Project
DANIO-CODE
Navigation
Downloads
ISMARA results DANIO-CODE
ISMARA - Integrated System for Motif Actitivity Response Analysis is a free online tool that recognizes most important transcription factors that are changing their activity in a set of samples.
All motifs sorted by activity significance
| Motif name | Z-value | Associated genes | Profile | Logo |
|---|---|---|---|---|
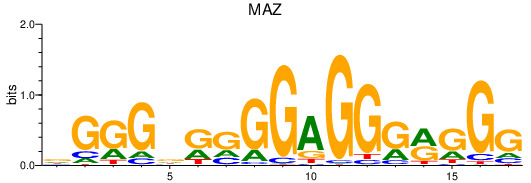
| MAZ | 6.68 |
|

|
|
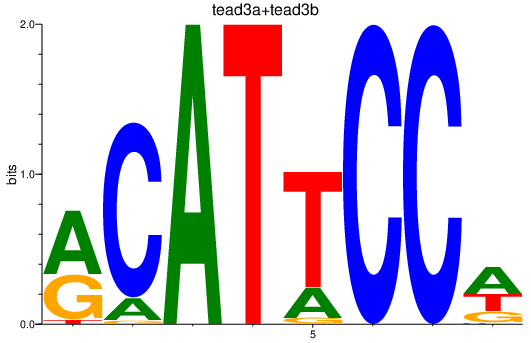
| tead3a+tead3b | 6.25 |
|

|
|
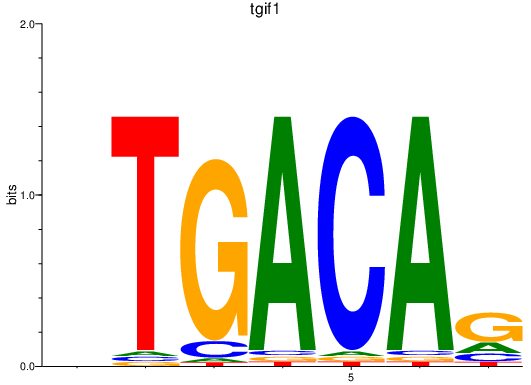
| tgif1_meis2a_meis1a | 6.12 |
|

|
|
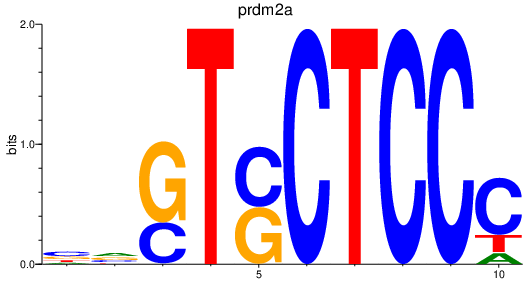
| prdm2a | 5.70 |
|

|
|
| creb1a+creb1b | 5.52 |
|

|

|
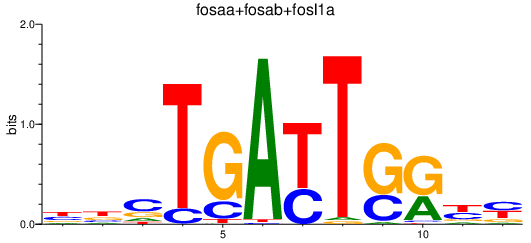
| fosaa+fosab+fosl1a_nfya_nfyal | 5.34 |
|

|
|
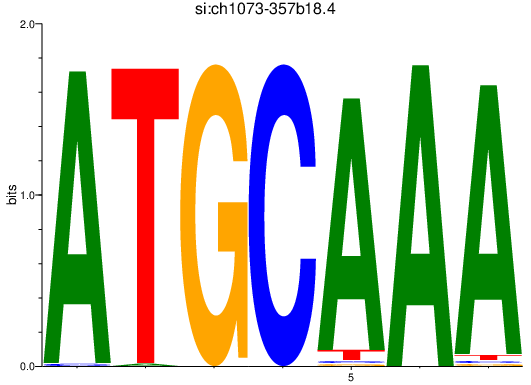
| si:ch1073-357b18.4 | 4.36 |
|

|
|
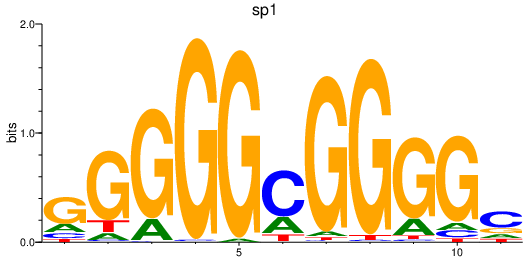
| sp1 | 4.19 |
|

|
|
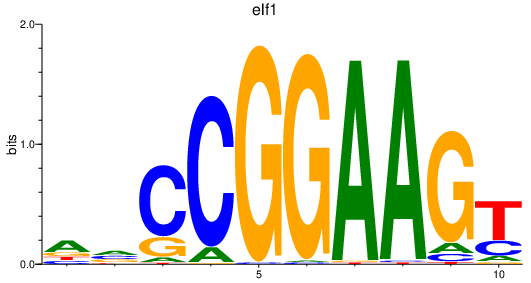
| elf1_etv1_elk3_elk1_erf+erfl3_etv5a+etv5b_gabpa_elk4 | 4.10 |
|

|
|
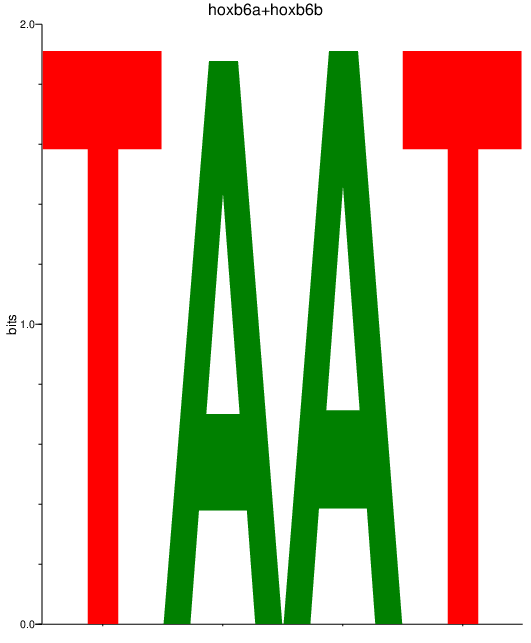
| hoxb6a+hoxb6b | 4.02 |
|

|
|
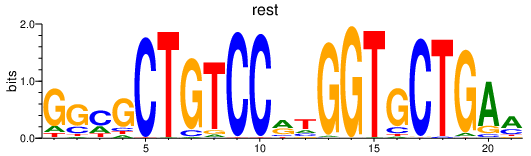
| rest | 3.70 |
|
|
|
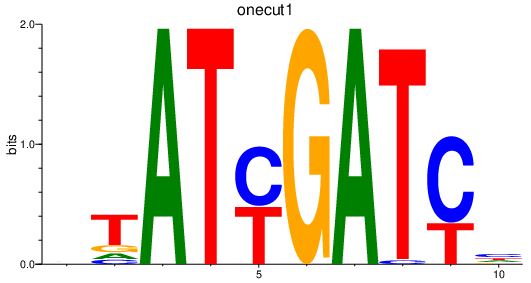
| onecut1 | 3.65 |
|

|
|
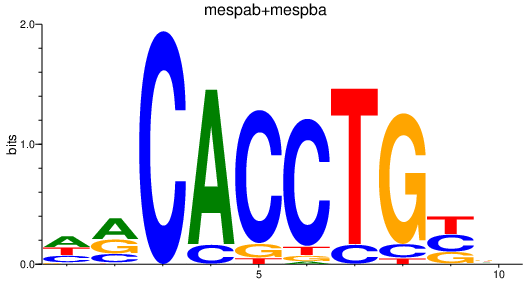
| mespab+mespba_id4 | 3.60 |
|

|
|
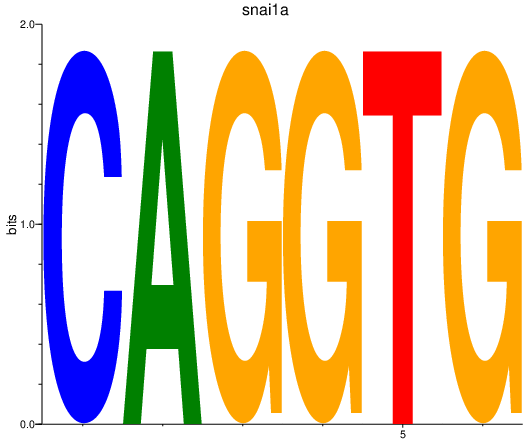
| snai1a | 3.51 |
|

|
|
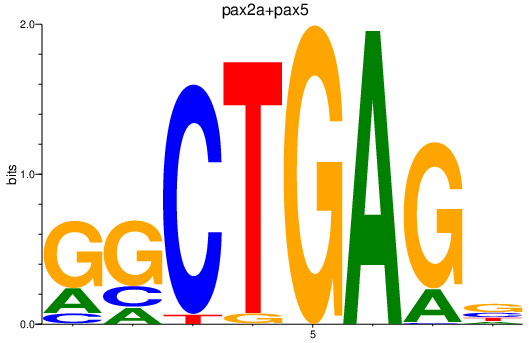
| pax2a+pax5 | 3.36 |
|
|
|
| elf2a+elf2b | 3.33 |
|

|

|
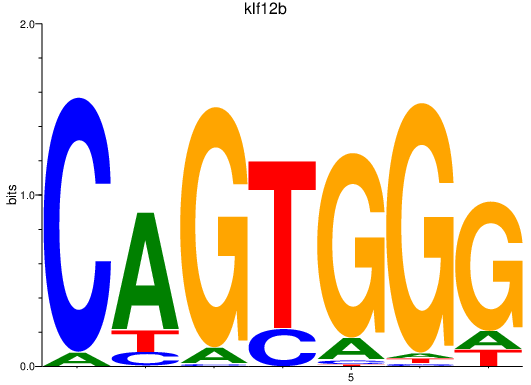
| klf12b | 3.27 |
|

|
|
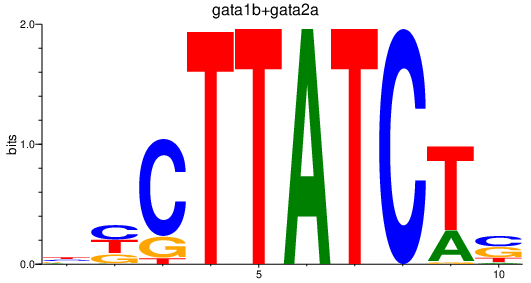
| gata1b+gata2a | 3.21 |
|

|
|
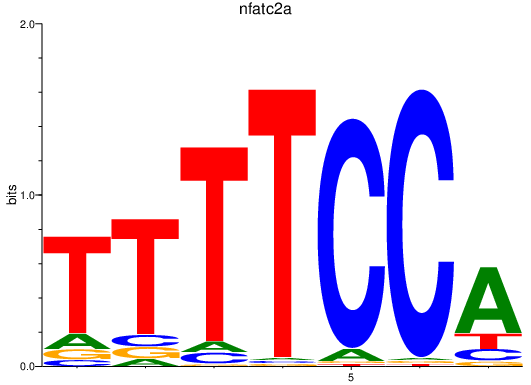
| nfatc2a | 3.16 |
|

|
|
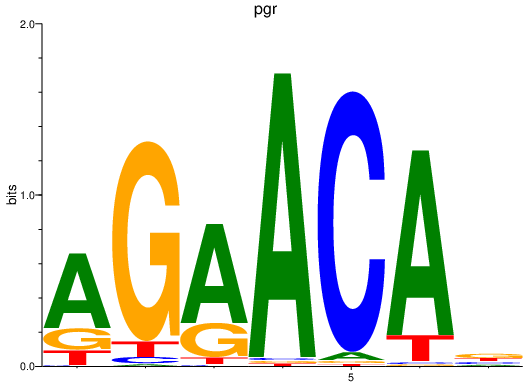
| pgr | 3.14 |
|
|
|
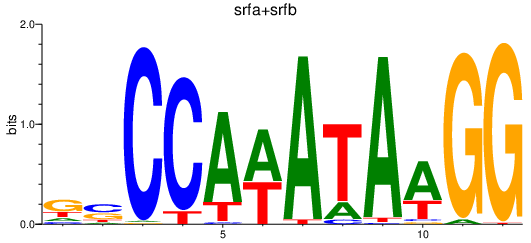
| srfa+srfb | 3.12 |
|
|
|
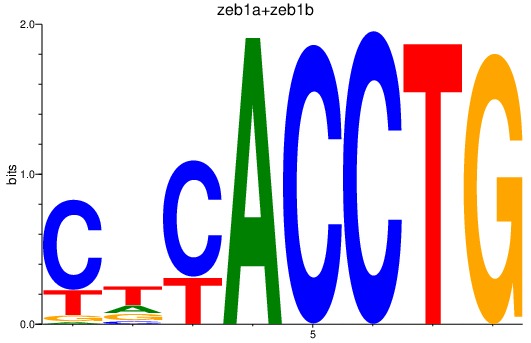
| zeb1a+zeb1b | 3.11 |
|

|
|
| foxp3b | 3.10 |
|

|

|
| atf2_atf1_atf3_crema+cremb | 3.06 |
|

|
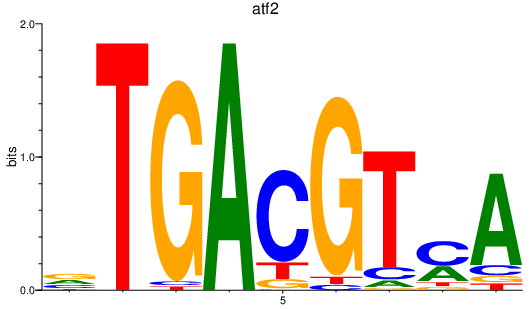
|
| mef2d_mef2ca+mef2cb | 3.04 |
|

|
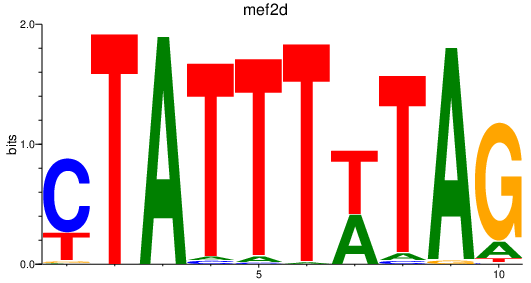
|
| pbx1a+pbx2+pbx3a+pbx3b | 3.01 |
|

|

|
| lef1 | 2.96 |
|

|
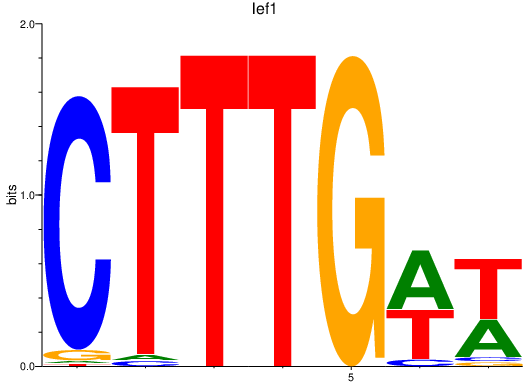
|
| sox19a+sox19b+sox3 | 2.92 |
|

|
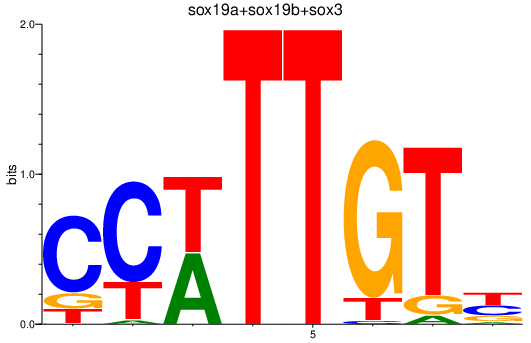
|
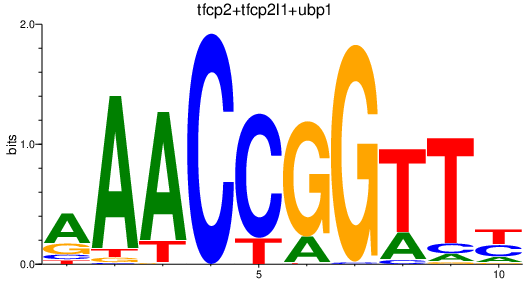
| tfcp2+tfcp2l1+ubp1 | 2.91 |
|

|
|
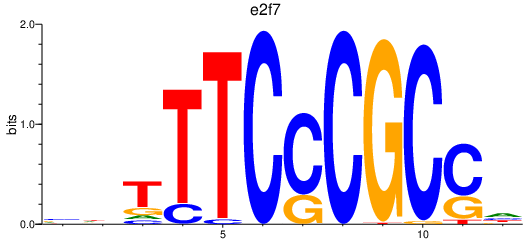
| e2f7_e2f1 | 2.85 |
|

|
|
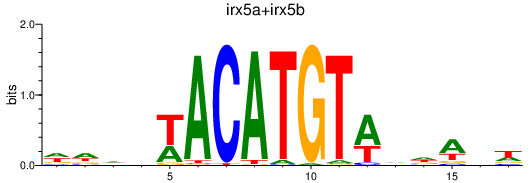
| irx5a+irx5b_irx3b | 2.75 |
|

|
|
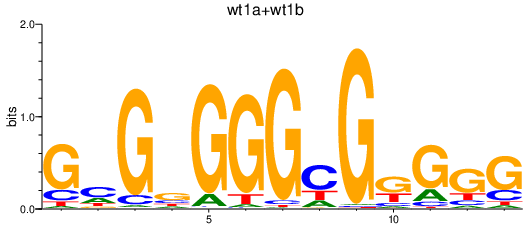
| wt1a+wt1b | 2.74 |
|

|
|
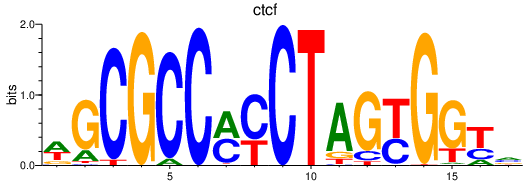
| ctcf | 2.73 |
|

|
|
| si:ch211-69l10.4+tbx15_tbx4_tbx1_mgaa | 2.71 |
|

|

|
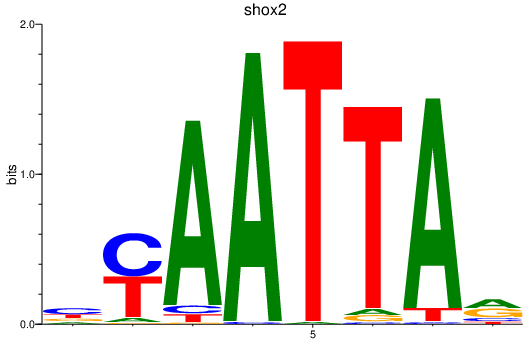
| shox2_uncx4.1_lhx2a_vsx1_shox | 2.71 |
|

|
|
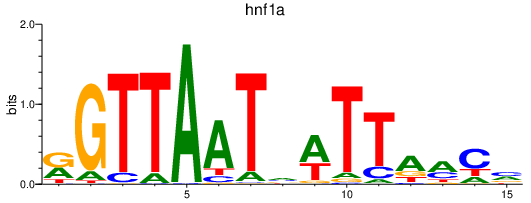
| hnf1a | 2.70 |
|

|
|
| hnf1ba+hnf1bb | 2.65 |
|
|
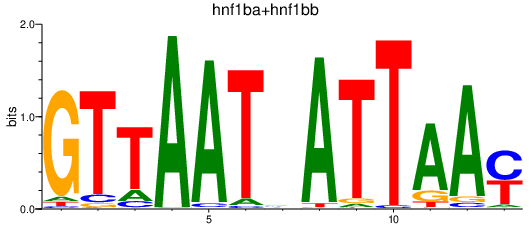
|
| barhl1a+barhl1b | 2.61 |
|

|
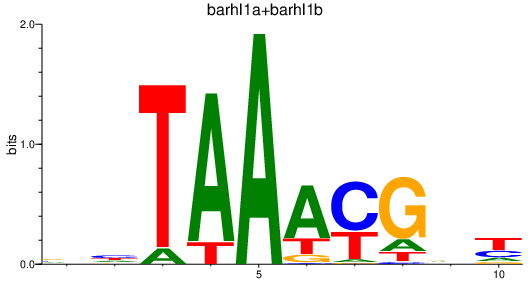
|
| mybl2b | 2.61 |
|

|
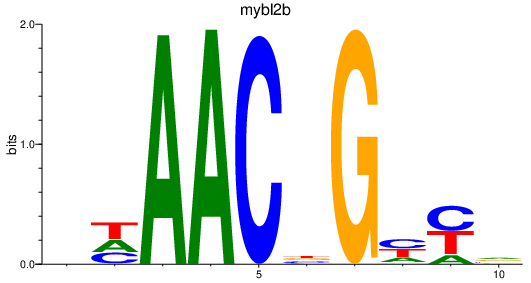
|
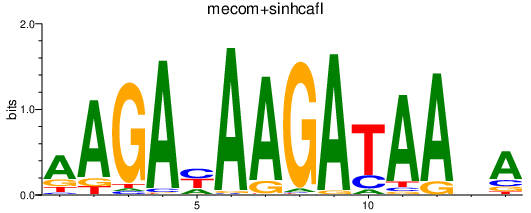
| mecom+sinhcafl | 2.59 |
|
|
|
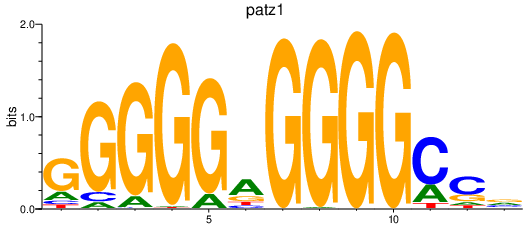
| patz1 | 2.58 |
|

|
|
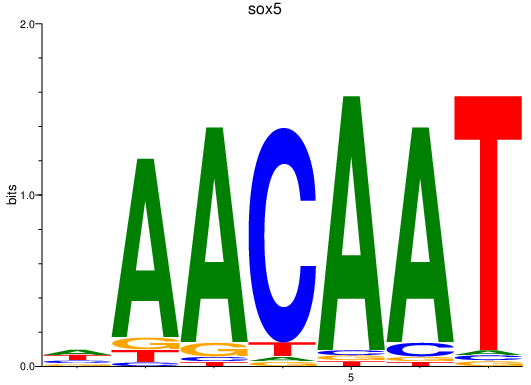
| sox5 | 2.57 |
|

|
|
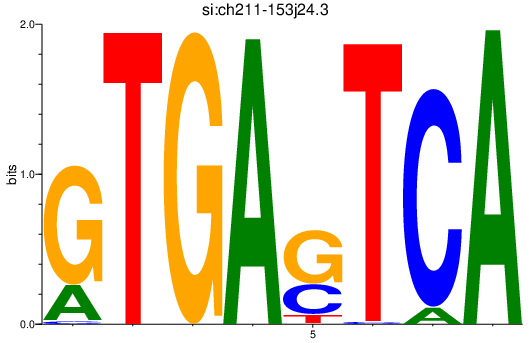
| si:ch211-153j24.3 | 2.56 |
|

|
|
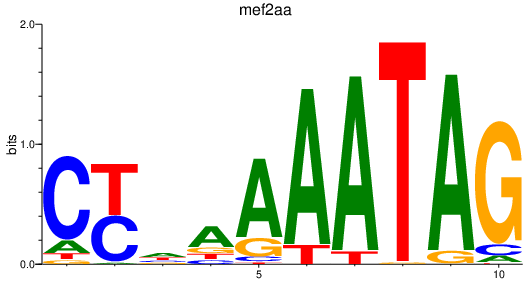
| mef2aa | 2.55 |
|
|
|
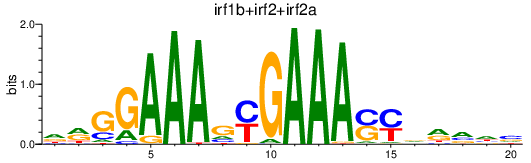
| irf1b+irf2+irf2a | 2.53 |
|
|
|
| CABZ01079241.1+zfx+znf711 | 2.53 |
|

|

|
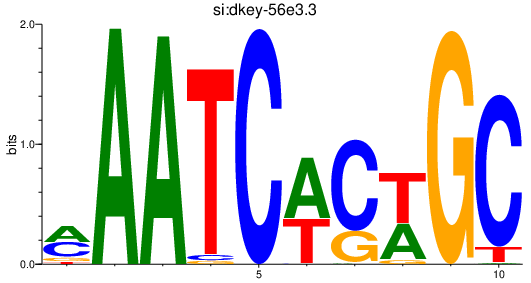
| si:dkey-56e3.3 | 2.52 |
|

|
|
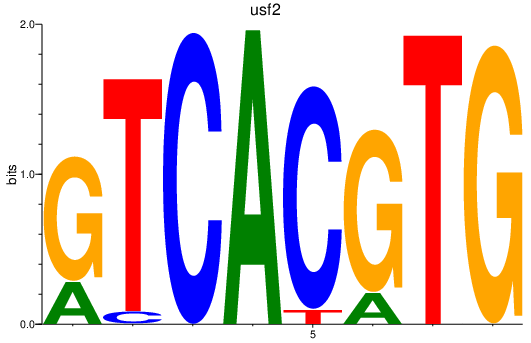
| usf2_srebf1_tfeb_max+si:ch73-59f11.3_mlxipl_usf1l_usf1_bhlhe41_mitfa+mitfb_mnta_mycn_mlx_mxi1_tfec | 2.52 |
|

