
|
NM_001127396
NM_006949
|
STXBP2
|
syntaxin binding protein 2
|
|
chr12_-_83830670
|
1.092
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr17_+_36599664
|
1.090
|
NM_001190460
|
KRTAP9-1
|
keratin associated protein 9-1
|
|
chr5_+_70918856
|
1.090
|
NM_022132
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr1_+_181421984
|
1.088
|
|
LAMC2
|
laminin, gamma 2
|
|
chr17_-_41625921
|
1.087
|
NM_001193466
|
KIAA1267
|
KIAA1267
|
|
chr1_+_61320213
|
1.087
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr16_-_21221872
|
1.075
|
NM_001014444
NM_001888
|
CRYM
|
crystallin, mu
|
|
chr17_-_18848713
|
1.074
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr2_+_219751230
|
1.073
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr8_-_110726201
|
1.066
|
NM_001099753
NM_001099754
NM_001099755
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr2_-_178645718
|
1.066
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr1_+_3360880
|
1.064
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr6_-_47385470
|
1.059
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr6_-_138231006
|
1.055
|
|
|
|
|
chr11_-_116168316
|
1.051
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr3_+_160002571
|
1.044
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr6_+_7486862
|
1.040
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr19_-_12747421
|
1.039
|
NM_001100176
NM_013312
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr6_-_47385605
|
1.038
|
NM_014452
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr17_+_45911179
|
1.037
|
NM_018346
|
RSAD1
|
radical S-adenosyl methionine domain containing 1
|
|
chr6_+_7486836
|
1.033
|
|
DSP
|
desmoplakin
|
|
chr6_-_47385599
|
1.030
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr12_-_63439449
|
1.030
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr16_+_56616782
|
1.029
|
NM_002428
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted)
|
|
chr17_+_45911216
|
1.029
|
|
RSAD1
|
radical S-adenosyl methionine domain containing 1
|
|
chr1_-_27113018
|
1.028
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr6_-_35996915
|
1.027
|
NM_003137
|
SRPK1
|
SRSF protein kinase 1
|
|
chr6_-_47385604
|
1.027
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr6_+_87921980
|
1.023
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr17_-_42250961
|
1.023
|
NM_030753
|
WNT3
|
wingless-type MMTV integration site family, member 3
|
|
chr17_+_69939806
|
1.022
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr16_+_65471881
|
1.021
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chr20_+_46971618
|
1.019
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr16_+_68353738
|
1.015
|
NM_007014
NM_199423
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr19_+_7608020
|
1.014
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr2_-_220116062
|
1.006
|
NM_001195731
|
CHPF
|
chondroitin polymerizing factor
|
|
chr20_+_43953331
|
1.002
|
|
CTSA
|
cathepsin A
|
|
chr5_+_70918888
|
0.998
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr3_+_10832867
|
0.998
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr11_+_76171780
|
0.992
|
NM_015516
|
TSKU
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis)
|
|
chr6_-_28999388
|
0.990
|
|
TRIM27
|
tripartite motif containing 27
|
|
chr22_-_36907638
|
0.989
|
NM_001004426
NM_003560
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent)
|
|
chr20_+_43028533
|
0.987
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr20_+_43953428
|
0.985
|
|
CTSA
|
cathepsin A
|
|
chr6_-_138862273
|
0.984
|
|
|
|
|
chr6_-_53517793
|
0.981
|
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr19_-_7607852
|
0.976
|
|
|
|
|
chr3_-_15875932
|
0.972
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr6_-_30151526
|
0.966
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr2_-_208342296
|
0.964
|
NM_003468
|
FZD5
|
frizzled homolog 5 (Drosophila)
|
|
chr1_-_24307514
|
0.961
|
|
MYOM3
|
myomesin family, member 3
|
|
chr6_-_35996867
|
0.959
|
|
SRPK1
|
SRSF protein kinase 1
|
|
chr6_+_32229771
|
0.959
|
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr12_-_63439456
|
0.955
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr3_-_69674412
|
0.952
|
|
FRMD4B
|
FERM domain containing 4B
|
|
chr1_+_27541049
|
0.951
|
NM_001193308
NM_032872
|
SYTL1
|
synaptotagmin-like 1
|
|
chr2_+_219751207
|
0.950
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr10_+_5478521
|
0.949
|
NM_005863
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr12_-_63439415
|
0.948
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr2_+_74610039
|
0.948
|
NM_013247
NM_145074
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr1_+_27541086
|
0.947
|
|
SYTL1
|
synaptotagmin-like 1
|
|
chr16_+_29726962
|
0.946
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr2_-_74522513
|
0.945
|
|
RTKN
|
rhotekin
|
|
chr16_+_57106883
|
0.944
|
NM_001160305
NM_024860
|
SETD6
|
SET domain containing 6
|
|
chrX_+_130984912
|
0.943
|
NM_001042453
NM_016542
|
MST4
|
serine/threonine protein kinase MST4
|