|
|
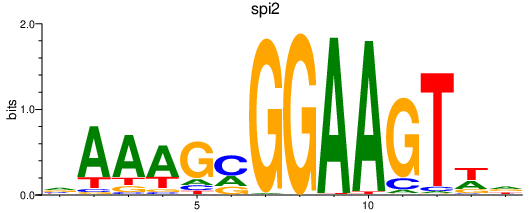
| spi2 | 2.48 |
|

|
|
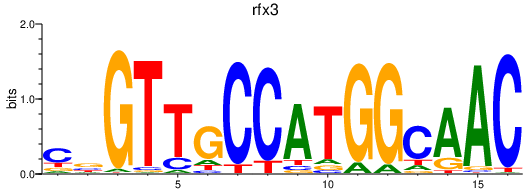
| rfx3_rfx1a_rfx2 | 2.45 |
|

|
|
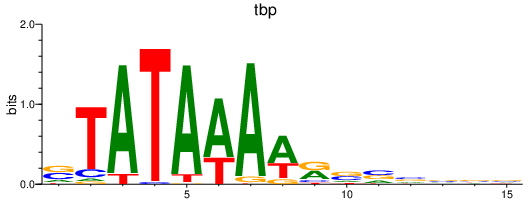
| tbp | 2.45 |
|

|
|
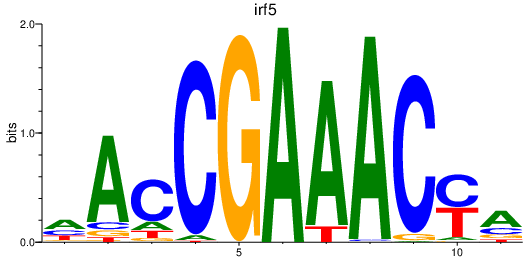
| irf5 | 2.42 |
|

|
|
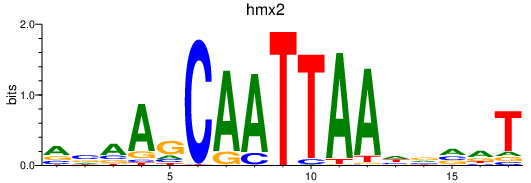
| hmx2_hmx3a | 2.42 |
|

|
|
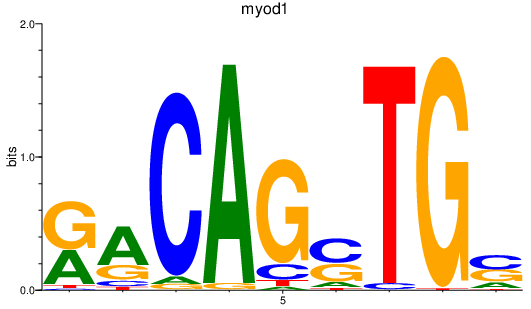
| myod1 | 2.41 |
|
|
|
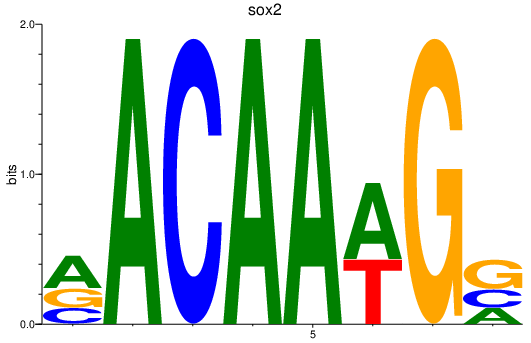
| sox2 | 2.37 |
|

|
|
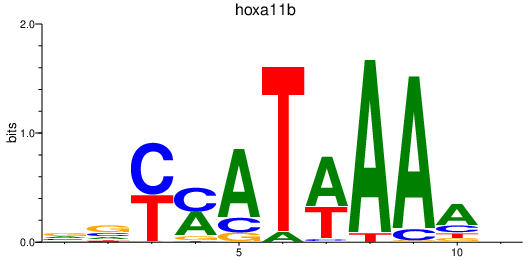
| hoxa11b | 2.30 |
|

|
|
| tfdp1a | 2.29 |
|
|
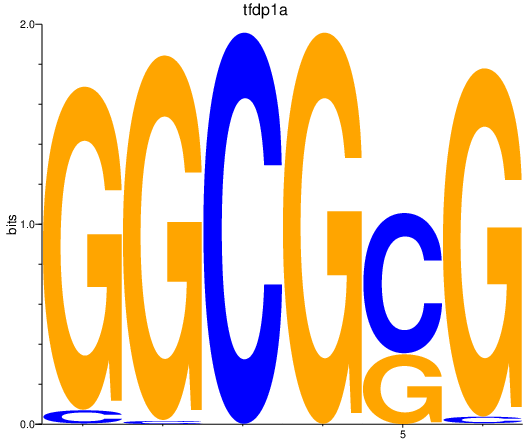
|
| zbtb14 | 2.26 |
|

|
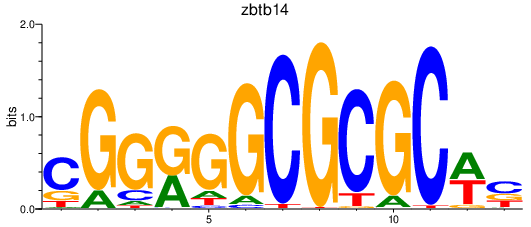
|
| e2f5_si:ch211-160f23.5 | 2.15 |
|

|
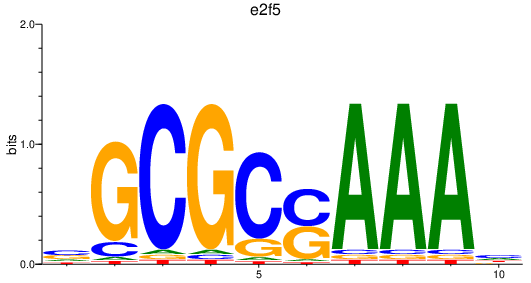
|
| clocka+clockb | 2.12 |
|

|
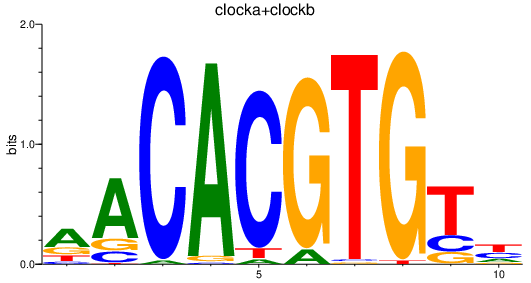
|
| foxa1_foxa_foxa2 | 2.11 |
|

|
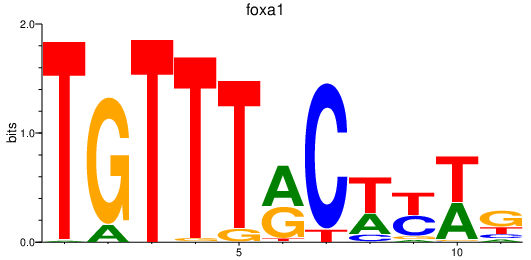
|
| atf5a+atf5b | 2.10 |
|

|
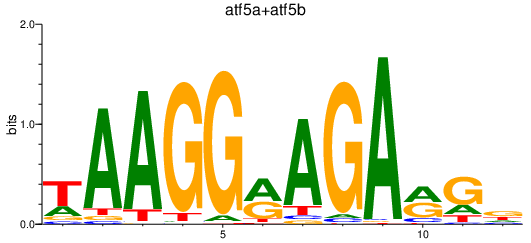
|
| GMEB2 | 2.10 |
|

|
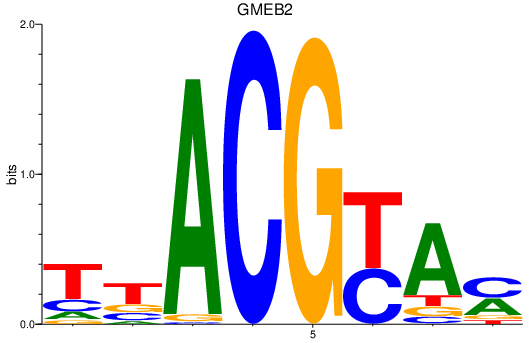
|
| hoxd13a | 2.05 |
|

|
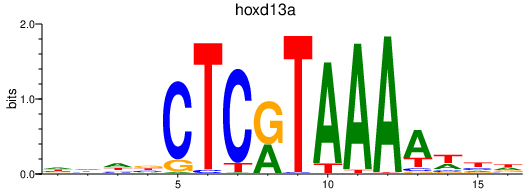
|
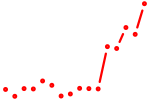
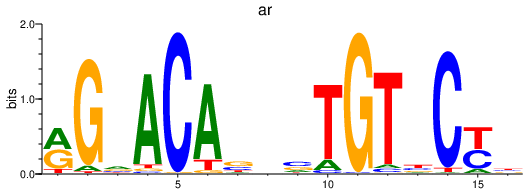
| ar_nr3c1 | 2.04 |
|
|
|
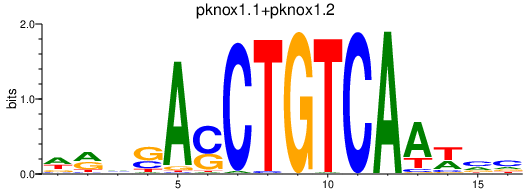
| pknox1.1+pknox1.2_meis2b | 2.04 |
|

|
|
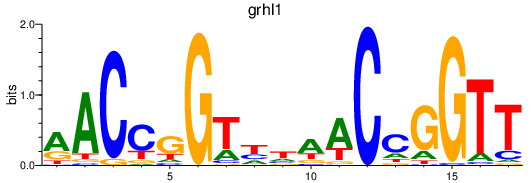
| grhl1 | 2.01 |
|

|
|
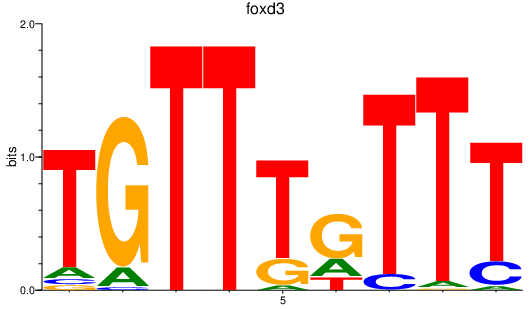
| foxd3 | 1.97 |
|

|
|
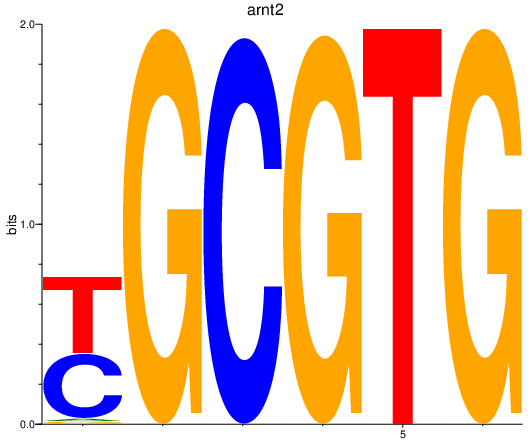
| arnt2 | 1.96 |
|

|
|
| sp2 | 1.93 |
|

|

|
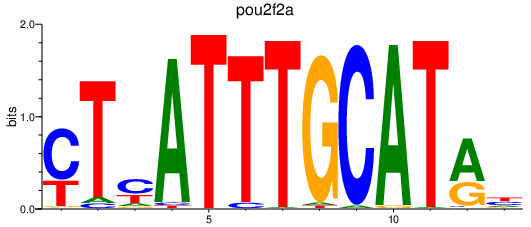
| pou2f2a | 1.90 |
|

|
|
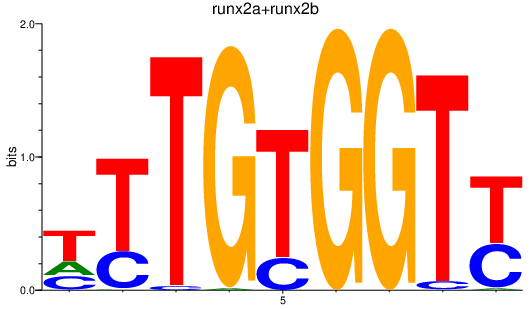
| runx2a+runx2b | 1.89 |
|

|
|
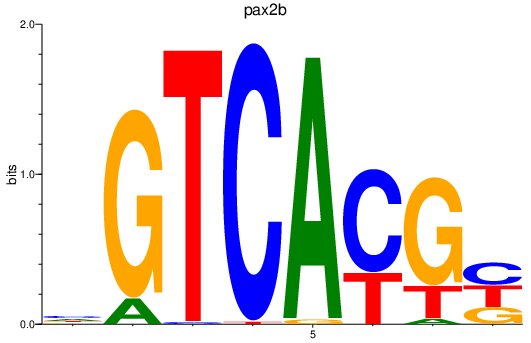
| pax2b | 1.85 |
|
|
|
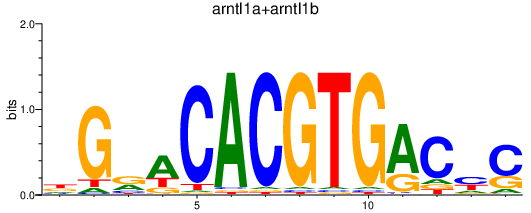
| arntl1a+arntl1b | 1.84 |
|
|
|
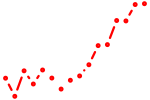
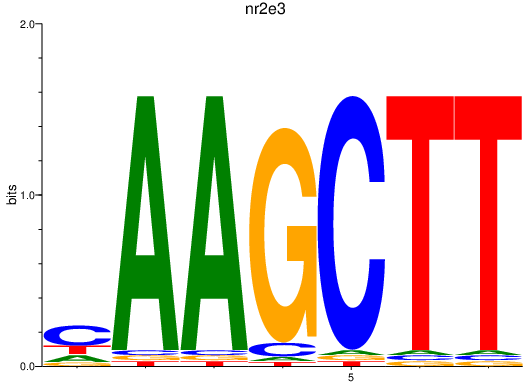
| nr2e3 | 1.83 |
|
|
|
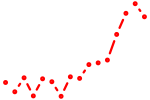
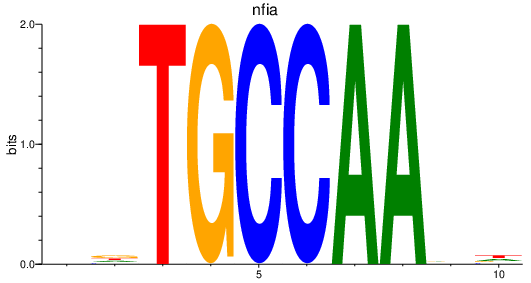
| nfia | 1.82 |
|
|
|
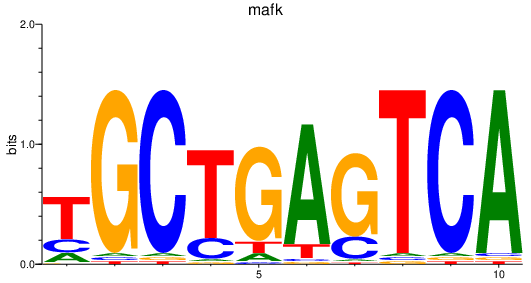
| mafk | 1.82 |
|

|
|
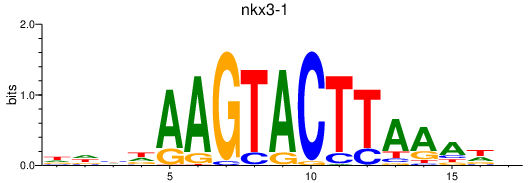
| nkx3-1 | 1.80 |
|

|
|
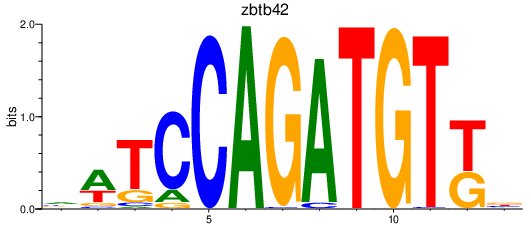
| zbtb42 | 1.78 |
|

|
|
| si:dkey-229b18.3+snai2 | 1.77 |
|

|
|
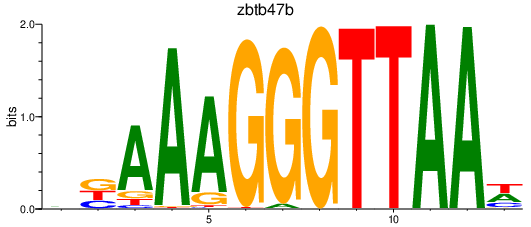
| zbtb47b | 1.76 |
|

|
|
| myf6 | 1.74 |
|
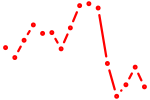
|
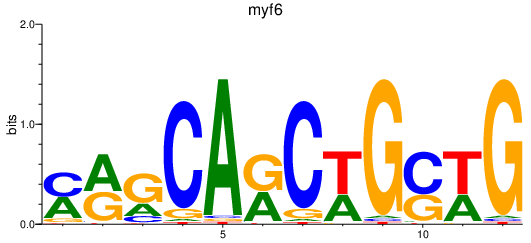
|
| znf740a+znf740b | 1.73 |
|
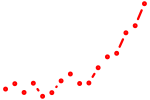
|
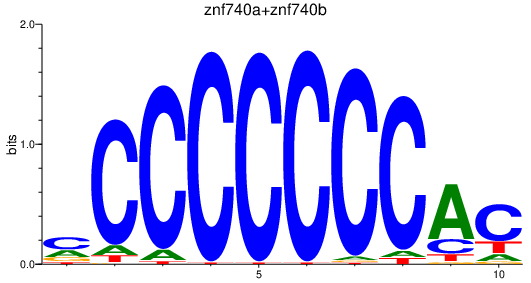
|
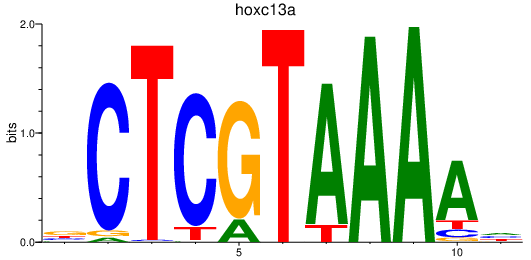
| hoxc13a | 1.73 |
|

|
|
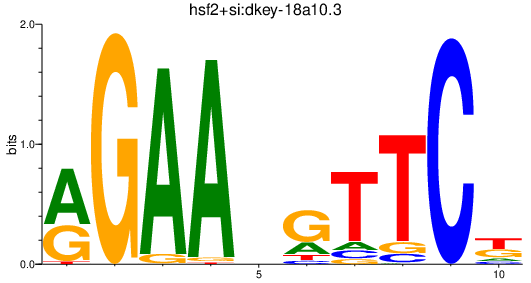
| hsf2+si:dkey-18a10.3 | 1.72 |
|
|
|
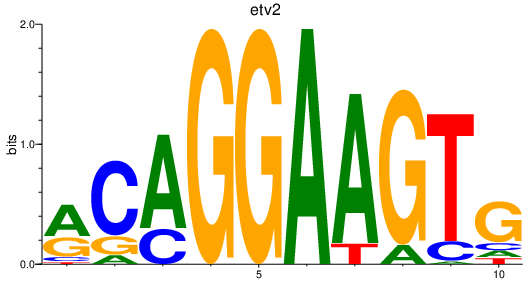
| etv2_erg_fev+fli1a | 1.71 |
|

|
|
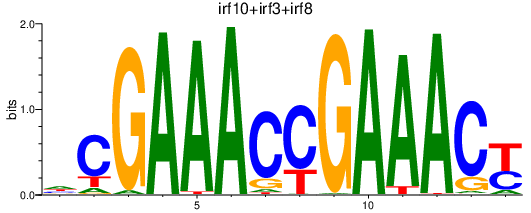
| irf10+irf3+irf8 | 1.71 |
|

|
|
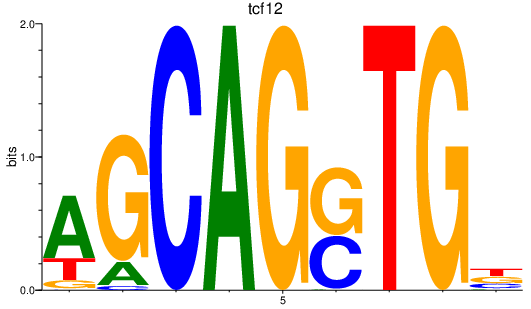
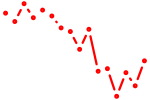
| tcf12 | 1.67 |
|

|
|
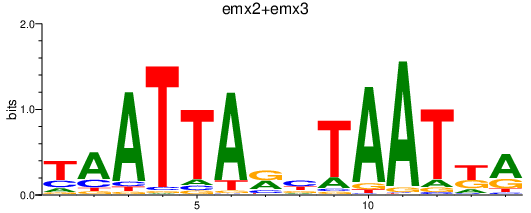
| emx2+emx3 | 1.67 |
|
|
|
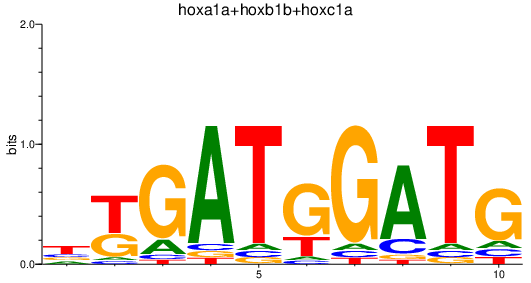
| hoxa1a+hoxb1b+hoxc1a | 1.66 |
|

|
|
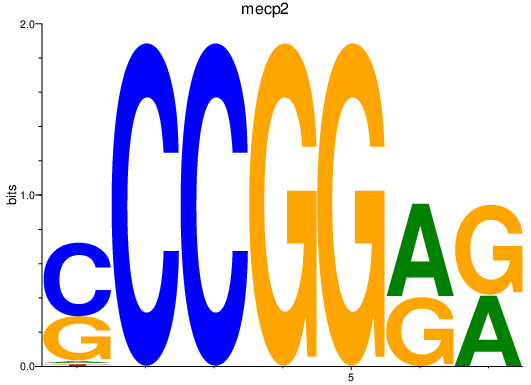
| mecp2 | 1.65 |
|
|
|
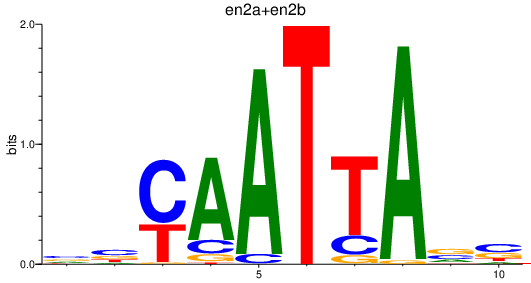
| en2a+en2b_gbx1_en1b_emx1 | 1.64 |
|

|
|
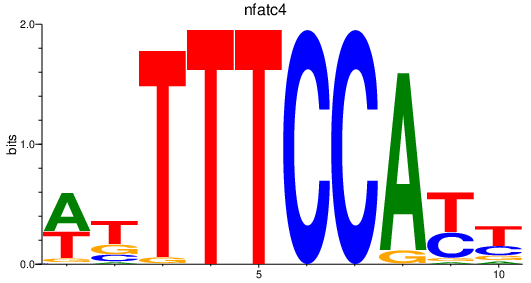
| nfatc4 | 1.63 |
|

|
|
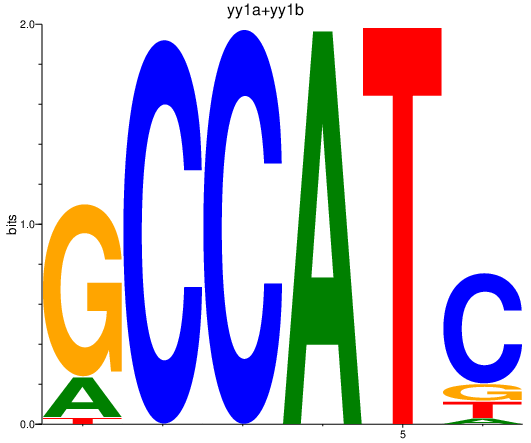
| yy1a+yy1b | 1.63 |
|
|
|
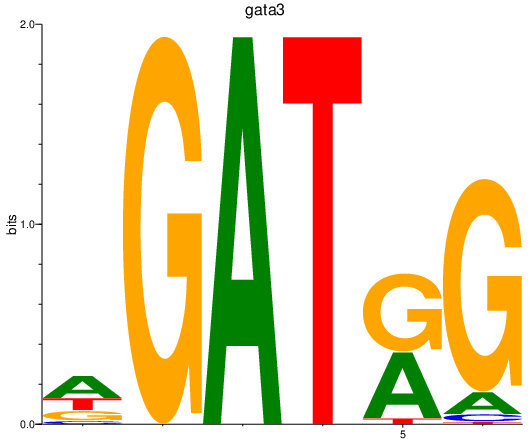
| gata3 | 1.62 |
|

|
|
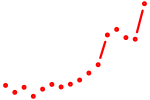
| ikzf2 | 1.62 |
|
|
|
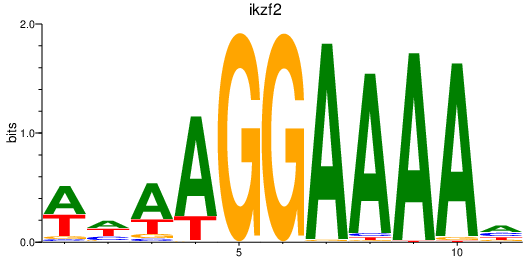
| hmx4 | 1.61 |
|

|
|
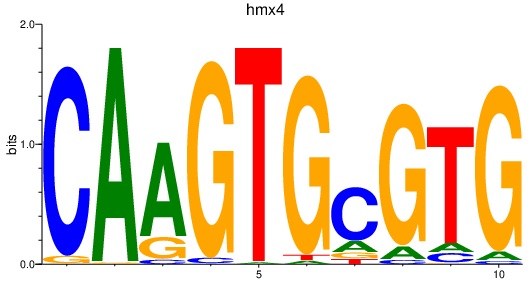
| tp53 | 1.61 |
|

|
|
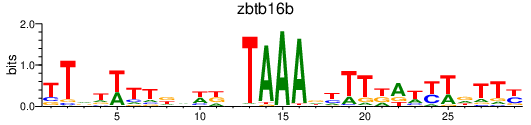
| zbtb16b | 1.60 |
|

|
|
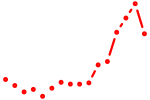
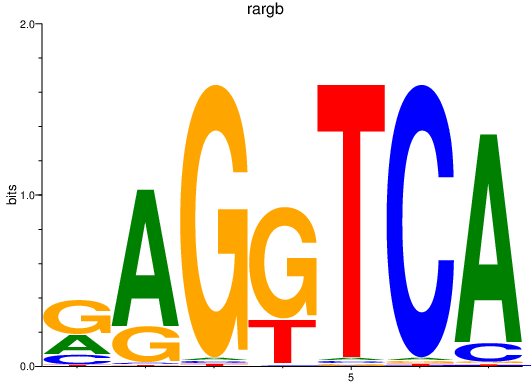
| rargb_pparab | 1.60 |
|
|
|
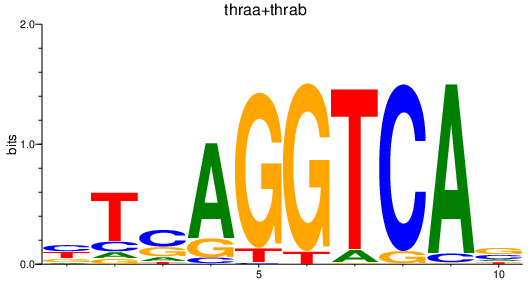
| thraa+thrab | 1.59 |
|

|
|
| esrrb_esrra+esrrgb_esrrd+esrrga | 1.59 |
|
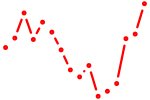
|
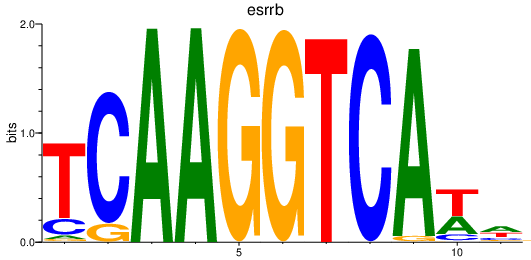
|
| mafb+nrl | 1.59 |
|

|
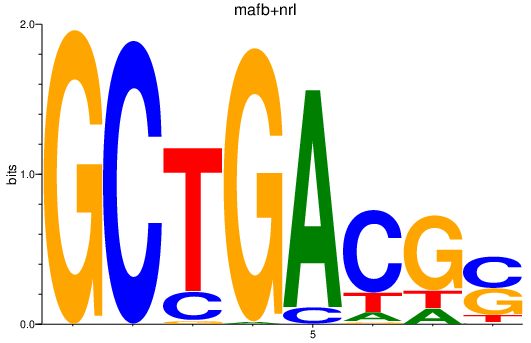
|
| ebf3b | 1.57 |
|

|
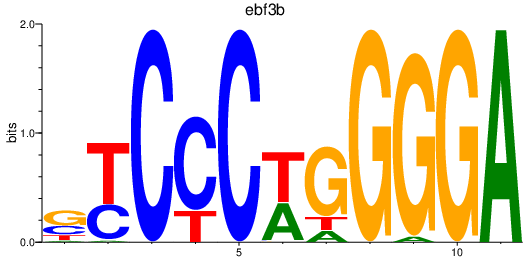
|
| si:dkey-43p13.5+uncx | 1.57 |
|

|

|
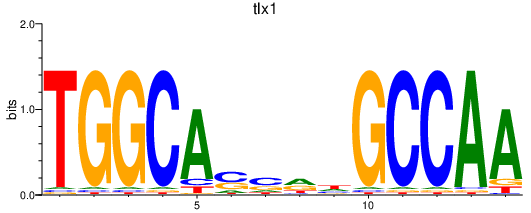
| tlx1 | 1.56 |
|

|
|
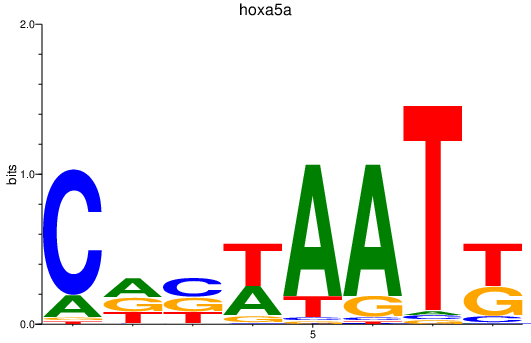
| hoxa5a | 1.55 |
|

|
|
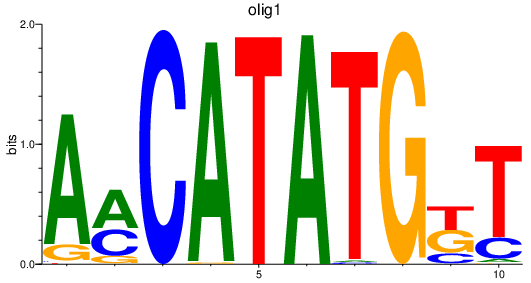
| olig1_olig2+olig4 | 1.55 |
|

|
|
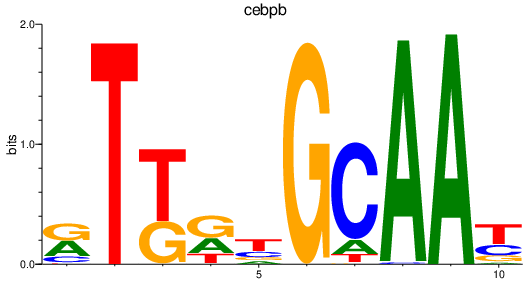
| cebpb | 1.54 |
|
|
|
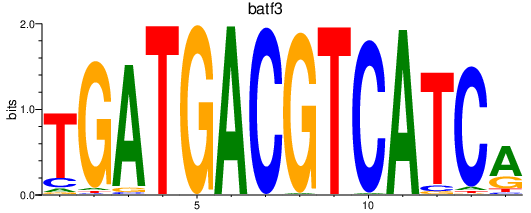
| batf3 | 1.53 |
|

|
|
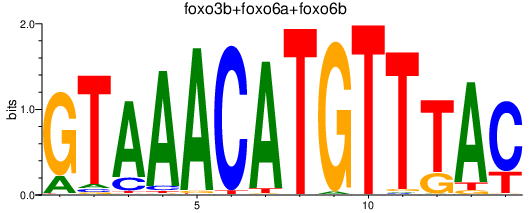
| foxo3b+foxo6a+foxo6b | 1.48 |
|

|
|
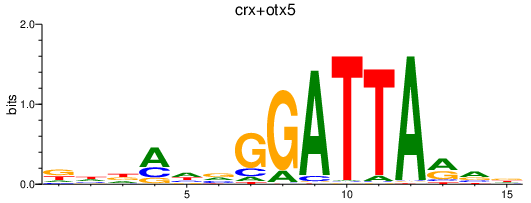
| crx+otx5 | 1.48 |
|
|
|
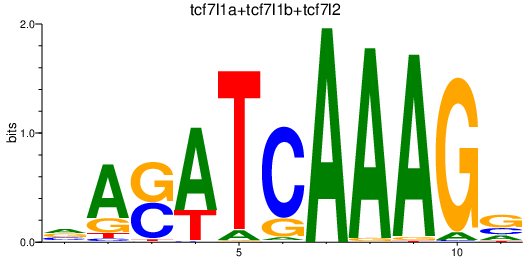
| tcf7l1a+tcf7l1b+tcf7l2 | 1.48 |
|

|
|
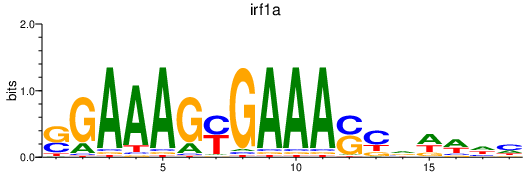
| irf1a | 1.47 |
|

|
|
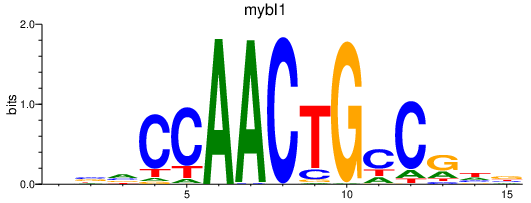
| mybl1 | 1.46 |
|

|
|
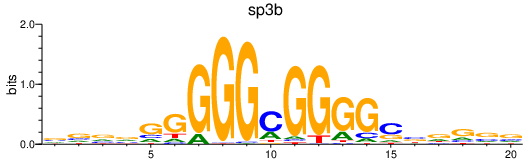
| sp3b | 1.45 |
|

|
|
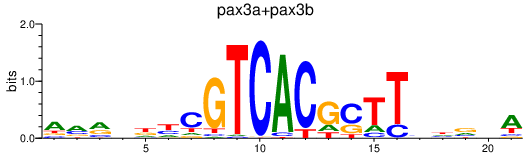
| pax3a+pax3b | 1.45 |
|

|
|
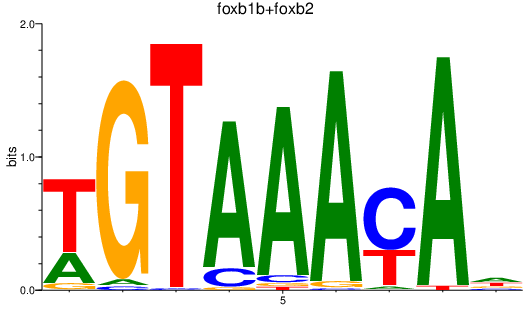
| foxb1b+foxb2 | 1.44 |
|

|
|
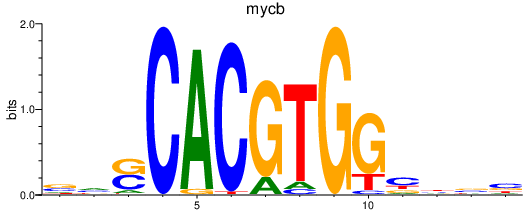
| mycb | 1.42 |
|
|
|
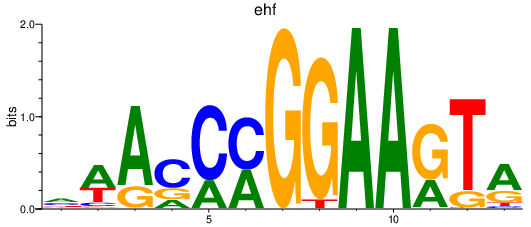
| ehf | 1.41 |
|

|
|
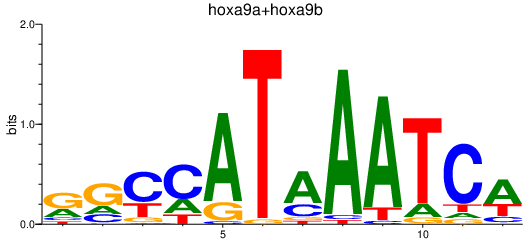
| hoxa9a+hoxa9b | 1.41 |
|

|
|
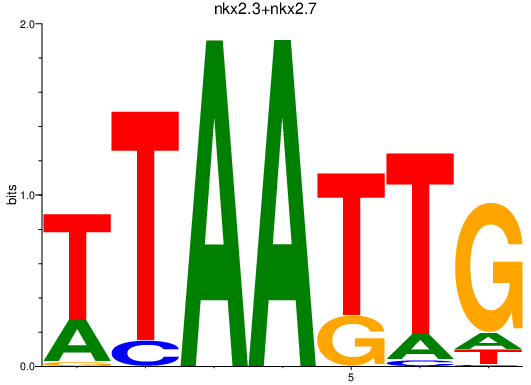
| nkx2.3+nkx2.7 | 1.40 |
|

|
|
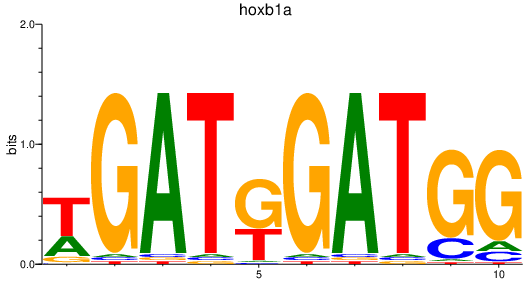
| hoxb1a | 1.40 |
|

|
|
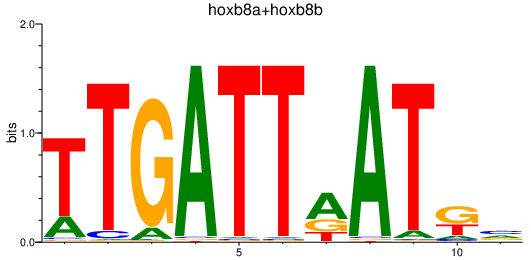
| hoxb8a+hoxb8b | 1.40 |
|
|
|
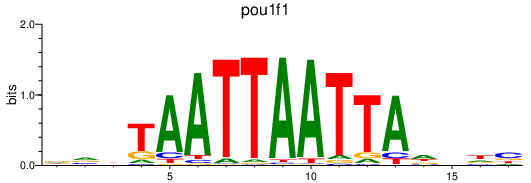
| pou1f1 | 1.38 |
|

|
|
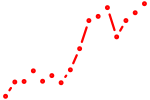
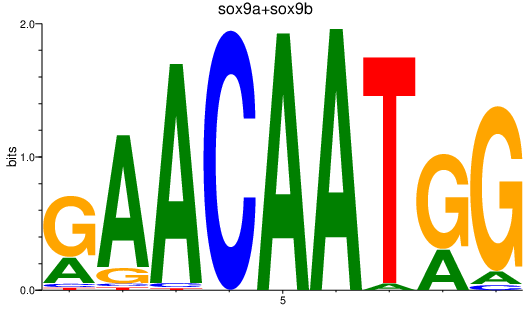
| sox9a+sox9b | 1.36 |
|
|
|
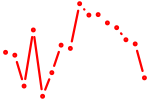
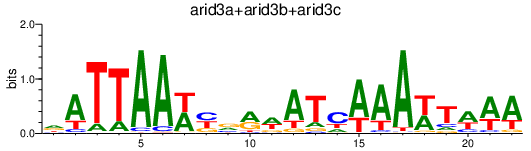
| arid3a+arid3b+arid3c | 1.35 |
|
|
|
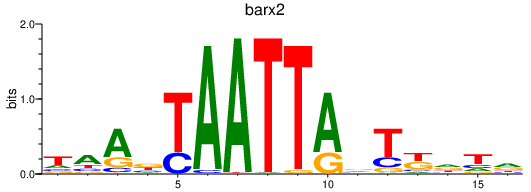
| barx2 | 1.34 |
|

|
|
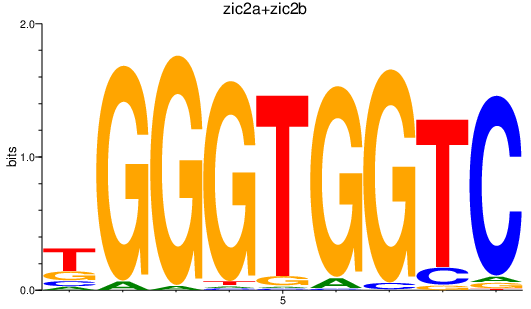
| zic2a+zic2b | 1.33 |
|

|
|
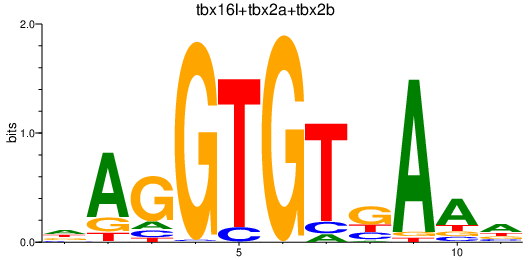
| tbx16l+tbx2a+tbx2b | 1.33 |
|
|
|
| nfil3-5+nfil3-6+si:dkey-60d5.3 | 1.33 |
|

|

|
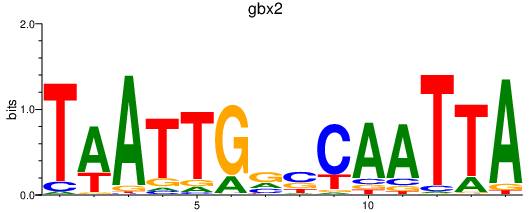
| gbx2 | 1.32 |
|

|
|
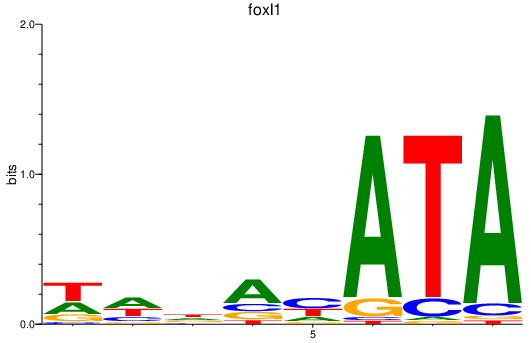
| foxl1 | 1.32 |
|
|
|
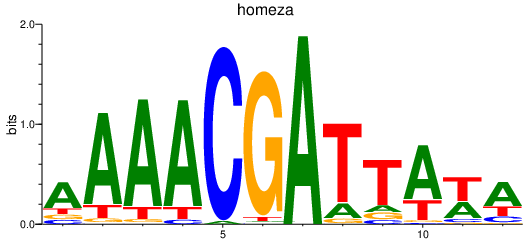
| homeza | 1.32 |
|

|
|
| hoxb3a | 1.31 |
|
|

|
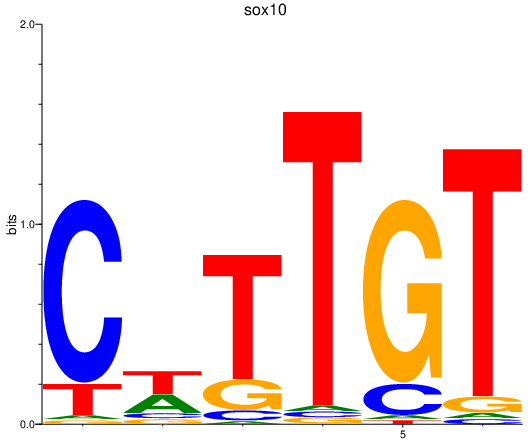
| sox10 | 1.31 |
|

|
|
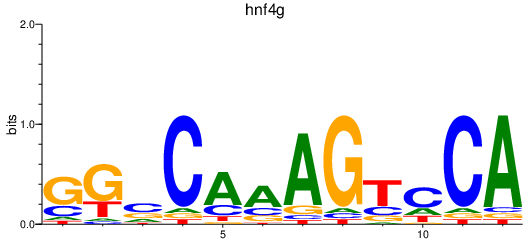
| hnf4g | 1.29 |
|

|
|
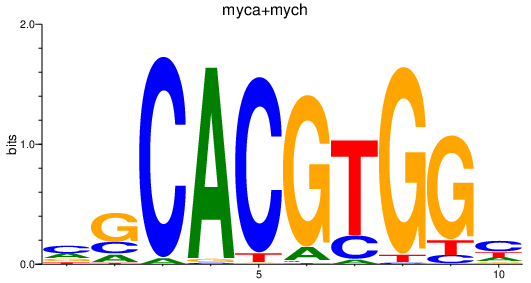
| myca+mych | 1.28 |
|
|
|
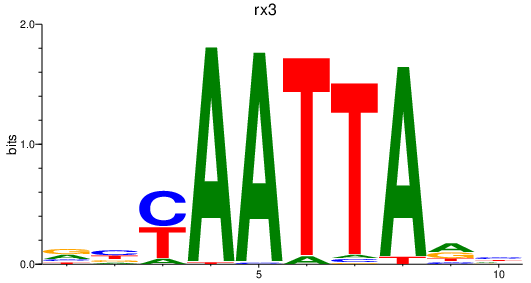
| rx3 | 1.28 |
|

|
|
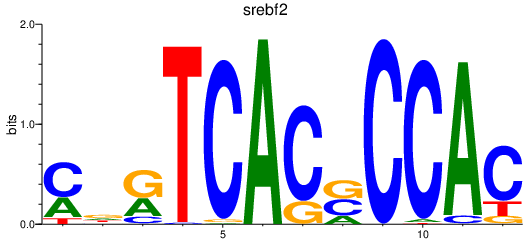
| srebf2 | 1.28 |
|
|
|
| arxb_drgx | 1.26 |
|
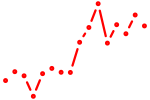
|
|
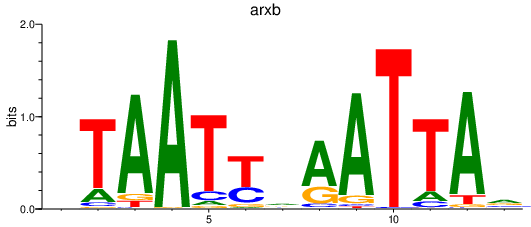
| egr4 | 1.26 |
|

|
|
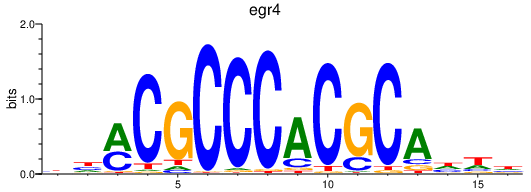
| foxo4 | 1.26 |
|

|
|
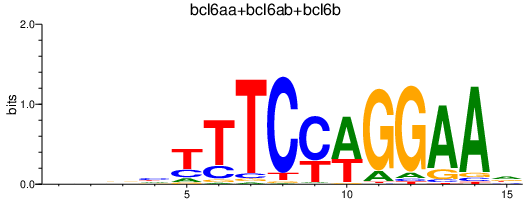
| bcl6aa+bcl6ab+bcl6b | 1.26 |
|

|
|
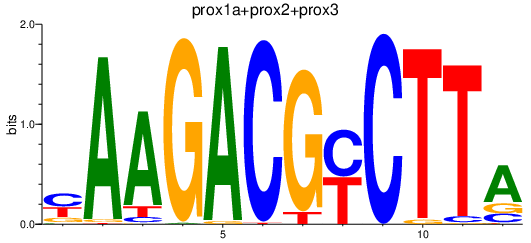
| prox1a+prox2+prox3 | 1.23 |
|

|
|
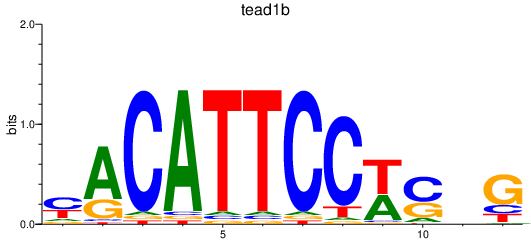
| tead1b_tead1a | 1.23 |
|

|
|
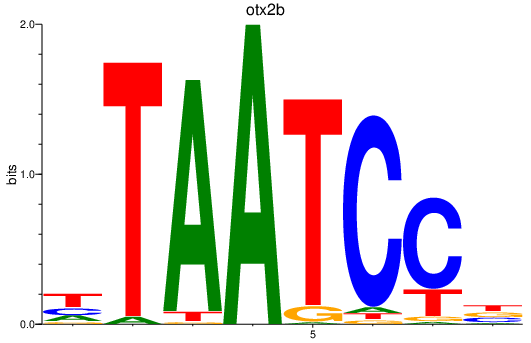
| otx2b | 1.22 |
|
|
|
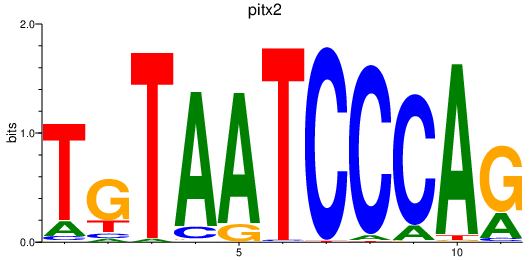
| pitx2 | 1.21 |
|

|
|
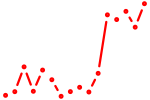
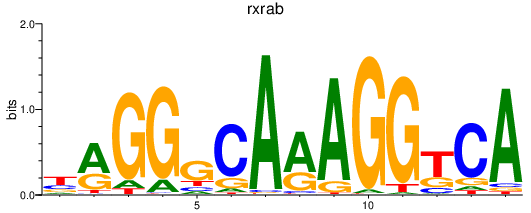
| rxrab | 1.21 |
|
|
|
| tbr1a+tbr1b+tbx21 | 1.21 |
|

|

|
| prrx1a+prrx1b | 1.18 |
|

|

|
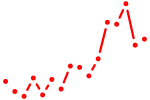
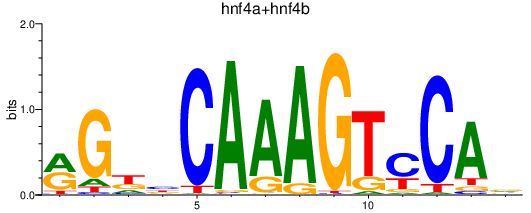
| hnf4a+hnf4b | 1.18 |
|
|
|
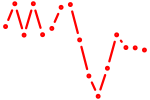
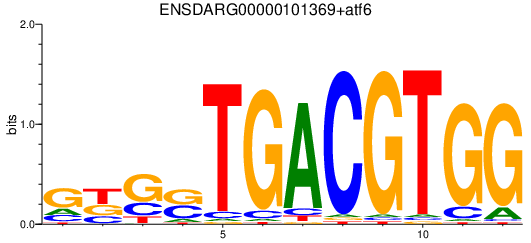
| ENSDARG00000101369+atf6 | 1.17 |
|
|
|
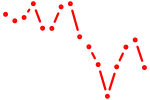
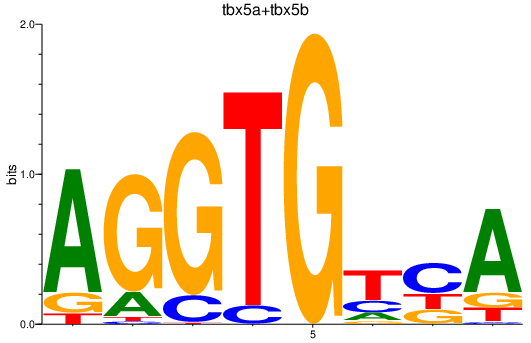
| tbx5a+tbx5b | 1.17 |
|
|
|
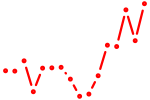
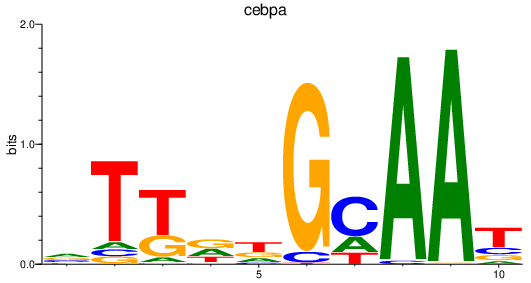
| cebpa | 1.17 |
|
|
|
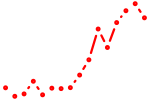
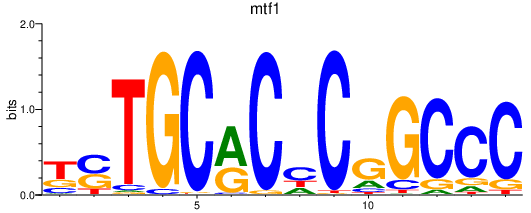
| mtf1 | 1.16 |
|
|
|
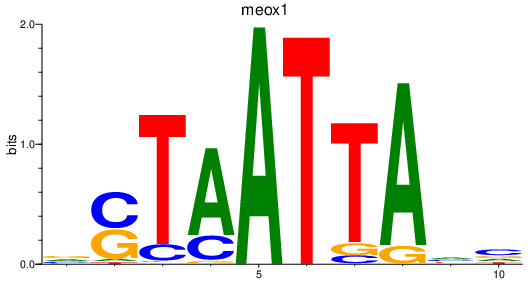
| meox1 | 1.16 |
|

|
|
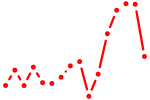
| scrt1a+scrt1b+scrt2 | 1.16 |
|
|

|
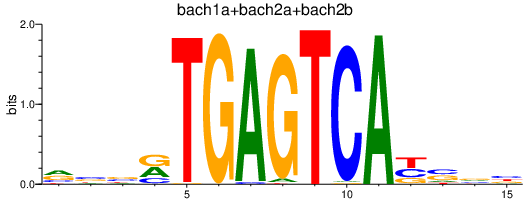
| bach1a+bach2a+bach2b | 1.16 |
|

|
|
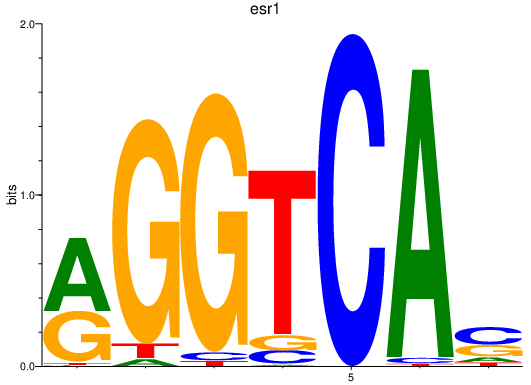
| esr1 | 1.16 |
|

|
|
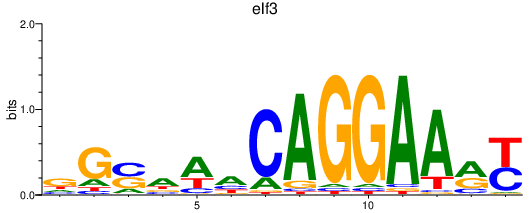
| elf3 | 1.15 |
|

|
|
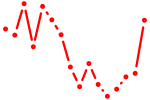
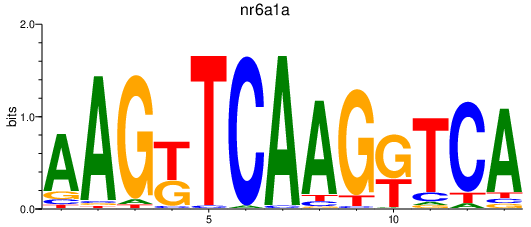
| nr6a1a | 1.14 |
|
|
|
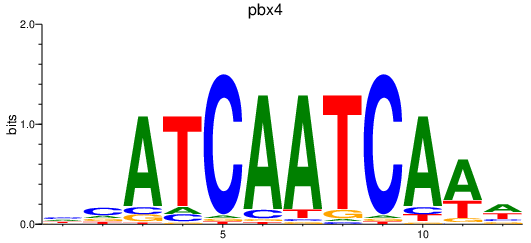
| pbx4 | 1.14 |
|
|
|
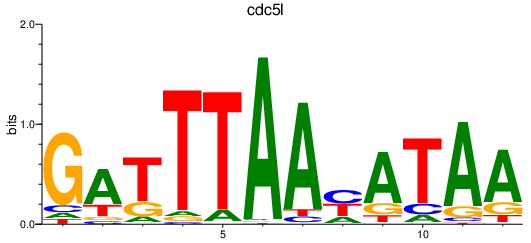
| cdc5l | 1.14 |
|

|
|
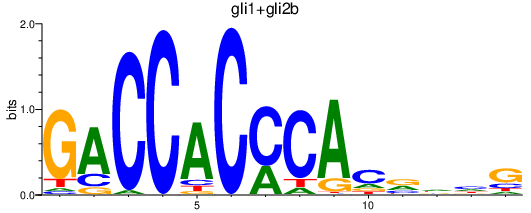
| gli1+gli2b | 1.14 |
|

|
|
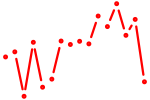
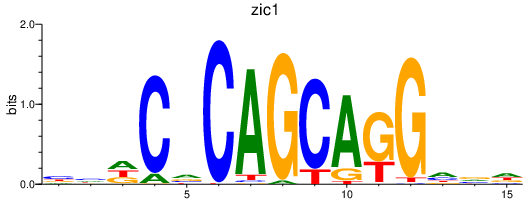
| zic1_si:dkey-149i17.7 | 1.13 |
|
|
|
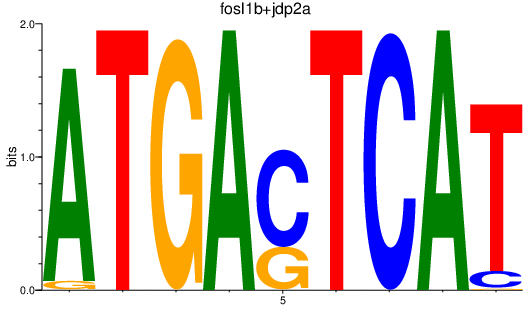
| fosl1b+jdp2a | 1.13 |
|

|
|
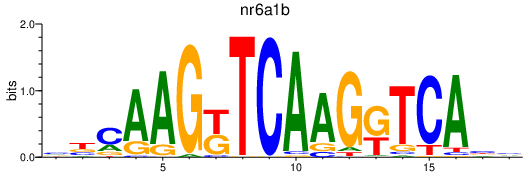
| nr6a1b | 1.13 |
|

|
|
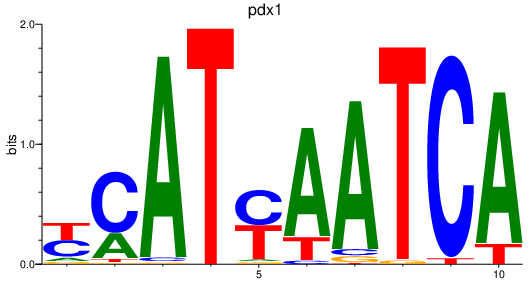
| pdx1 | 1.12 |
|
|
|
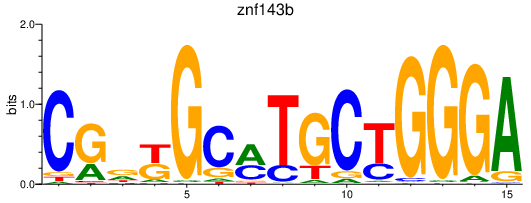
| znf143b | 1.12 |
|
|
|
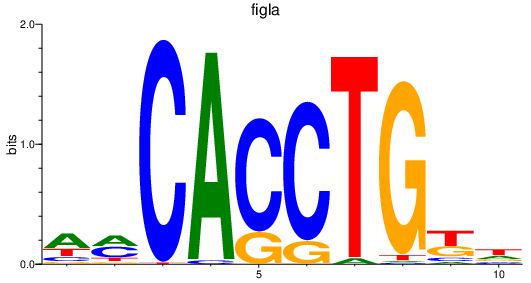
| figla | 1.12 |
|

|
|
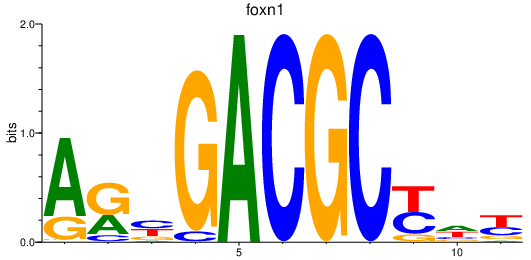
| foxn1 | 1.11 |
|
|
|
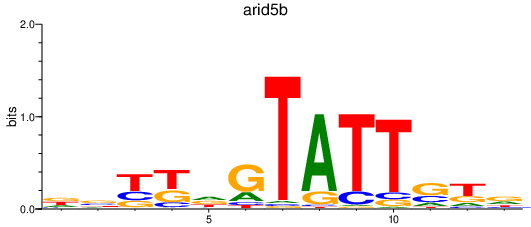
| arid5b | 1.11 |
|

|
|
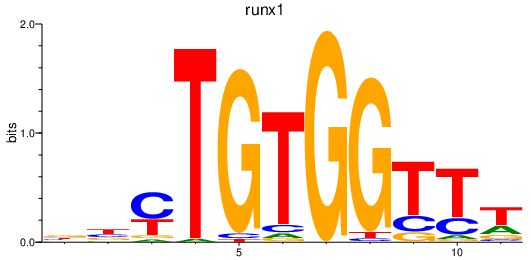
| runx1 | 1.10 |
|
|
|
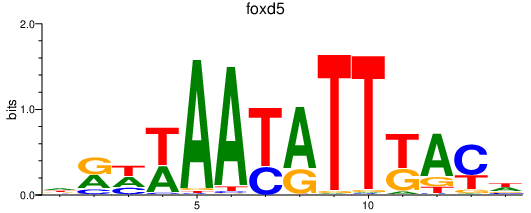
| foxd5_foxd2 | 1.10 |
|
|
|
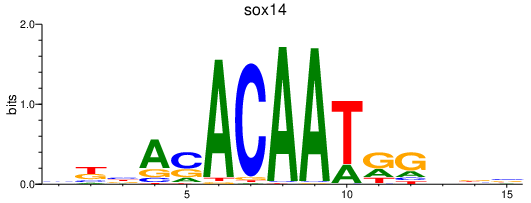
| sox14 | 1.10 |
|

|
|
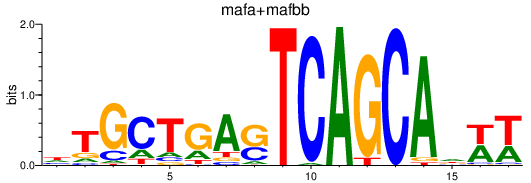
| mafa+mafbb | 1.10 |
|
|
|
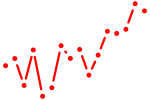
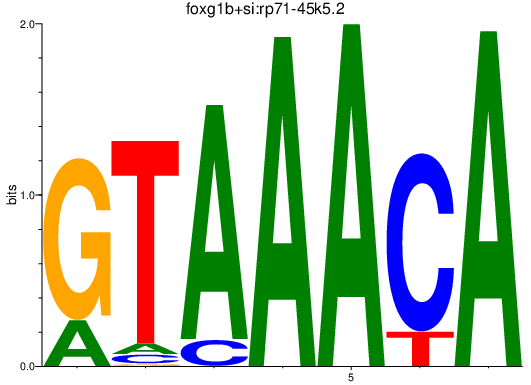
| foxg1b+si:rp71-45k5.2_foxd1+zgc:162612_foxp2_foxo1a+foxo1b | 1.09 |
|
|
|
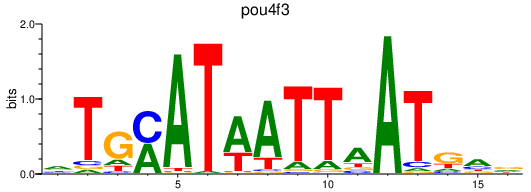
| pou4f3_zgc:158291 | 1.09 |
|

|
|
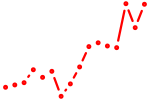
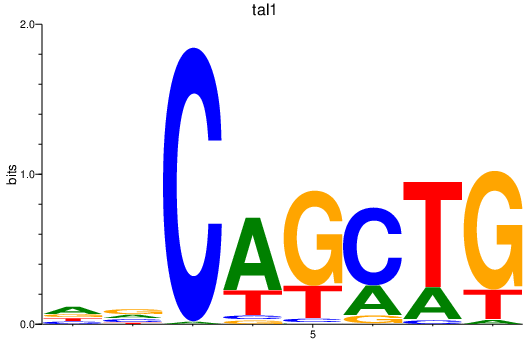
| tal1 | 1.09 |
|
|
|
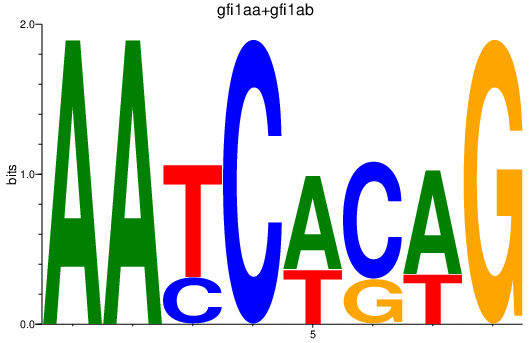
| gfi1aa+gfi1ab | 1.08 |
|
|
|
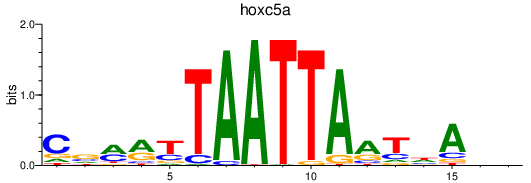
| hoxc5a | 1.08 |
|

|
|
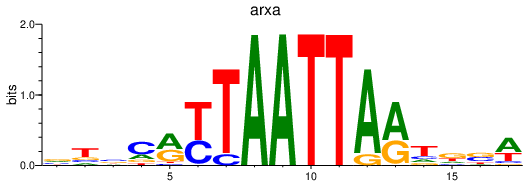
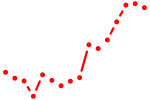
| arxa | 1.08 |
|

|
|
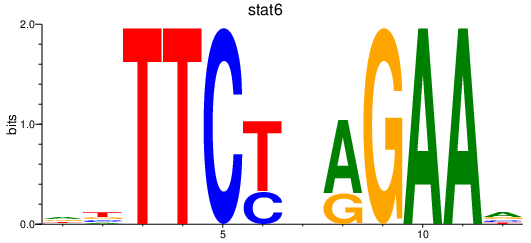
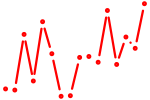
| stat6 | 1.07 |
|
|
|
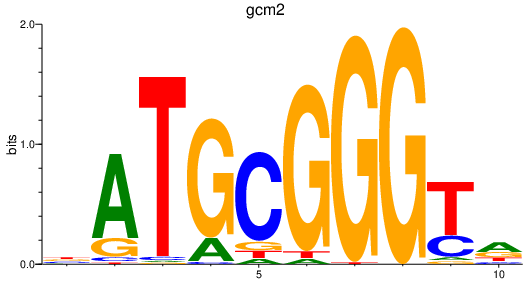
| gcm2 | 1.06 |
|
|
|
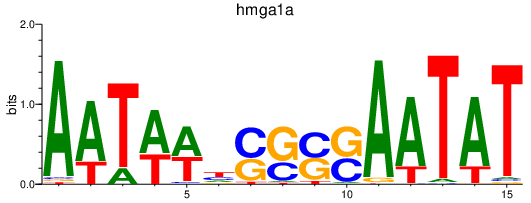
| hmga1a | 1.06 |
|

|
|
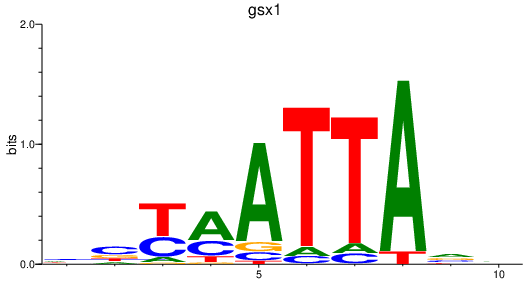
| gsx1 | 1.06 |
|

|
|
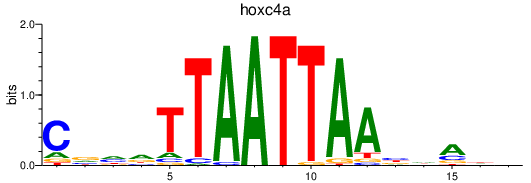
| hoxc4a | 1.06 |
|
|
|
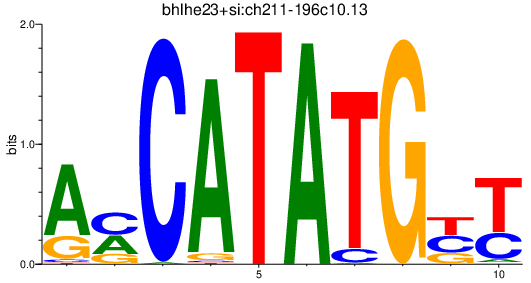
| bhlhe23+si:ch211-196c10.13_bhlhe22_neurod2_bhlha15 | 1.06 |
|

|
|
| sox21a+sox21b | 1.06 |
|
|

|
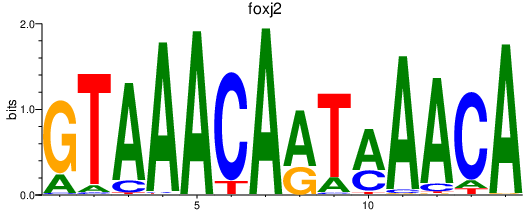
| foxj2 | 1.05 |
|
|
|
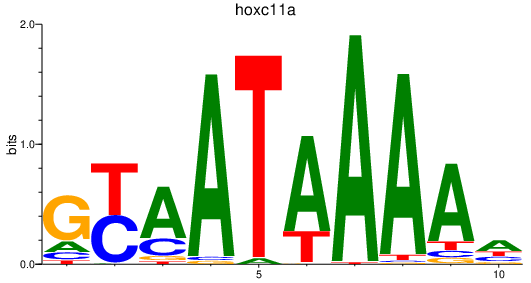
| hoxc11a | 1.05 |
|

|
|
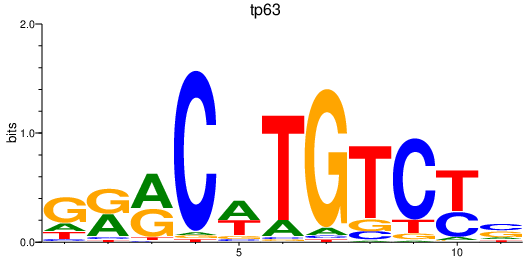
| tp63 | 1.04 |
|
|
|
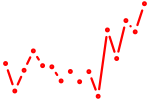
| rreb1a+rreb1b | 1.03 |
|
|
|
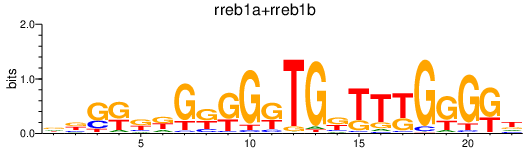
| lhx6 | 1.03 |
|

|
|
| nfe2l1b | 1.03 |
|
|

|
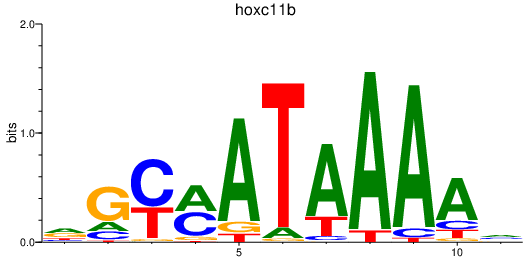
| hoxc11b | 1.03 |
|

|
|
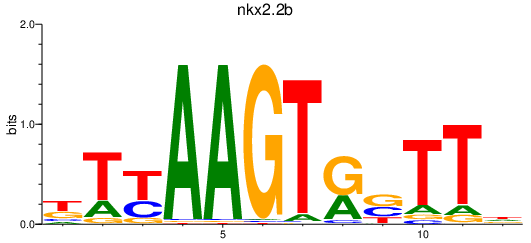
| nkx2.2b | 1.03 |
|
|
|
| ddit3 | 1.03 |
|
|
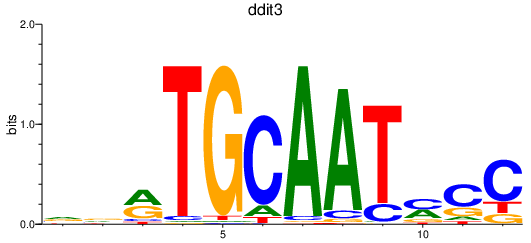
|
| arnt | 1.03 |
|

|

|
| smad1_si:dkey-222b8.1+smad4a+smad4b_smad2 | 1.02 |
|

|
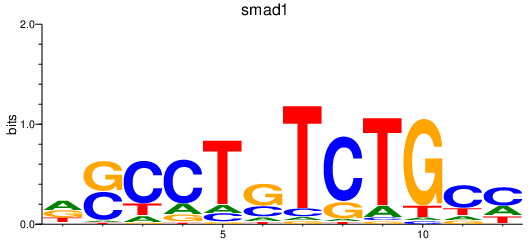
|
| mbd2 | 1.02 |
|
|

|
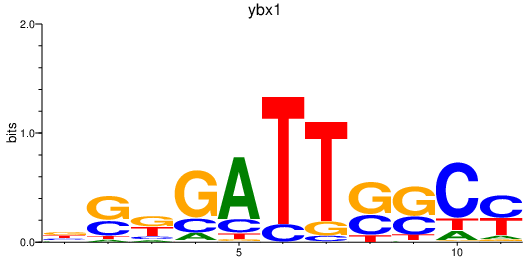
| ybx1 | 1.01 |
|
|
|
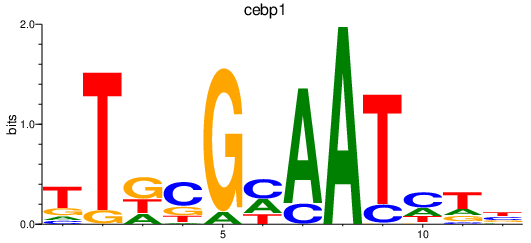
| cebp1 | 1.01 |
|
|
|
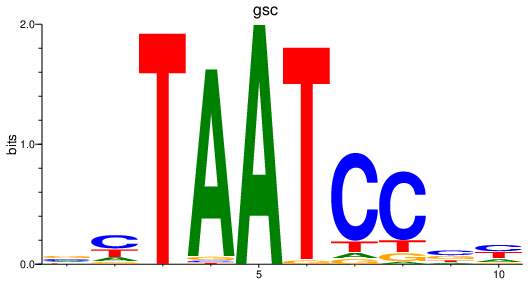
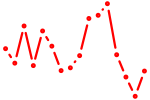
| gsc | 1.01 |
|
|
|
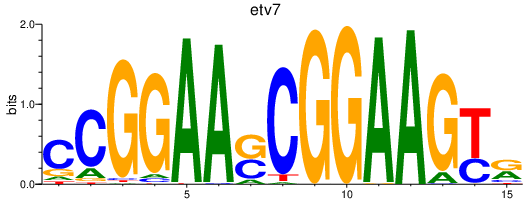
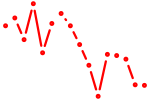
| etv7 | 1.00 |
|
|
|
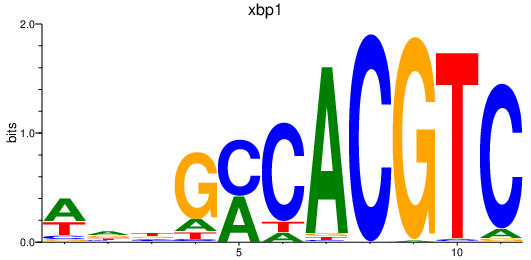
| xbp1 | 1.00 |
|
|
|
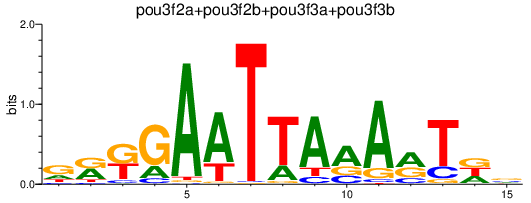
| pou3f2a+pou3f2b+pou3f3a+pou3f3b | 1.00 |
|

|
|
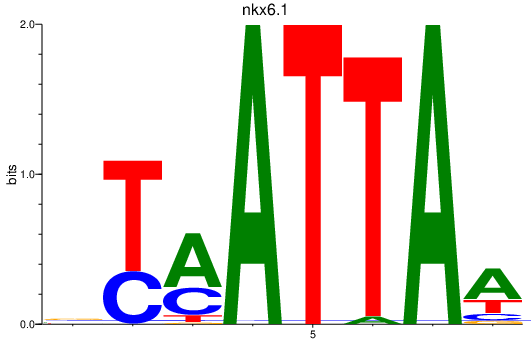
| nkx6.1 | 1.00 |
|
|
|
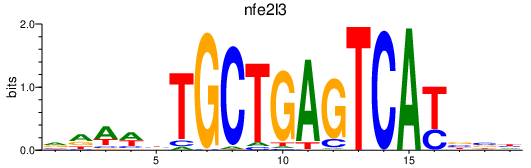
| nfe2l3 | 0.99 |
|
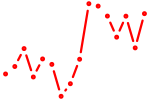
|
|
| pax10+pax4+pax6a+pax6b | 0.99 |
|

|
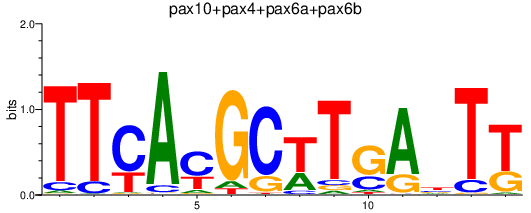
|
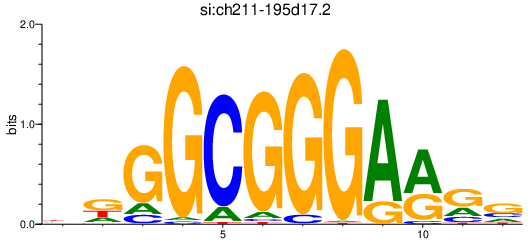
| si:ch211-195d17.2 | 0.99 |
|
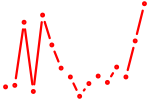
|
|
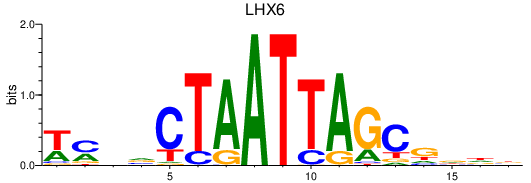
| LHX6 | 0.98 |
|

|
|
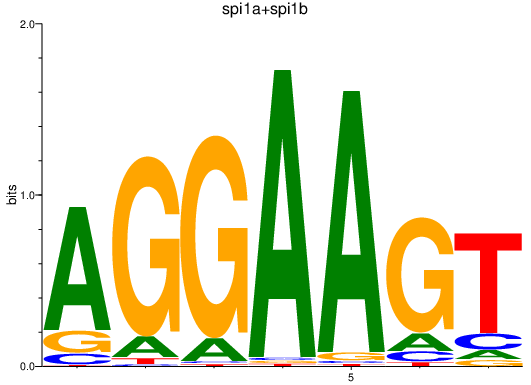
| spi1a+spi1b | 0.98 |
|

|
|
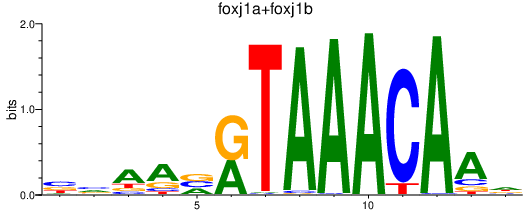
| foxj1a+foxj1b | 0.98 |
|

|
|
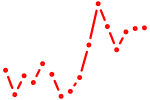
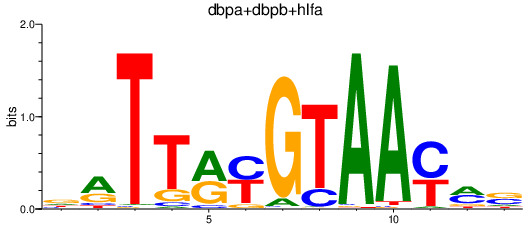
| dbpa+dbpb+hlfa | 0.97 |
|
|
|
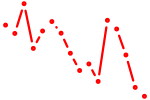
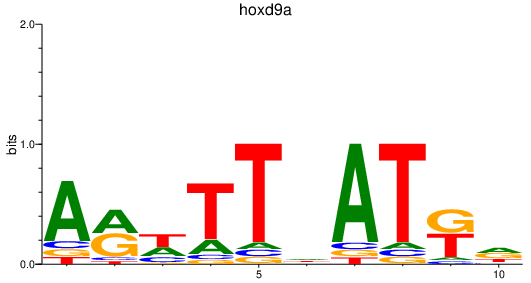
| hoxd9a | 0.97 |
|
|
|
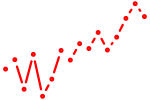
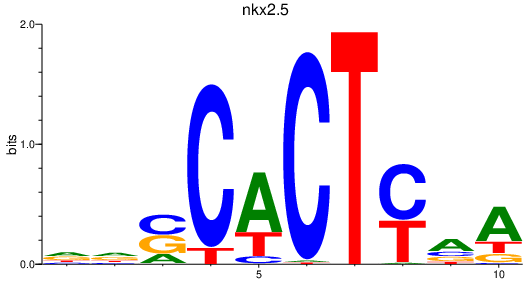
| nkx2.5 | 0.96 |
|
|
|
| sox1a+sox1b | 0.96 |
|

|

|
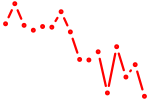
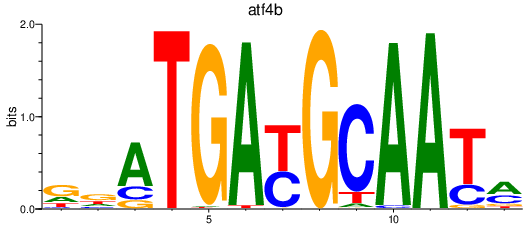
| atf4b | 0.96 |
|
|
|
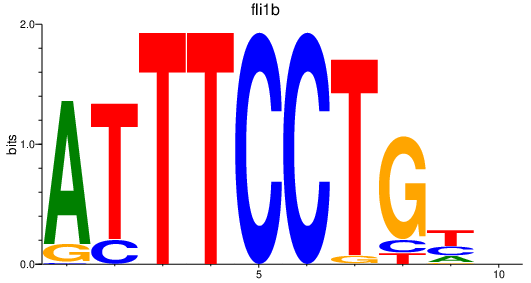
| fli1b | 0.95 |
|
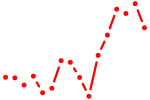
|
|
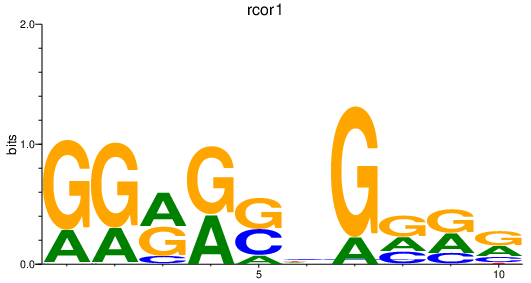
| rcor1 | 0.95 |
|

|
|
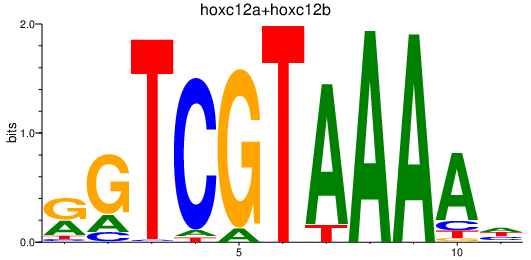
| hoxc12a+hoxc12b | 0.95 |
|

|
|
| alx1 | 0.95 |
|

|
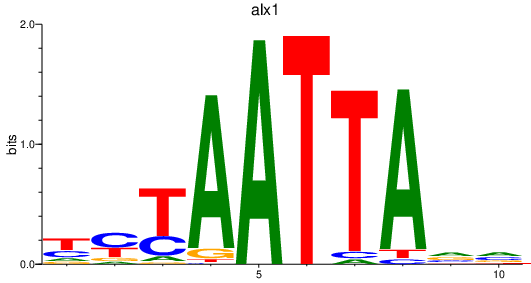
|
| LHX3+lhx4 | 0.95 |
|

|
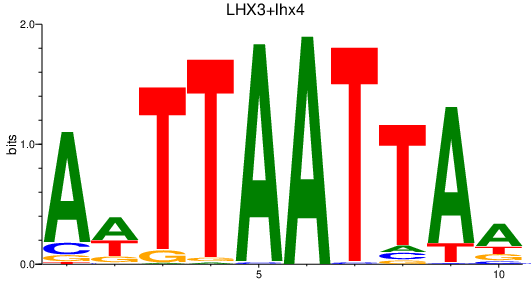
|
| arid6+zgc:77151 | 0.95 |
|

|
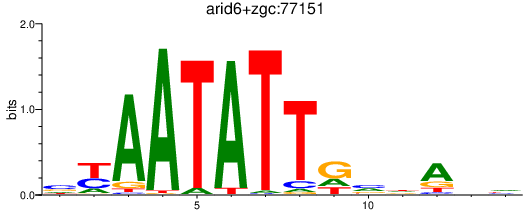
|
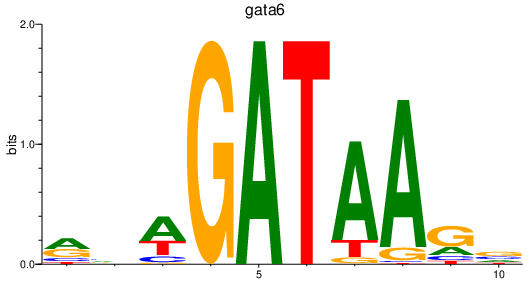
| gata6 | 0.93 |
|
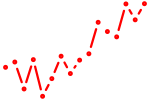
|
|
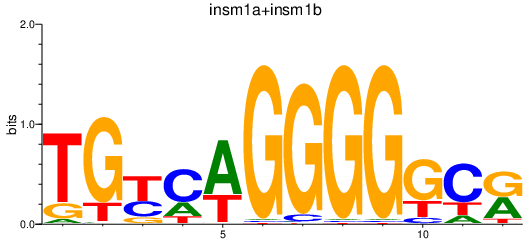
| insm1a+insm1b | 0.93 |
|
|
|
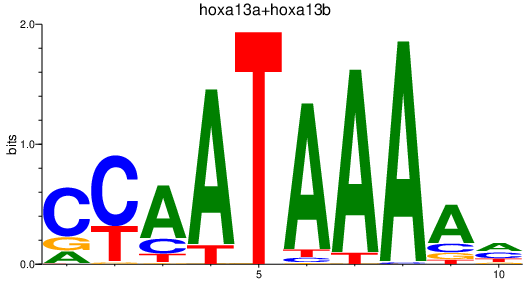
| hoxa13a+hoxa13b | 0.93 |
|
|
|
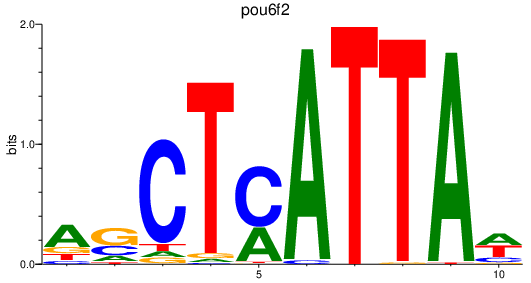
| pou6f2 | 0.93 |
|
|
|
| bsx+nanog | 0.92 |
|

|

|
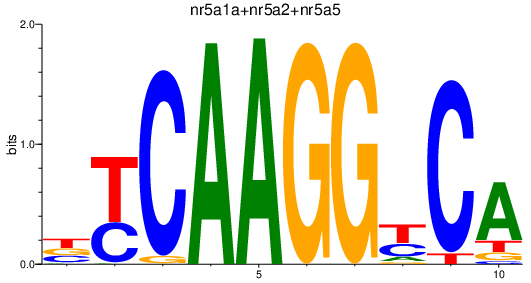
| nr5a1a+nr5a2+nr5a5 | 0.92 |
|
|
|
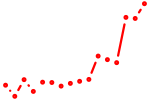
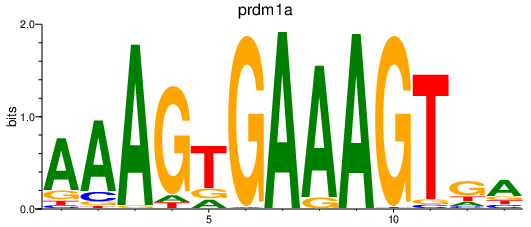
| prdm1a | 0.92 |
|
|
|
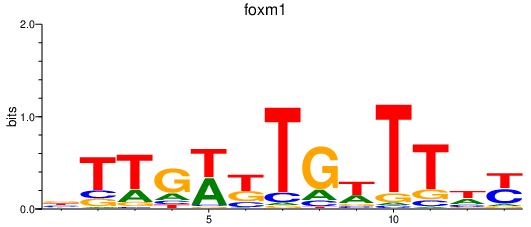
| foxm1 | 0.91 |
|

|
|
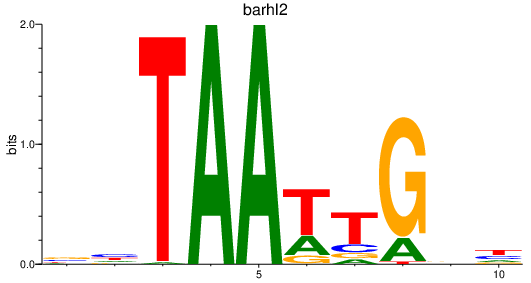
| barhl2 | 0.91 |
|

|
|
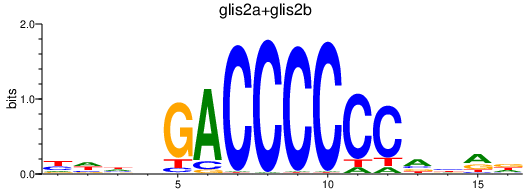
| glis2a+glis2b | 0.91 |
|

|
|
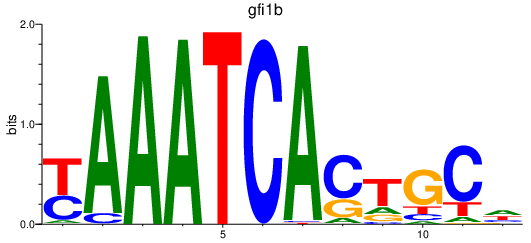
| gfi1b | 0.90 |
|
|
|
| nr4a2a | 0.89 |
|
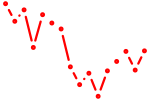
|

|
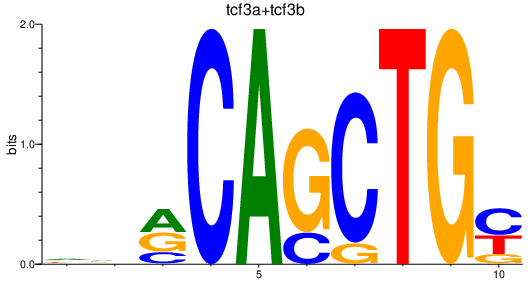
| tcf3a+tcf3b | 0.89 |
|
|
|
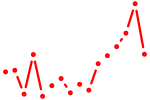
| tbxta+tbxtb | 0.89 |
|

|

|
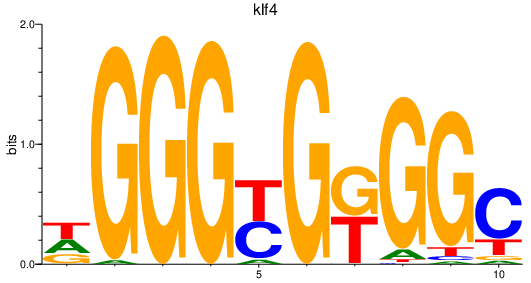
| klf4_ENSDARG00000005846+sp3a_sp4_sp8a+sp8b | 0.89 |
|

|
|
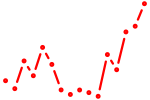
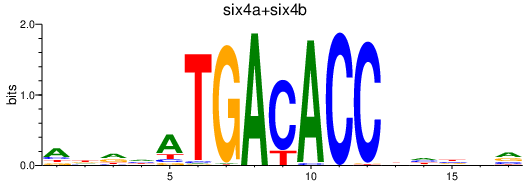
| six4a+six4b | 0.88 |
|
|
|
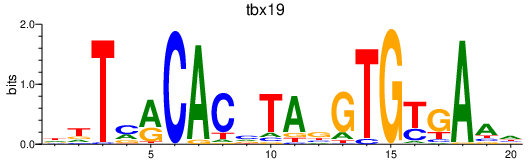
| tbx19 | 0.88 |
|

|
|
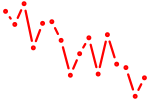
| hoxd12a | 0.88 |
|
|

|
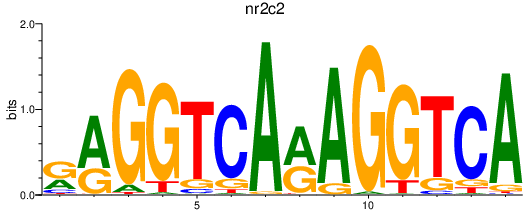
| nr2c2 | 0.88 |
|
|
|
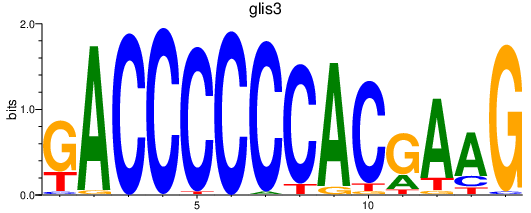
| glis3 | 0.88 |
|

|
|
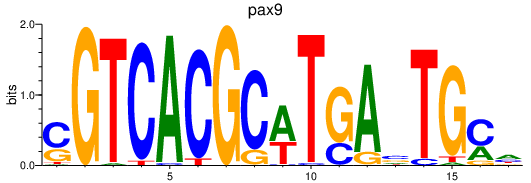
| pax9_pax1a+pax1b | 0.86 |
|

|
|
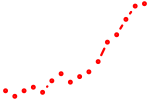
| irx6a | 0.86 |
|
|

|
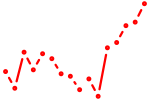
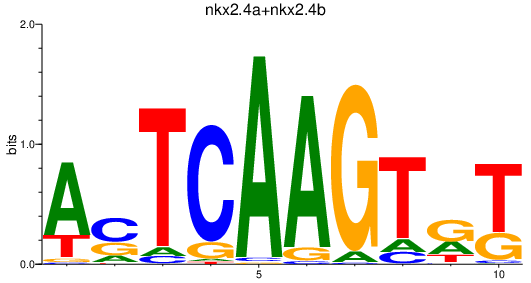
| nkx2.4a+nkx2.4b | 0.86 |
|
|
|
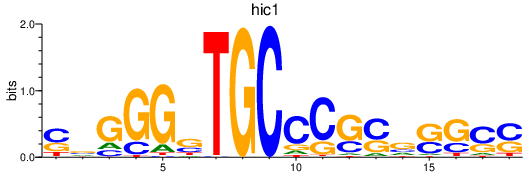
| hic1 | 0.85 |
|

|
|
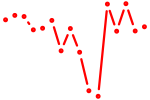
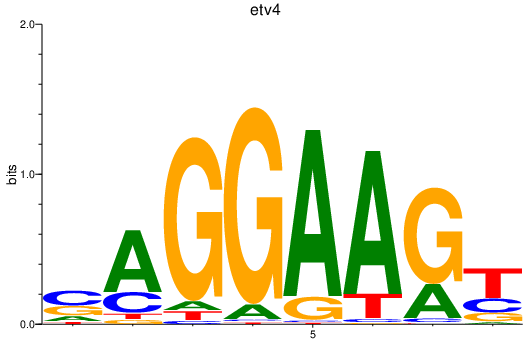
| etv4 | 0.85 |
|
|
|
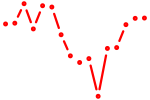
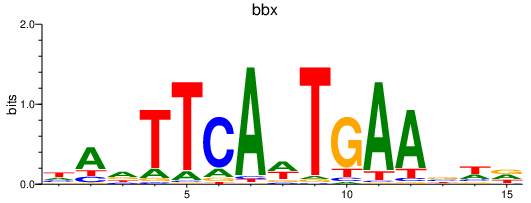
| bbx | 0.85 |
|
|
|
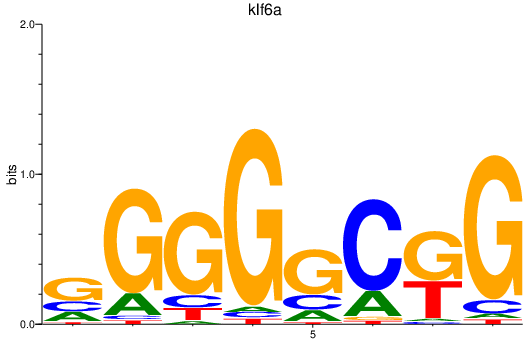
| klf6a | 0.85 |
|

|
|
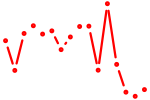
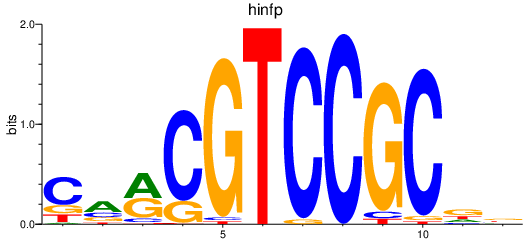
| hinfp | 0.85 |
|
|
|
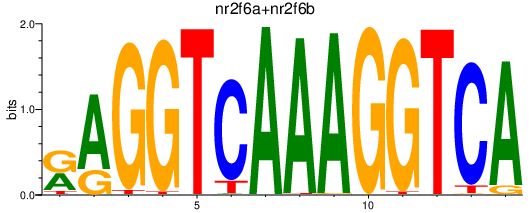
| nr2f6a+nr2f6b | 0.85 |
|

|
|
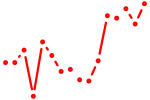
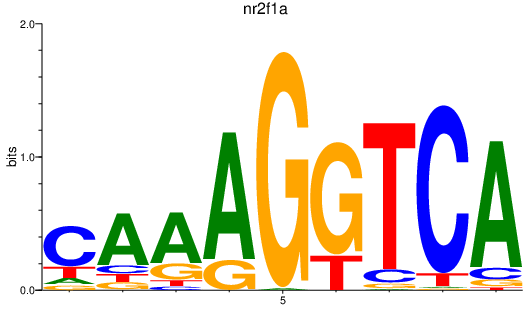
| nr2f1a | 0.85 |
|
|
|
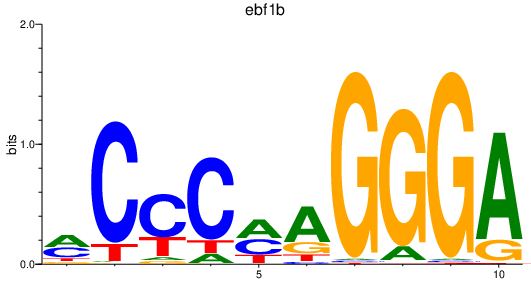
| ebf1b | 0.84 |
|

|
|
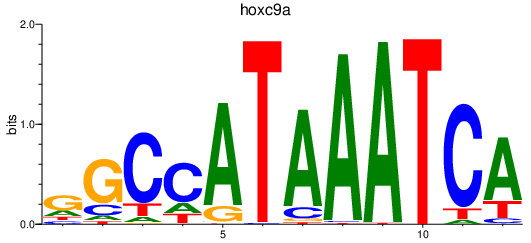
| hoxc9a | 0.84 |
|

|
|
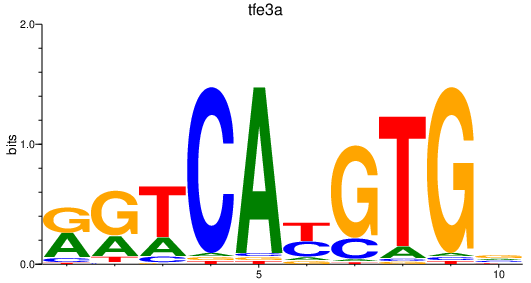
| tfe3a | 0.84 |
|

|
|
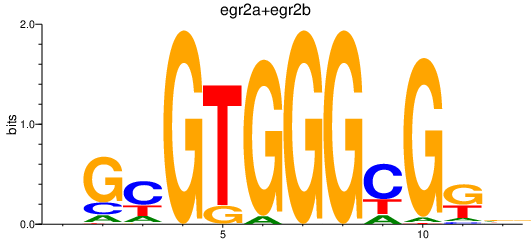
| egr2a+egr2b | 0.84 |
|
|
|
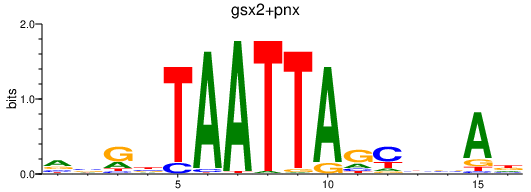
| gsx2+pnx | 0.83 |
|
|
|
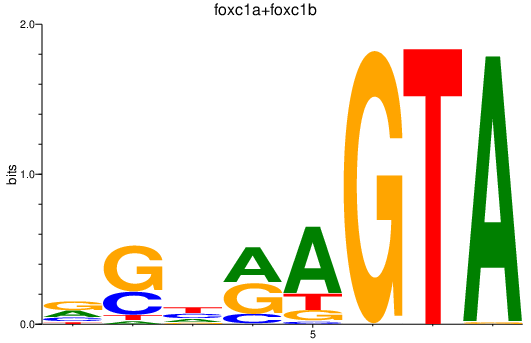
| foxc1a+foxc1b | 0.83 |
|
|
|
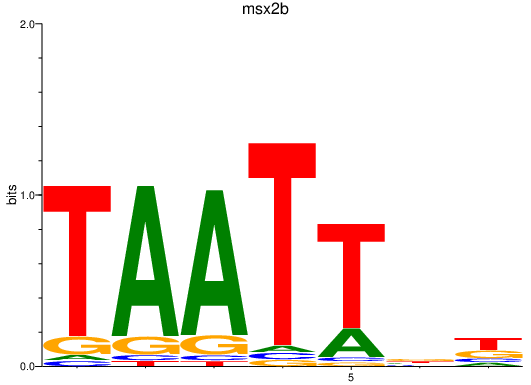
| msx2b | 0.83 |
|

|
|
| sox32 | 0.83 |
|
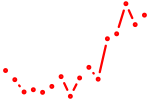
|
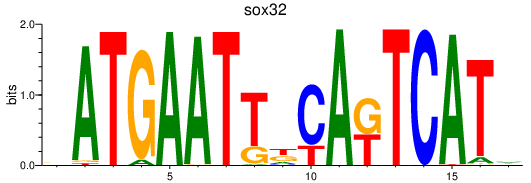
|
| ets2 | 0.82 |
|

|

|
| bcl11aa | 0.82 |
|

|
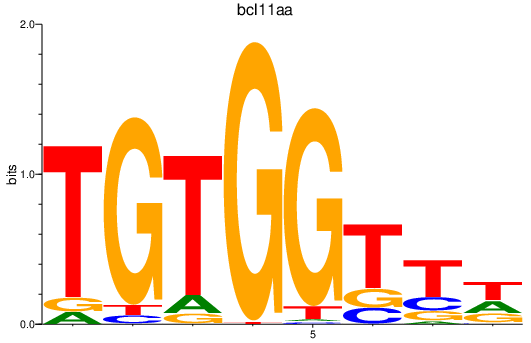
|
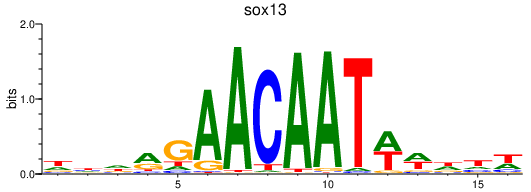
| sox13 | 0.82 |
|
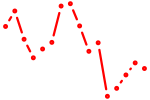
|
|
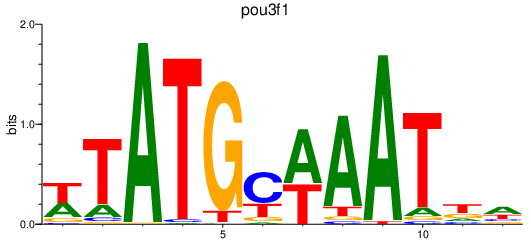
| pou3f1 | 0.82 |
|
|
|
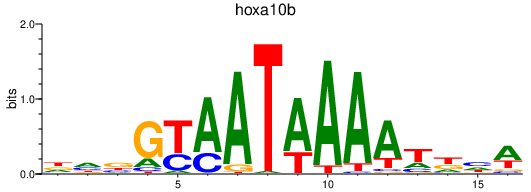
| hoxa10b | 0.82 |
|

|
|
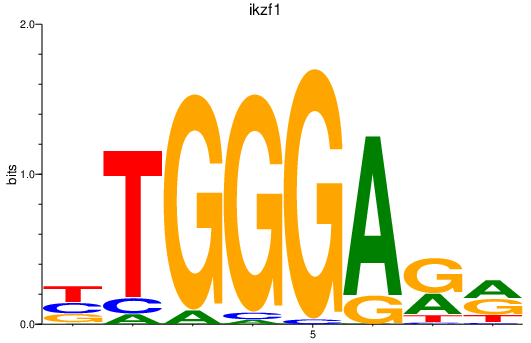
| ikzf1 | 0.82 |
|

|
|
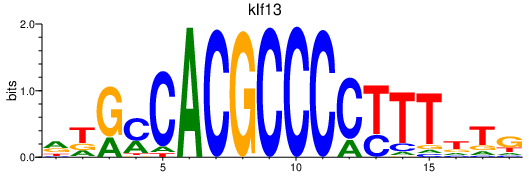
| klf13 | 0.81 |
|
|
|
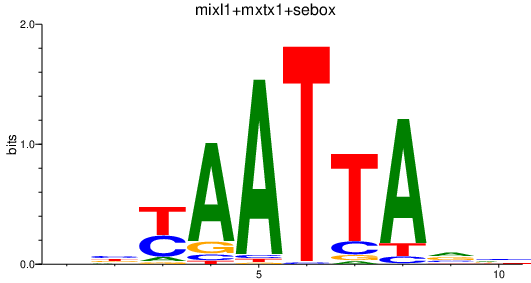
| mixl1+mxtx1+sebox | 0.81 |
|

|
|
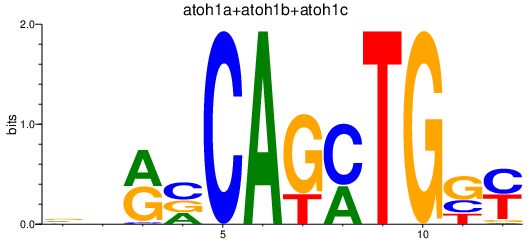
| atoh1a+atoh1b+atoh1c | 0.81 |
|

|
|
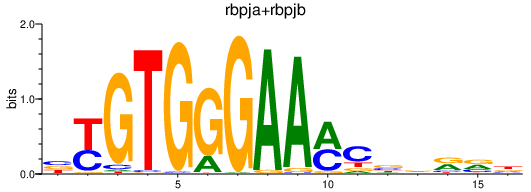
| rbpja+rbpjb | 0.81 |
|
|
|
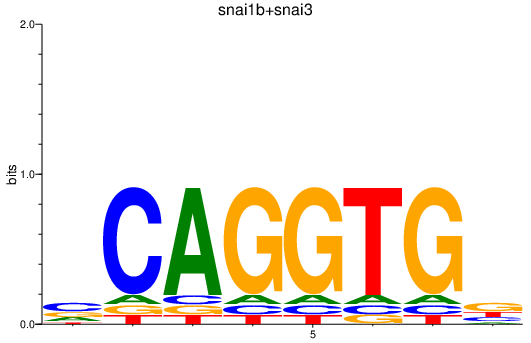
| snai1b+snai3 | 0.81 |
|

|
|
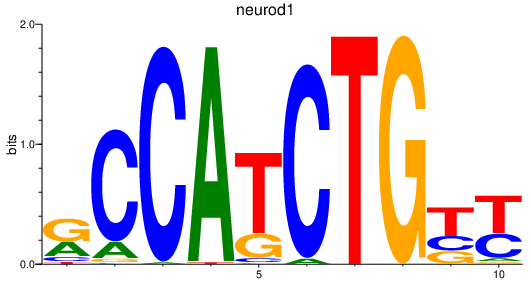
| neurod1 | 0.81 |
|

|
|
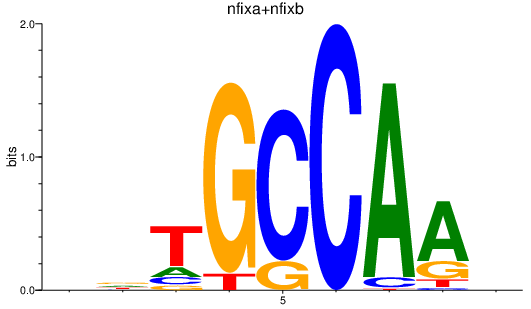
| nfixa+nfixb | 0.81 |
|

|
|
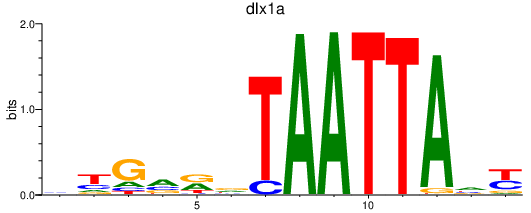
| dlx1a | 0.81 |
|

|
|
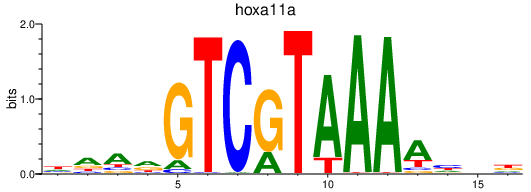
| hoxa11a | 0.80 |
|

|
|
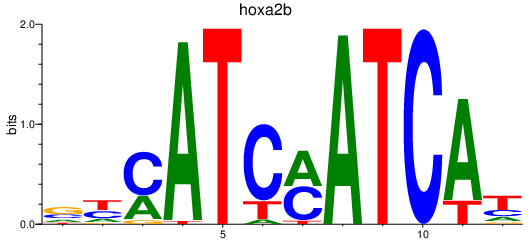
| hoxa2b | 0.80 |
|
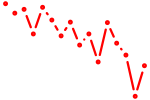
|
|
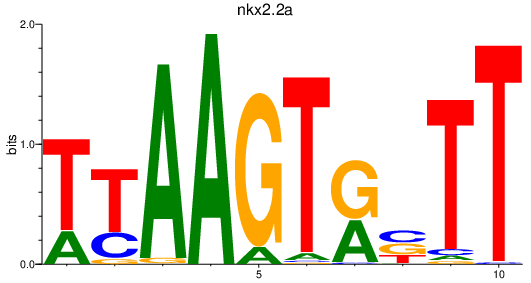
| nkx2.2a | 0.80 |
|

|
|
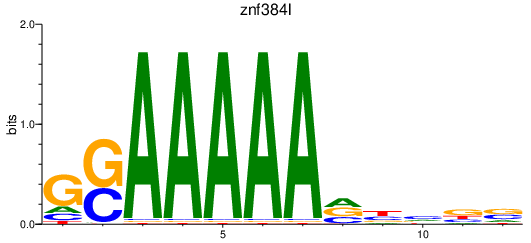
| znf384l | 0.80 |
|
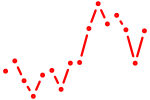
|
|
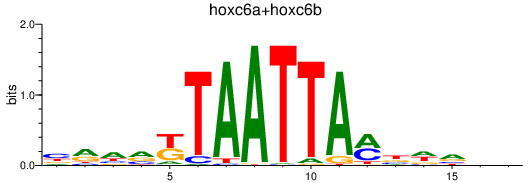
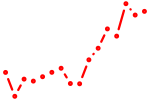
| hoxc6a+hoxc6b | 0.79 |
|

|
|
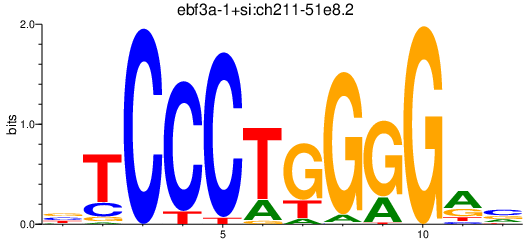
| ebf3a-1+si:ch211-51e8.2 | 0.79 |
|
|
|
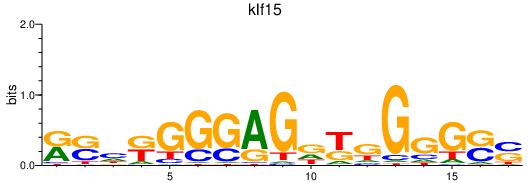
| klf15 | 0.79 |
|

|
|
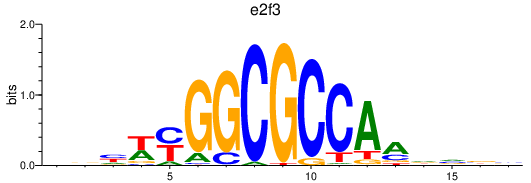
| e2f3 | 0.79 |
|

|
|
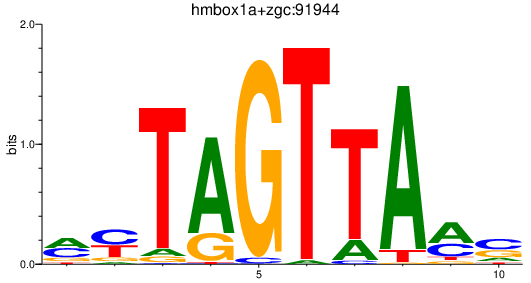
| hmbox1a+zgc:91944 | 0.78 |
|
|
|
| jund_batf | 0.78 |
|
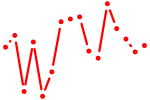
|
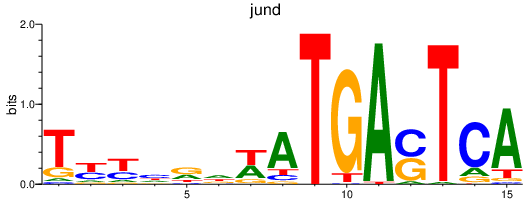
|
| junba+junbb | 0.78 |
|
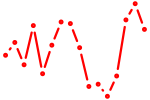
|
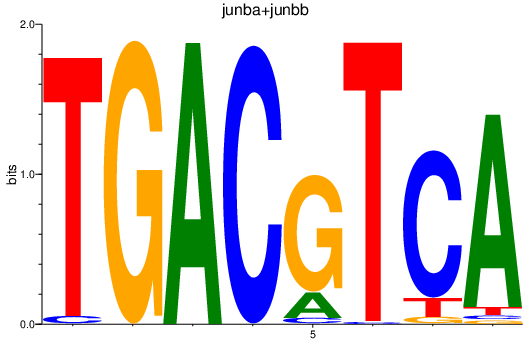
|
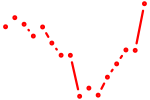
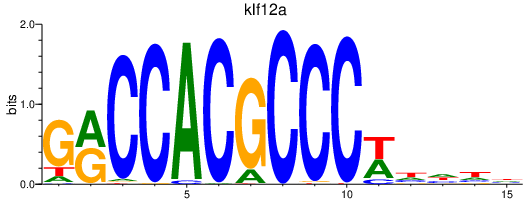
| klf12a | 0.78 |
|
|
|
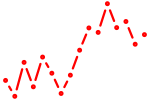
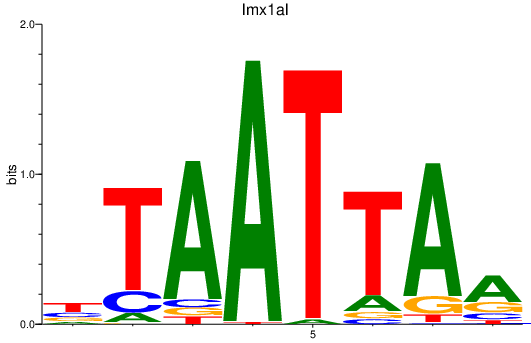
| lmx1al | 0.78 |
|
|
|
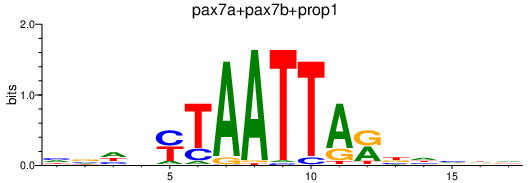
| pax7a+pax7b+prop1 | 0.78 |
|

|
|
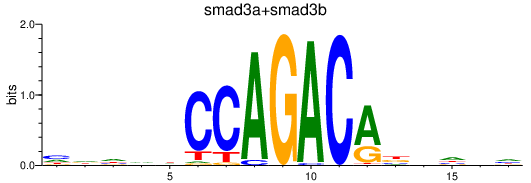
| smad3a+smad3b | 0.78 |
|

|
|
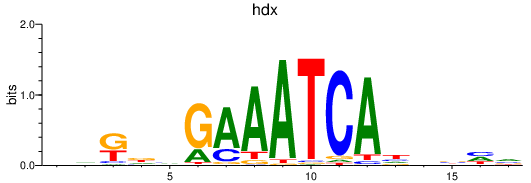
| hdx | 0.78 |
|
|
|
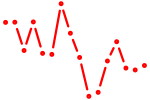
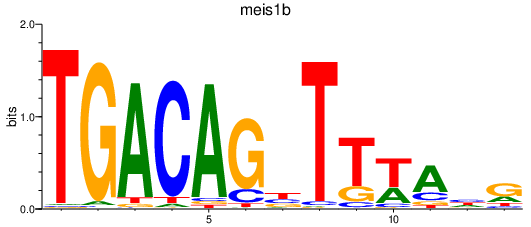
| meis1b | 0.77 |
|
|
|
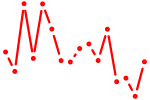
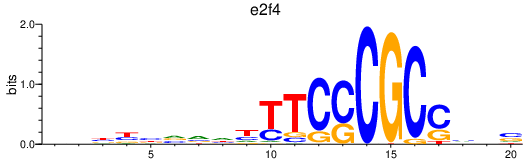
| e2f4 | 0.77 |
|
|
|
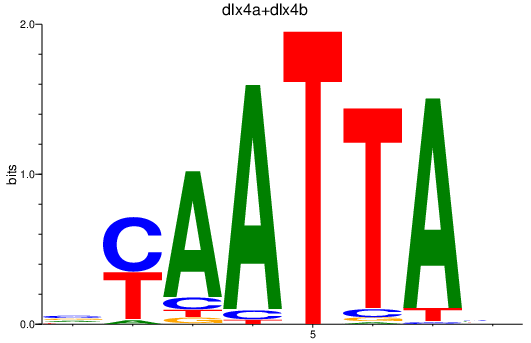
| dlx4a+dlx4b_dlx2a+dlx2b | 0.77 |
|

|
|
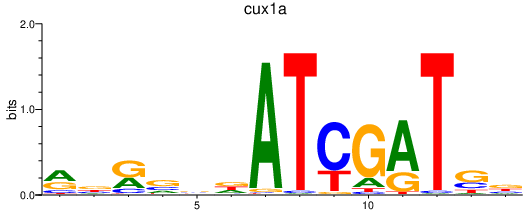
| cux1a | 0.76 |
|
|
|
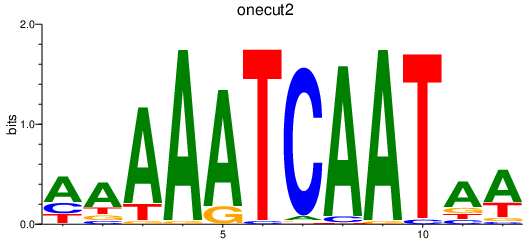
| onecut2 | 0.75 |
|

|
|
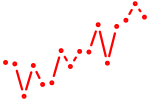
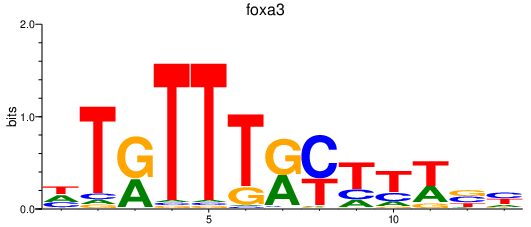
| foxa3 | 0.75 |
|
|
|
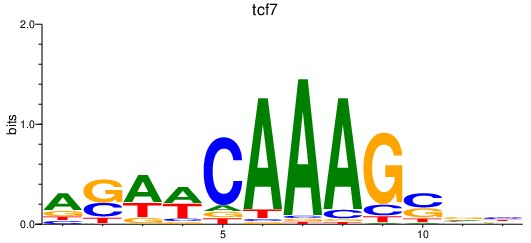
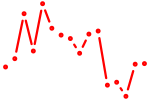
| tcf7 | 0.75 |
|

|
|
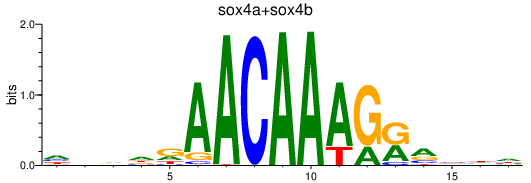
| sox4a+sox4b | 0.74 |
|
|
|
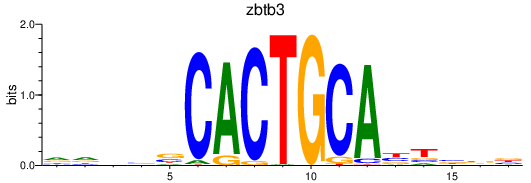
| zbtb3 | 0.74 |
|

|
|
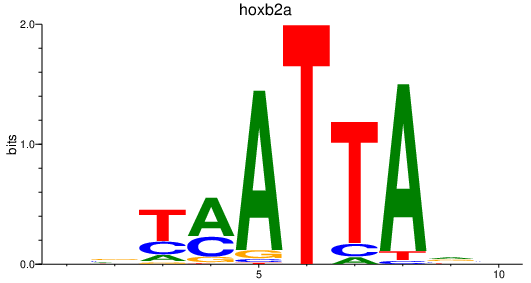
| hoxb2a | 0.74 |
|
|
|
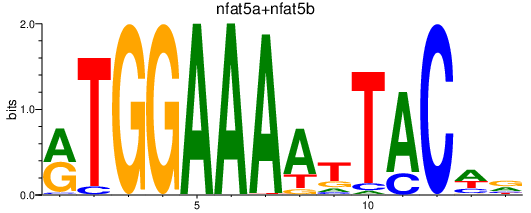
| nfat5a+nfat5b | 0.74 |
|
|
|
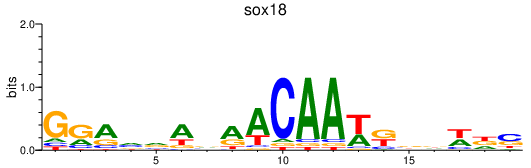
| sox18 | 0.73 |
|

|
|
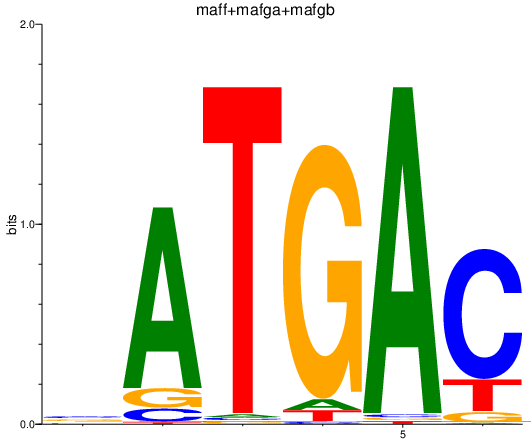
| maff+mafga+mafgb | 0.73 |
|
|
|
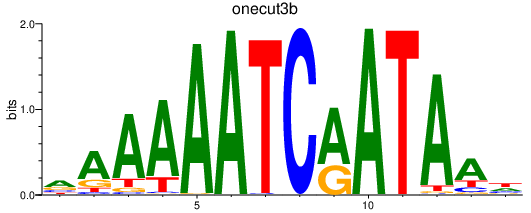
| onecut3b | 0.73 |
|
|
|
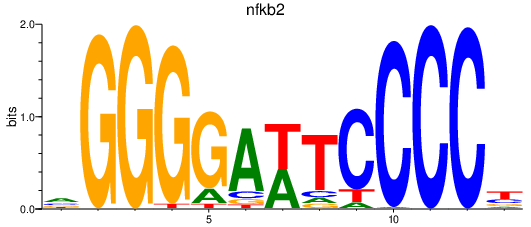
| nfkb2 | 0.73 |
|

|
|
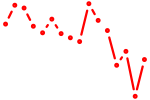
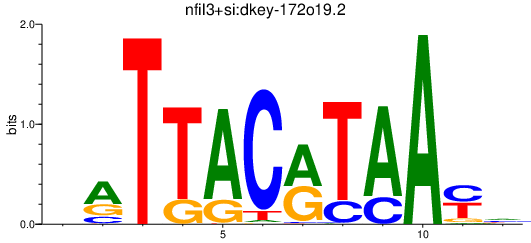
| nfil3+si:dkey-172o19.2 | 0.73 |
|
|
|
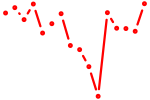
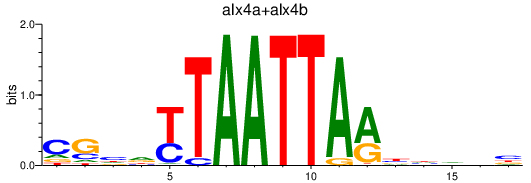
| alx4a+alx4b | 0.73 |
|
|
|
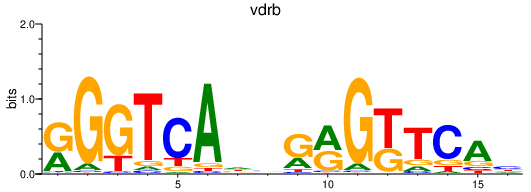
| vdrb_nr1i2 | 0.72 |
|
|
|
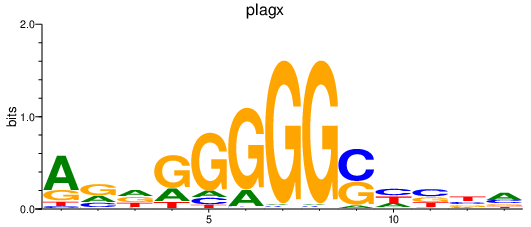
| plagx | 0.72 |
|

|
|
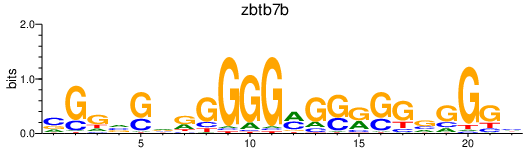
| zbtb7b | 0.72 |
|

|
|
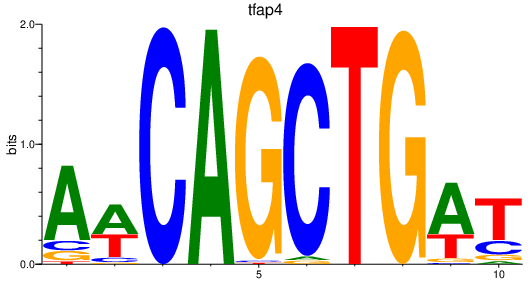
| tfap4 | 0.71 |
|
|
|
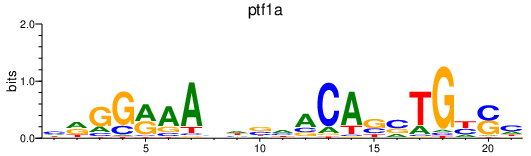
| ptf1a | 0.71 |
|

|
|
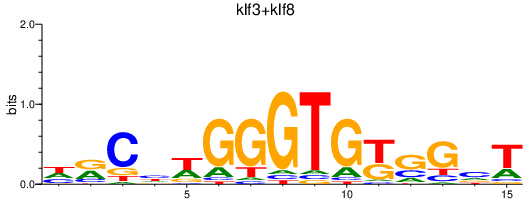
| klf3+klf8 | 0.71 |
|

|
|
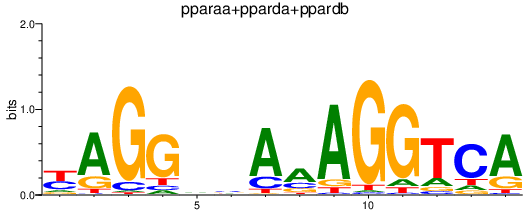
| pparaa+pparda+ppardb | 0.71 |
|

|
|
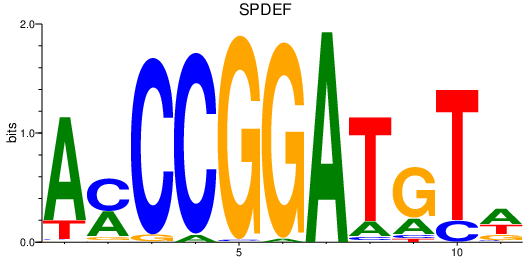
| SPDEF | 0.71 |
|
|
|
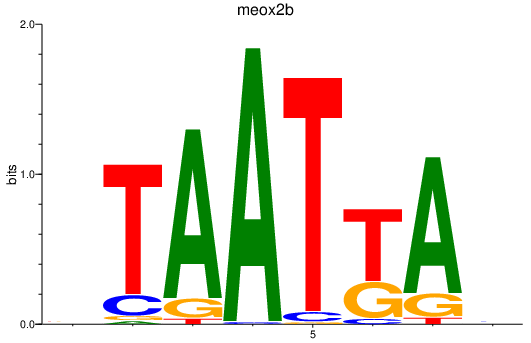
| meox2b | 0.71 |
|
|
|
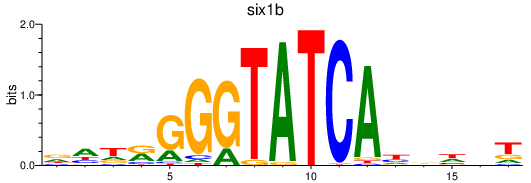
| six1b_six1a+six9_six3a+six3b+six7 | 0.71 |
|

|
|
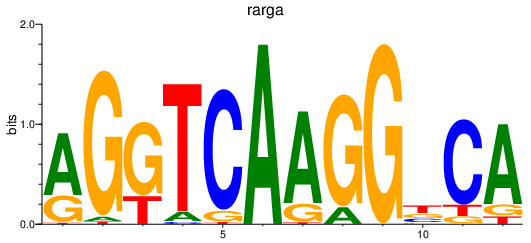
| rarga | 0.71 |
|

|
|
| zbtb38 | 0.71 |
|
|
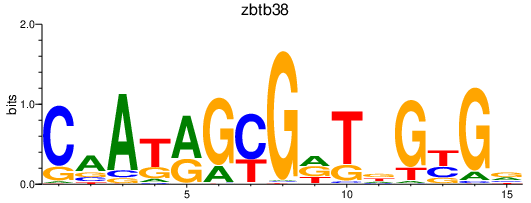
|
| tbx3a+tbx3b | 0.70 |
|
|
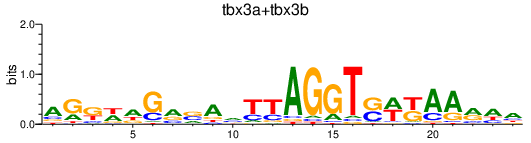
|
| myb_mybl2a | 0.70 |
|

|
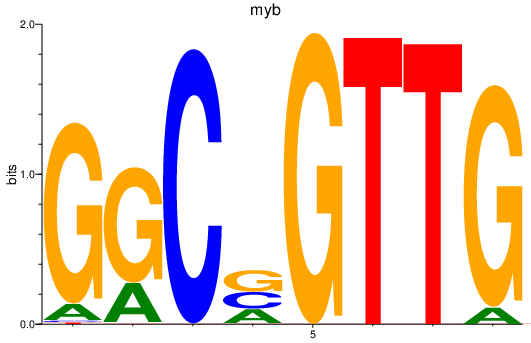
|
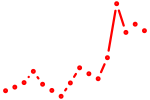
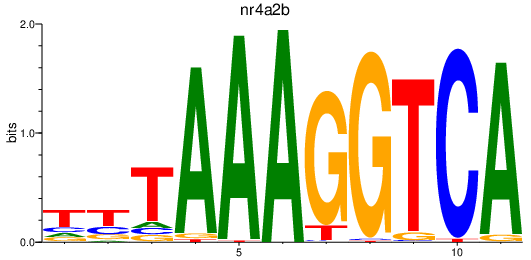
| nr4a2b_nr4a1 | 0.70 |
|
|
|
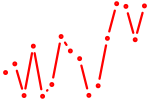
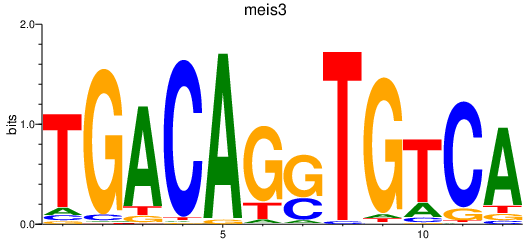
| meis3 | 0.70 |
|
|
|
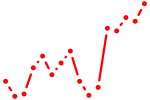
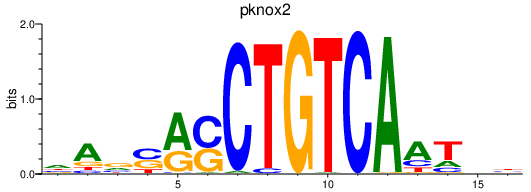
| pknox2 | 0.70 |
|
|
|
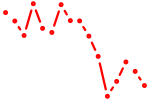
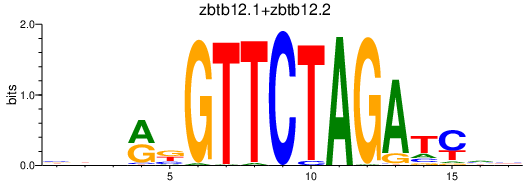
| zbtb12.1+zbtb12.2 | 0.70 |
|
|
|
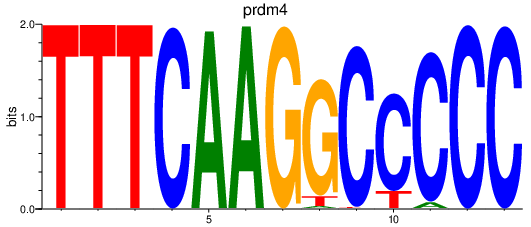
| prdm4 | 0.70 |
|

|
|
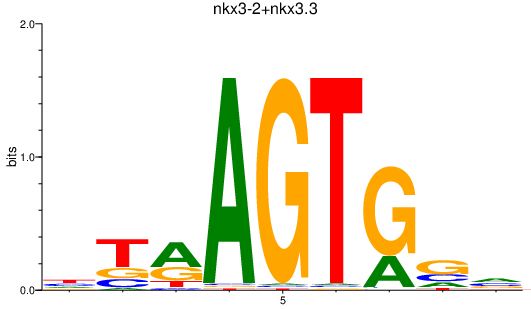
| nkx3-2+nkx3.3 | 0.69 |
|
|
|
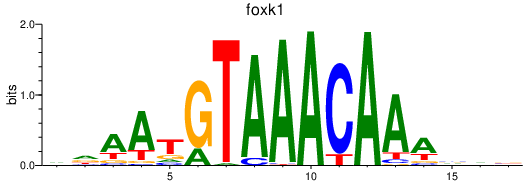
| foxk1_foxj3+si:dkey-68o6.6 | 0.69 |
|
|
|
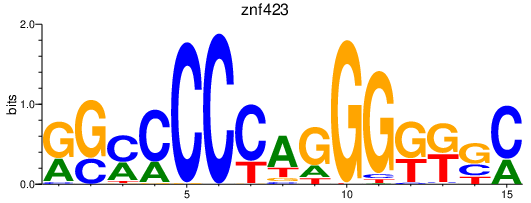
| znf423 | 0.69 |
|

|
|
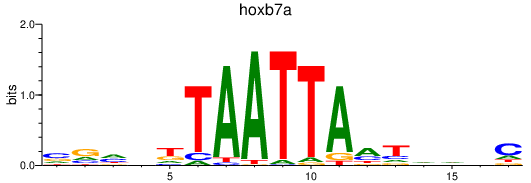
| hoxb7a | 0.69 |
|

|
|
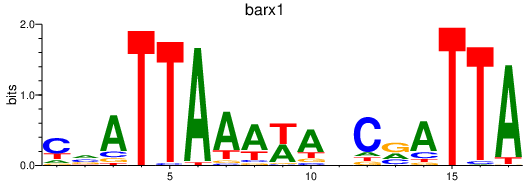
| barx1 | 0.68 |
|
|
|
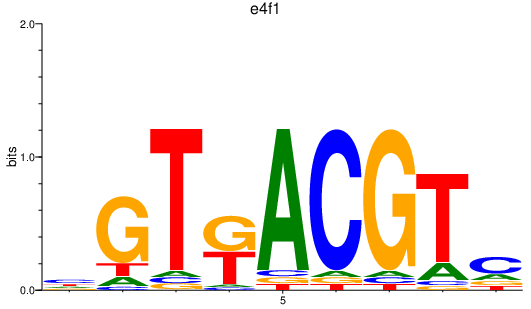
| e4f1 | 0.68 |
|

|
|
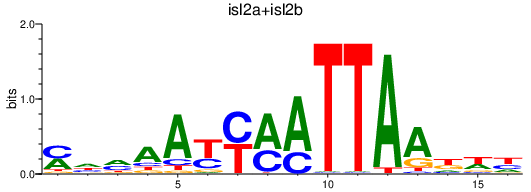
| isl2a+isl2b | 0.68 |
|
|
|
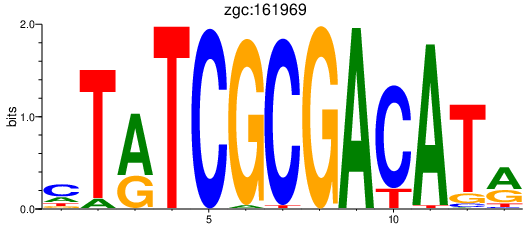
| zgc:161969 | 0.68 |
|

|
|
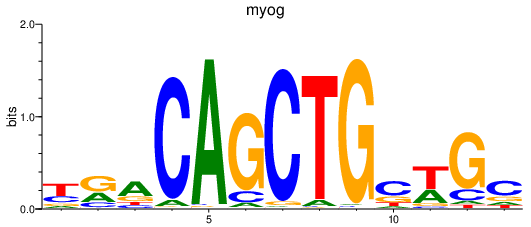
| myog | 0.67 |
|
|
|
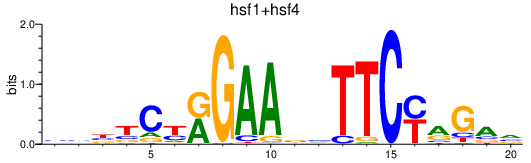
| hsf1+hsf4 | 0.67 |
|
|
|
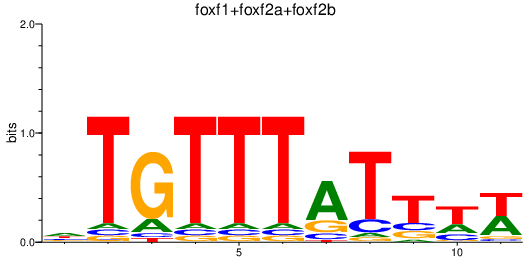
| foxf1+foxf2a+foxf2b | 0.67 |
|
|
|
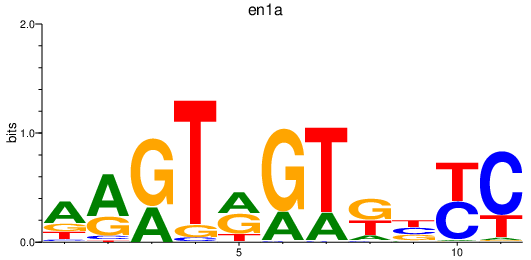
| en1a | 0.67 |
|

|
|
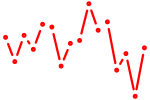
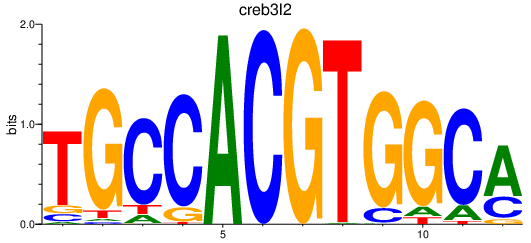
| creb3l2 | 0.67 |
|
|
|
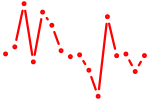
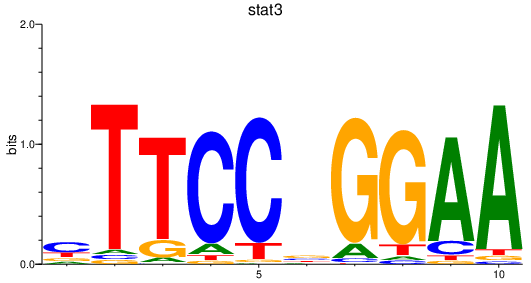
| stat3 | 0.67 |
|
|
|
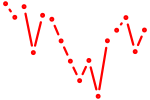
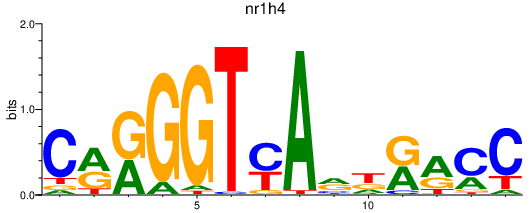
| nr1h4 | 0.67 |
|
|
|
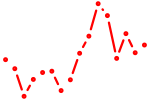
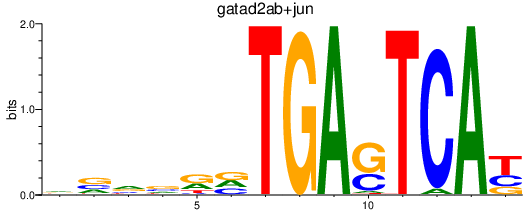
| gatad2ab+jun_fosl2_smarcc1b | 0.66 |
|
|
|
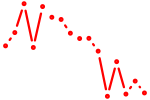
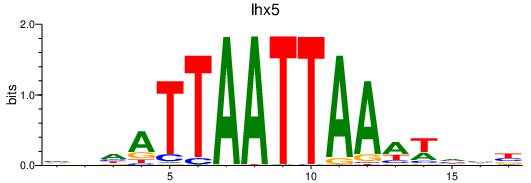
| lhx5 | 0.66 |
|
|
|
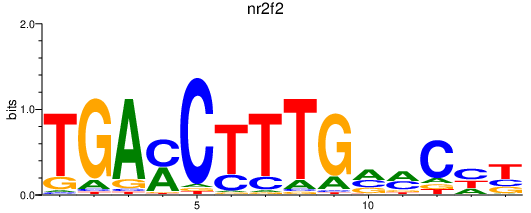
| nr2f2 | 0.66 |
|

|
|
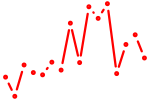
| rela_nfkb1 | 0.66 |
|
|

|
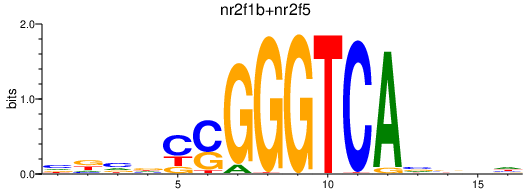
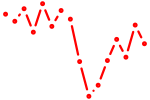
| nr2f1b+nr2f5 | 0.66 |
|

|
|
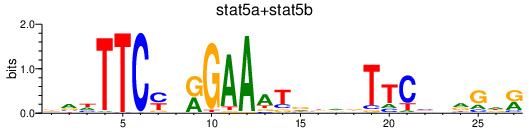
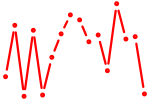
| stat5a+stat5b | 0.66 |
|
|
|
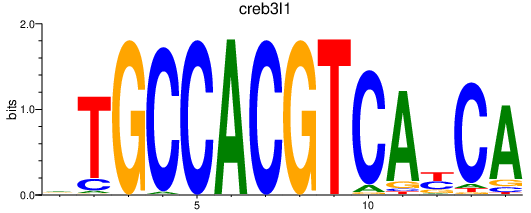
| creb3l1 | 0.66 |
|
|
|
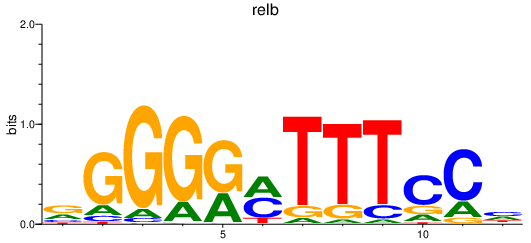
| relb | 0.66 |
|
|
|
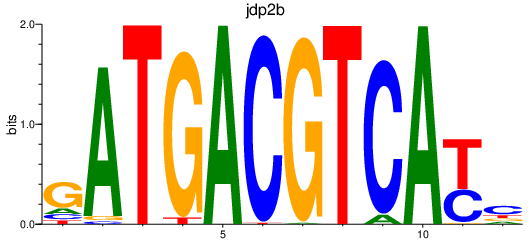
| jdp2b_june_atf7a+atf7b_creb5b | 0.66 |
|

|
|
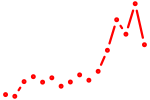
| foxg1a+foxg1c+foxg1d+zgc:113424 | 0.66 |
|
|
|
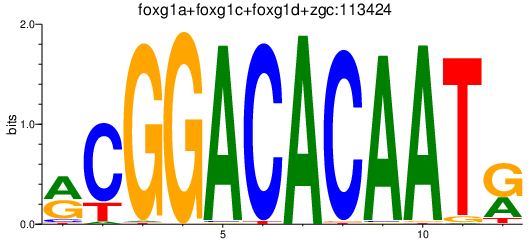
| hoxa4a+hoxc3a | 0.65 |
|

|
|
| raraa+rarab | 0.65 |
|
|

|
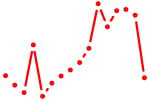
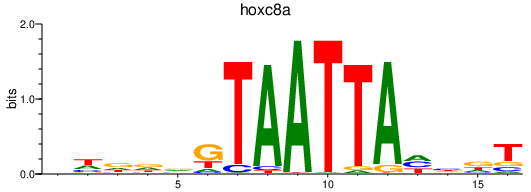
| hoxc8a | 0.65 |
|
|
|
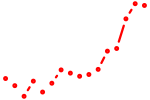
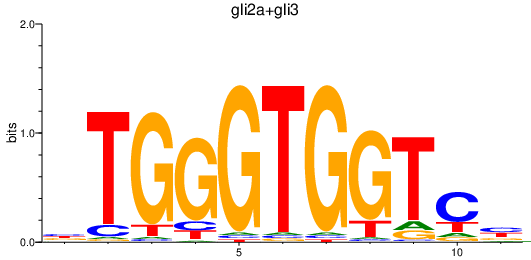
| gli2a+gli3 | 0.65 |
|
|
|
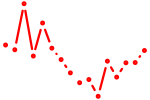
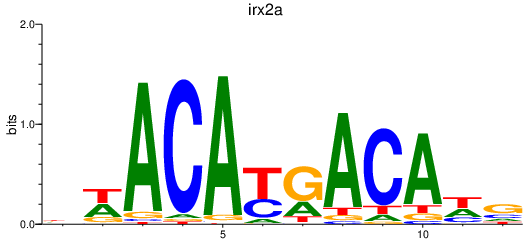
| irx2a | 0.65 |
|
|
|
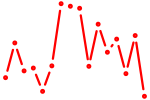
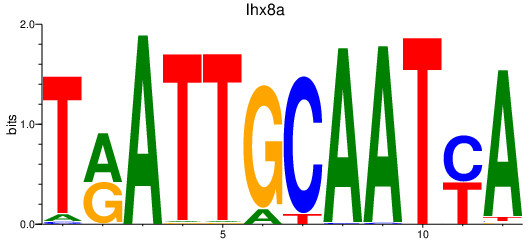
| lhx8a | 0.64 |
|
|
|
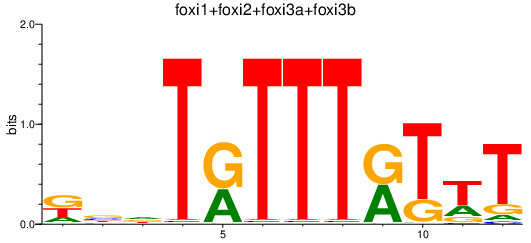
| foxi1+foxi2+foxi3a+foxi3b | 0.64 |
|

|
|
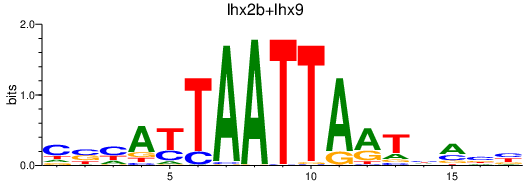
| lhx2b+lhx9 | 0.64 |
|
|
|
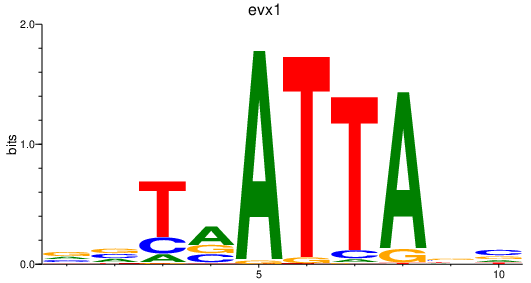
| evx1 | 0.63 |
|
|
|
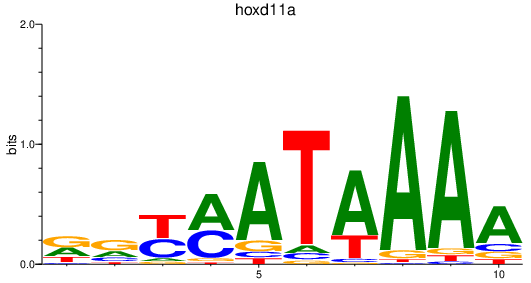
| hoxd11a | 0.63 |
|
|
|
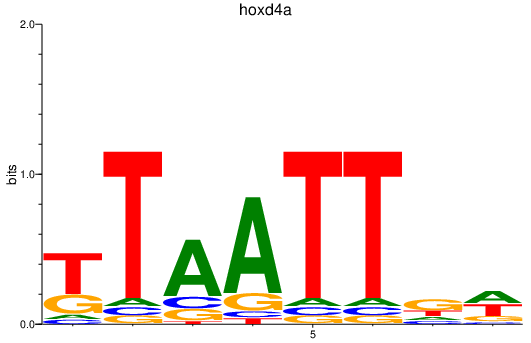
| hoxd4a | 0.60 |
|
|
|
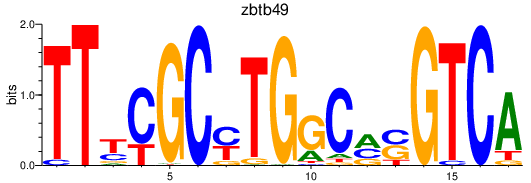
| zbtb49 | 0.60 |
|
|
|
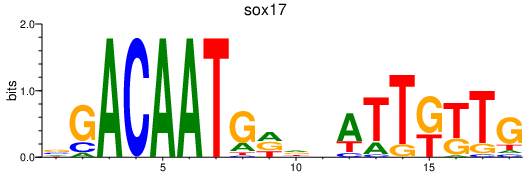
| sox17 | 0.59 |
|
|
|
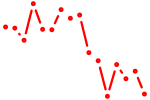
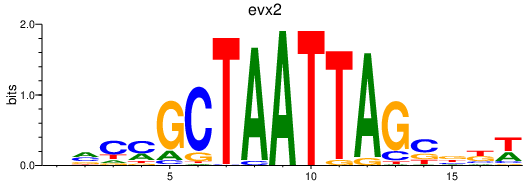
| evx2 | 0.59 |
|
|
|
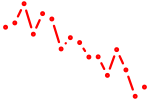
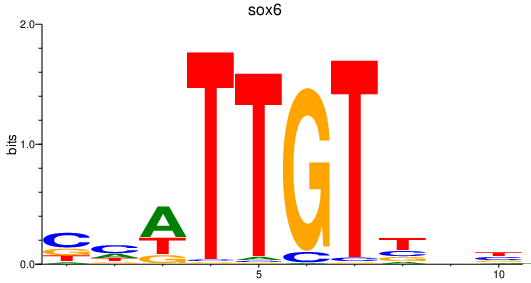
| sox6 | 0.59 |
|
|
|
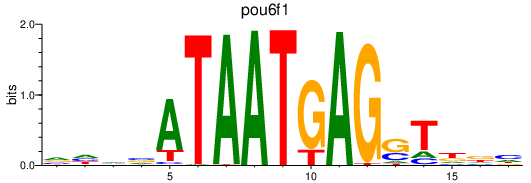
| pou6f1 | 0.58 |
|
|
|
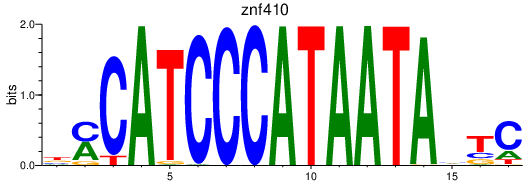
| znf410 | 0.57 |
|

|
|
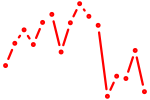
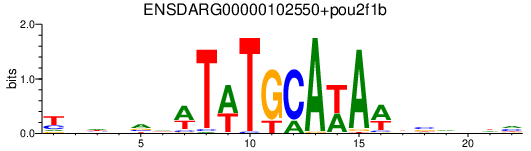
| ENSDARG00000102550+pou2f1b | 0.57 |
|
|
|
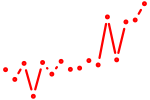
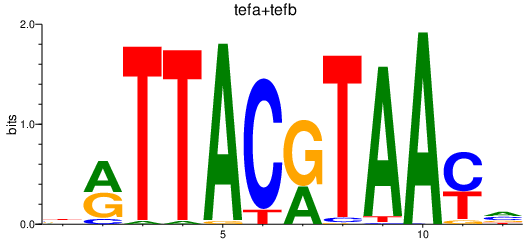
| tefa+tefb | 0.56 |
|
|
|
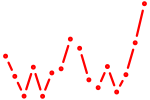
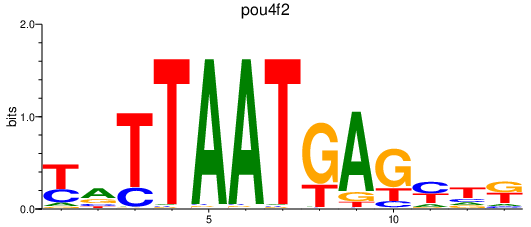
| pou4f2 | 0.56 |
|
|
|
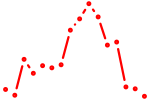
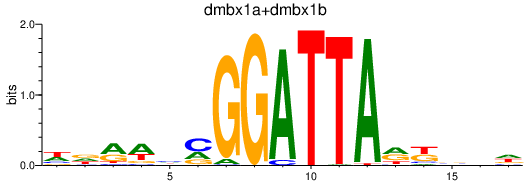
| dmbx1a+dmbx1b | 0.56 |
|
|
|
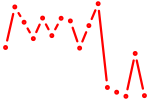
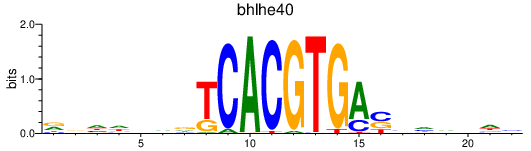
| bhlhe40 | 0.56 |
|
|
|
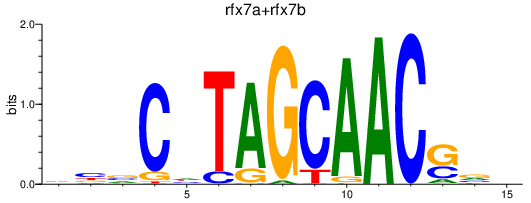
| rfx7a+rfx7b | 0.56 |
|

|
|
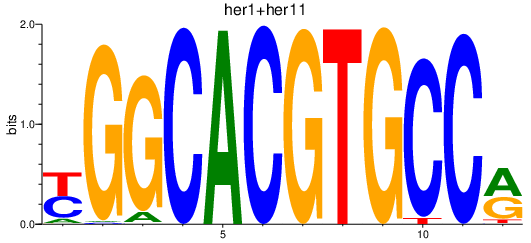
| her1+her11_her12+her15.2+her2 | 0.56 |
|

|
|
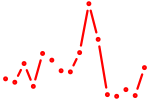
| tp73 | 0.56 |
|
|

|
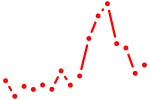
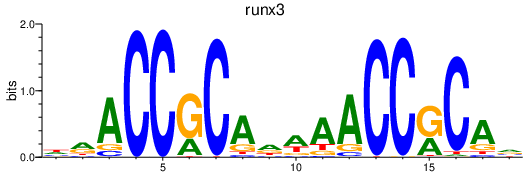
| runx3 | 0.55 |
|
|
|
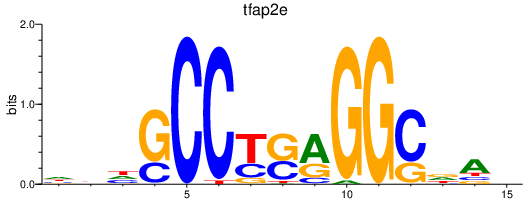
| tfap2e | 0.55 |
|

|
|
| foxb1a | 0.55 |
|
|

|
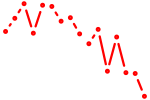
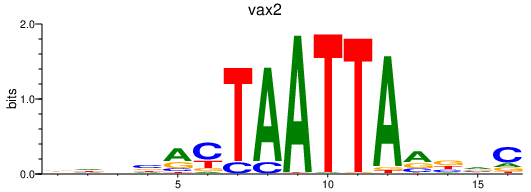
| vax2 | 0.55 |
|

|
|
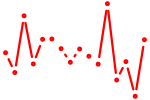
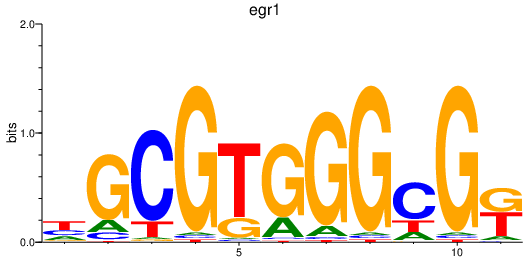
| egr1 | 0.55 |
|
|
|
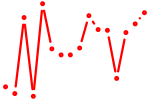
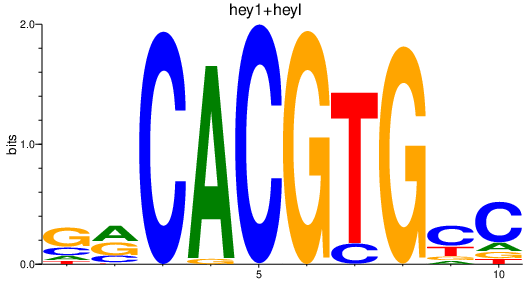
| hey1+heyl_hey2 | 0.55 |
|
|
|
| otpa+otpb | 0.54 |
|
|

|
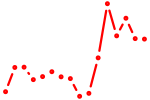
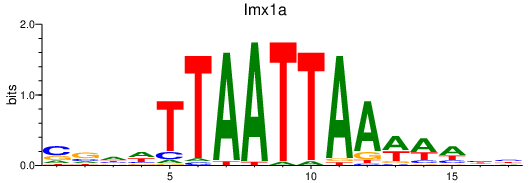
| lmx1a | 0.54 |
|

|
|
| nr4a3 | 0.54 |
|

|

|
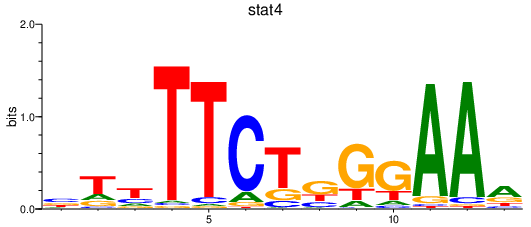
| stat4 | 0.54 |
|

|
|
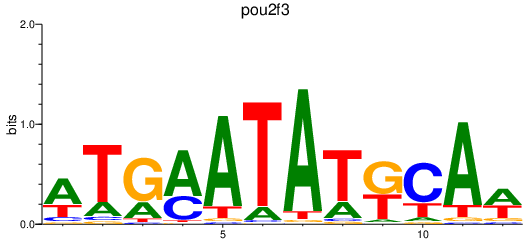
| pou2f3 | 0.54 |
|
|
|
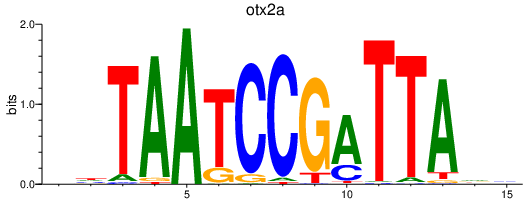
| otx2a_otx1 | 0.53 |
|
|
|
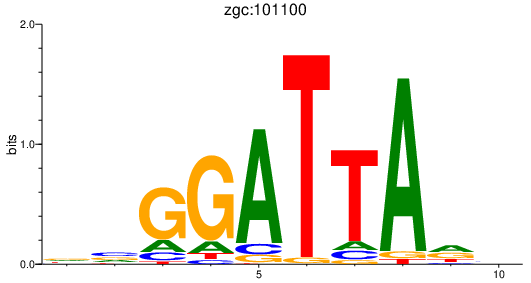
| zgc:101100 | 0.52 |
|
|
|
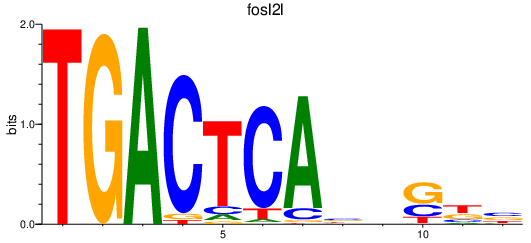
| fosl2l | 0.52 |
|
|
|
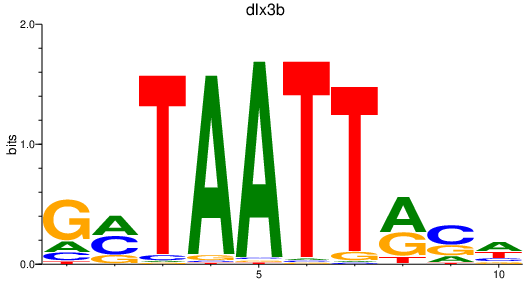
| dlx3b | 0.52 |
|
|
|
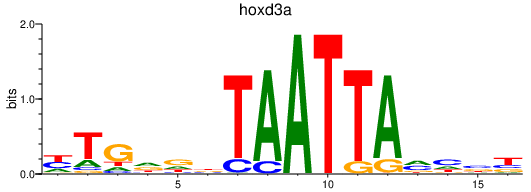
| hoxd3a | 0.52 |
|

|
|
| eomesa+eomesb | 0.52 |
|
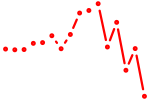
|
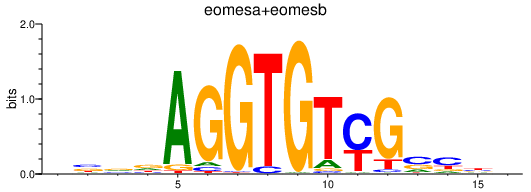
|
| egr3 | 0.51 |
|
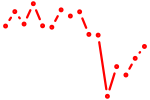
|
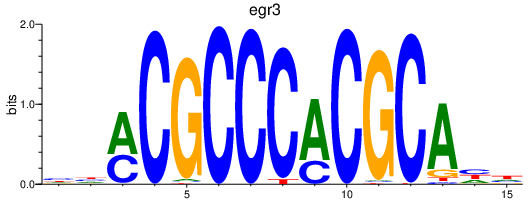
|
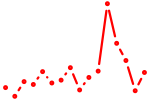
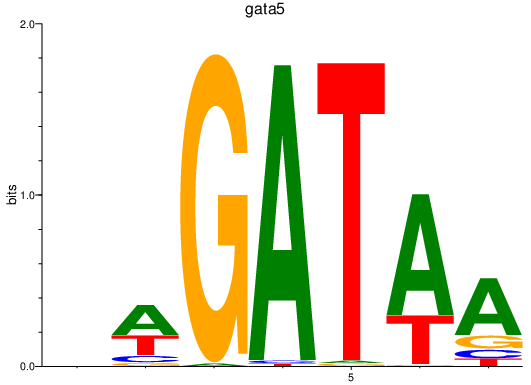
| gata5 | 0.51 |
|
|
|
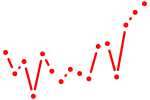
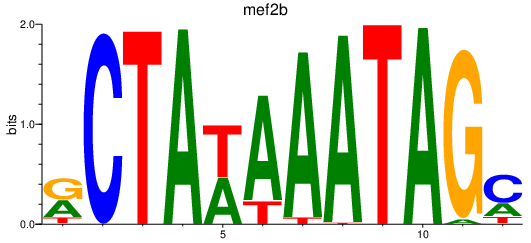
| mef2b | 0.51 |
|
|
|
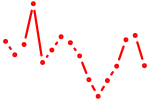
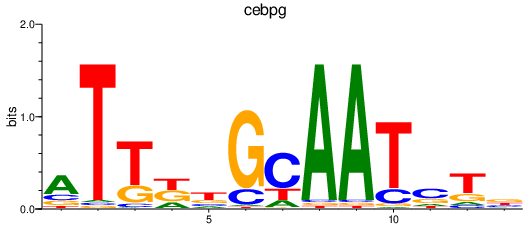
| cebpg | 0.50 |
|
|
|
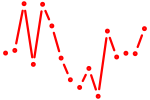
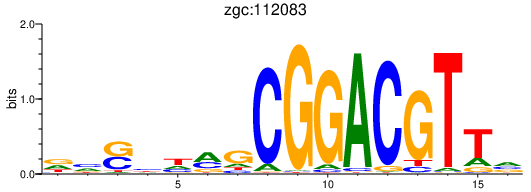
| zgc:112083 | 0.50 |
|
|
|
| rxraa+rxrba+rxrbb+rxrgb | 0.50 |
|
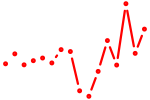
|
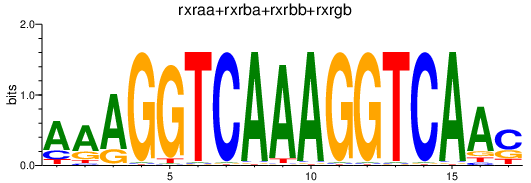
|
| e2f8 | 0.50 |
|
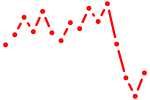
|
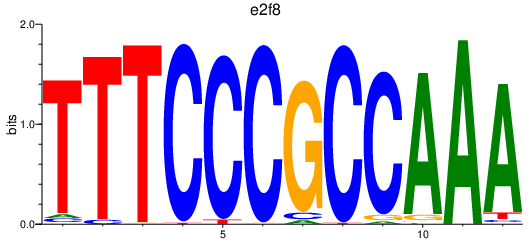
|
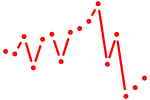
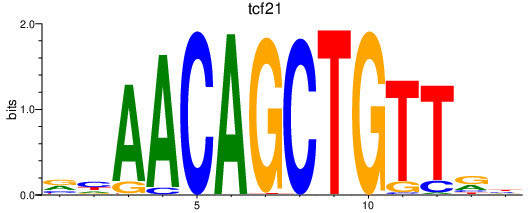
| tcf21 | 0.49 |
|
|
|
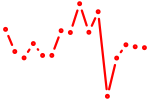
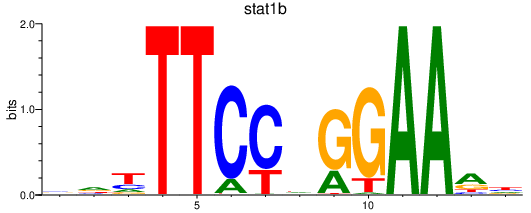
| stat1b_stat1a+stat2 | 0.49 |
|
|
|
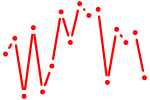
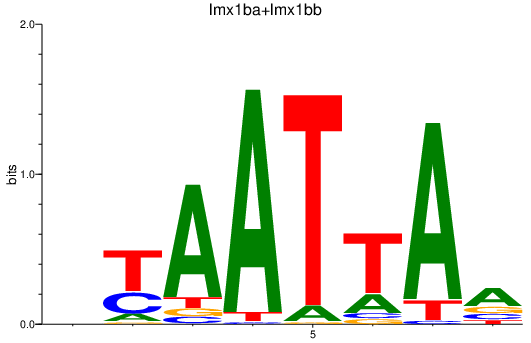
| lmx1ba+lmx1bb | 0.49 |
|
|
|
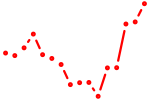
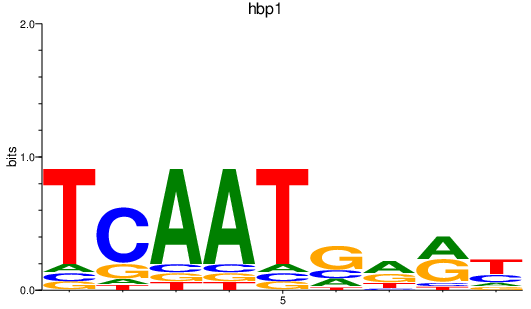
| hbp1 | 0.49 |
|
|
|
| nfe2l2a_nfe2+nfe2l1a | 0.48 |
|
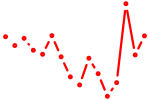
|
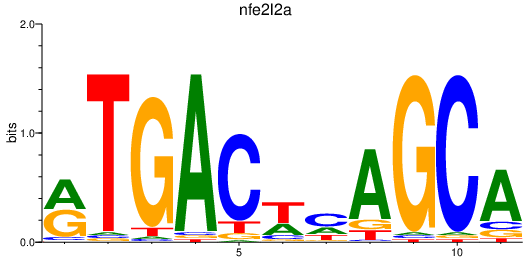
|
| irf6 | 0.47 |
|
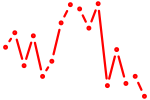
|
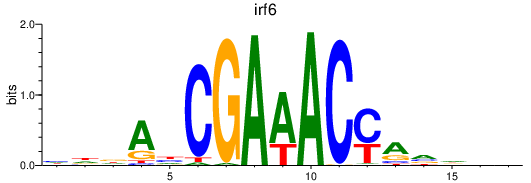
|
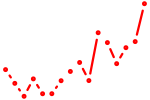
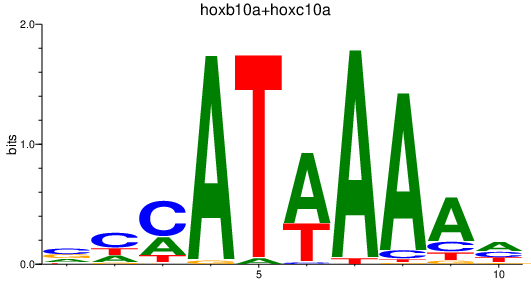
| hoxb10a+hoxc10a | 0.47 |
|
|
|
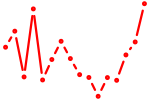
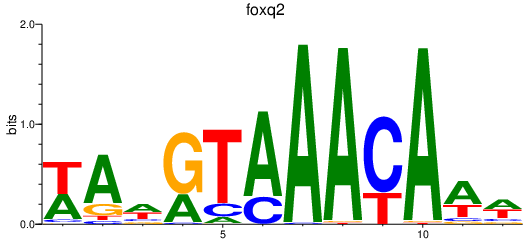
| foxq2 | 0.47 |
|
|
|
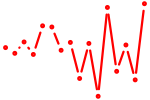
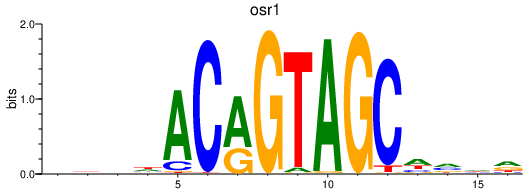
| osr1 | 0.46 |
|
|
|
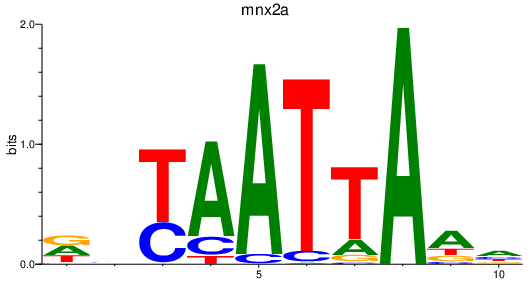
| mnx2a | 0.46 |
|

|
|
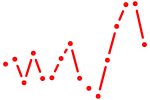
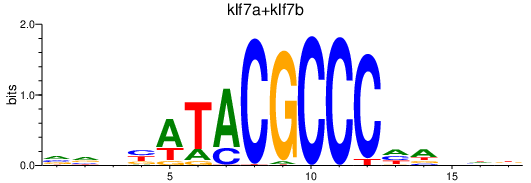
| klf7a+klf7b | 0.46 |
|
|
|
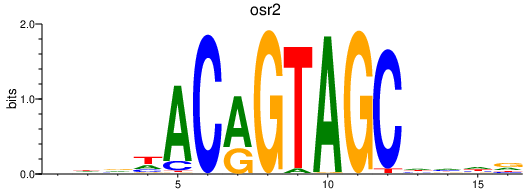
| osr2 | 0.46 |
|

|
|
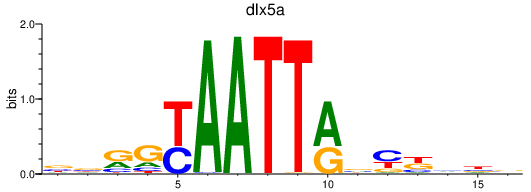
| dlx5a | 0.45 |
|

|
|
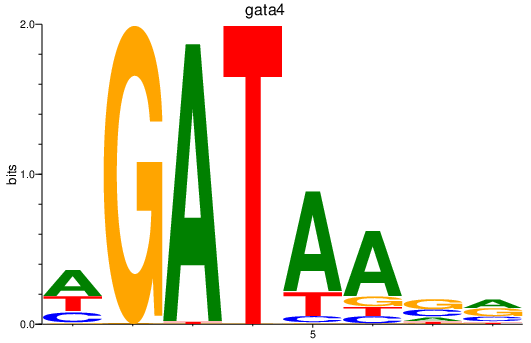
| gata4 | 0.45 |
|
|
|
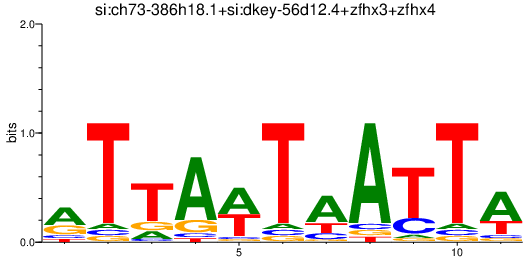
| si:ch73-386h18.1+si:dkey-56d12.4+zfhx3+zfhx4 | 0.45 |
|

|
|
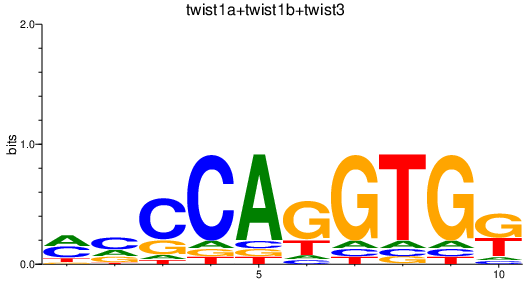
| twist1a+twist1b+twist3 | 0.45 |
|

|
|
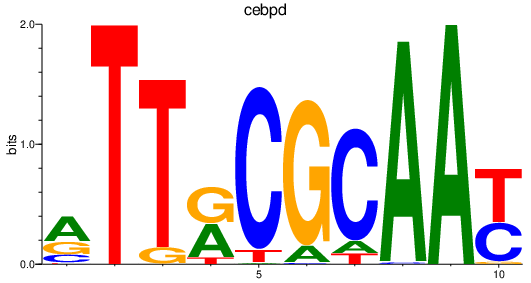
| cebpd | 0.45 |
|
|
|
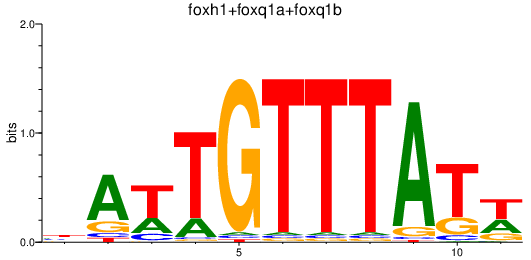
| foxh1+foxq1a+foxq1b | 0.45 |
|
|
|
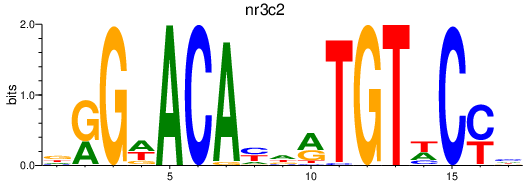
| nr3c2 | 0.44 |
|

|
|
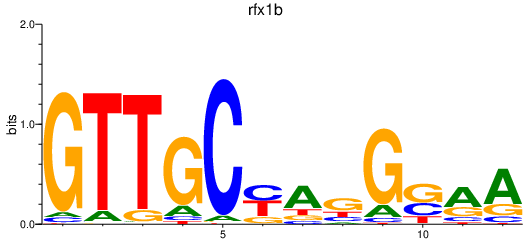
| rfx1b | 0.44 |
|

|
|
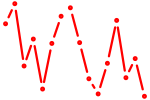
| hoxb9a | 0.43 |
|
|
|
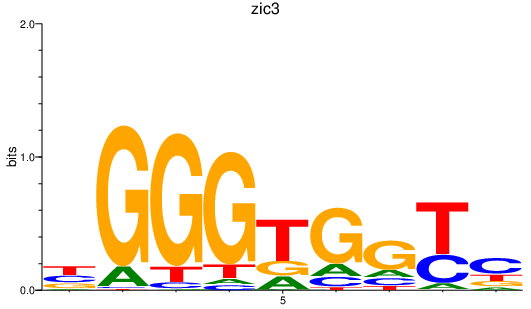
| zic3 | 0.43 |
|

|
|
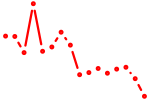
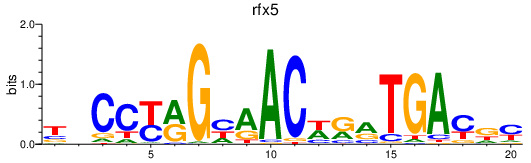
| rfx5 | 0.43 |
|
|
|
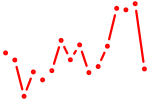
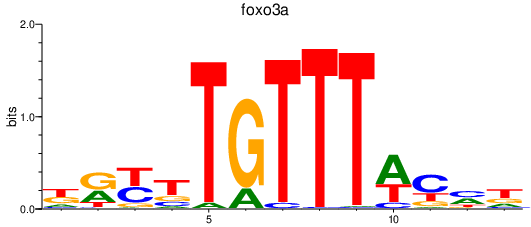
| foxo3a | 0.43 |
|
|
|
| znf148 | 0.42 |
|
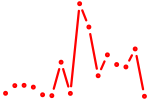
|
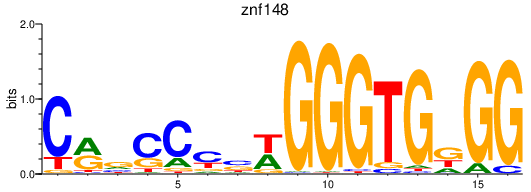
|
| vox | 0.42 |
|
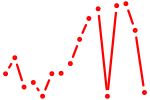
|
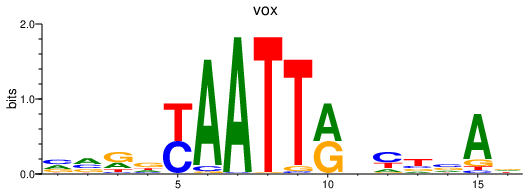
|
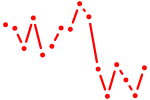
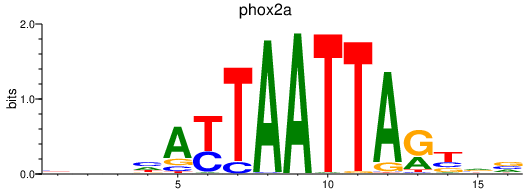
| phox2a | 0.42 |
|
|
|
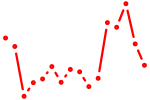
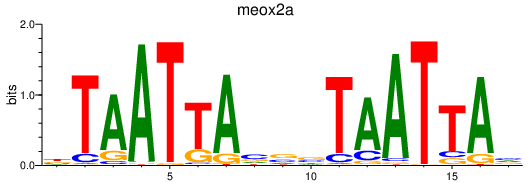
| meox2a | 0.42 |
|
|
|
| phox2ba | 0.41 |
|
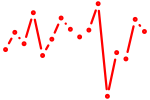
|

|
| irf4a+irf4b | 0.41 |
|
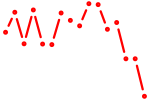
|
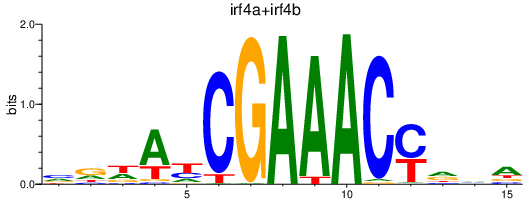
|
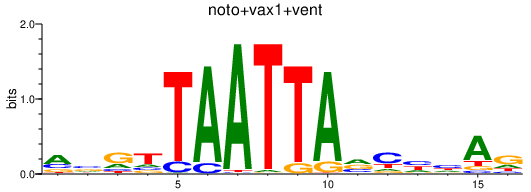
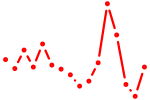
| noto+vax1+vent | 0.41 |
|
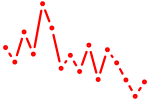
|
|
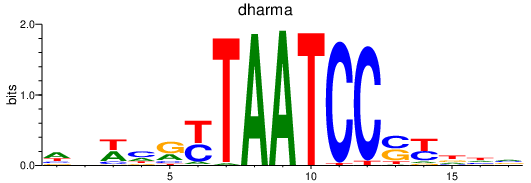
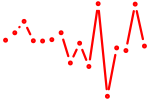
| dharma | 0.40 |
|
|
|
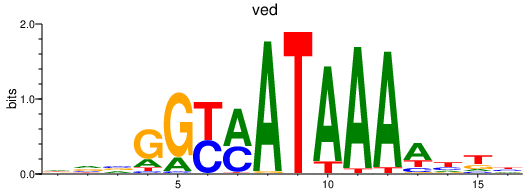
| ved | 0.40 |
|
|
|
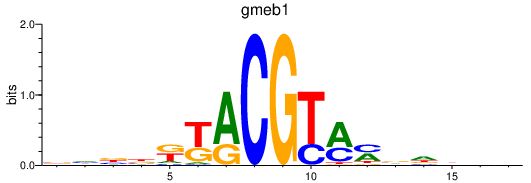
| gmeb1 | 0.40 |
|
|
|
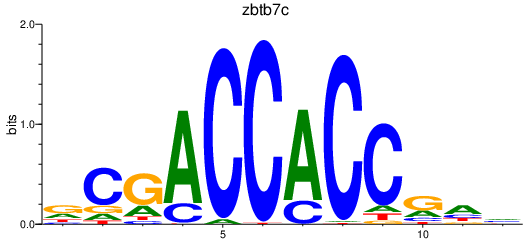
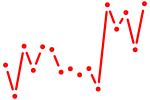
| zbtb7c | 0.40 |
|
|
|
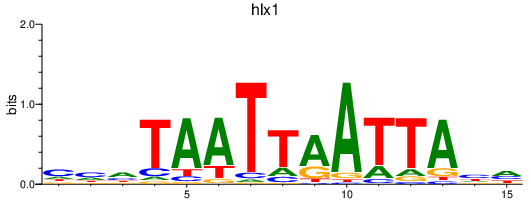
| hlx1 | 0.40 |
|

|
|
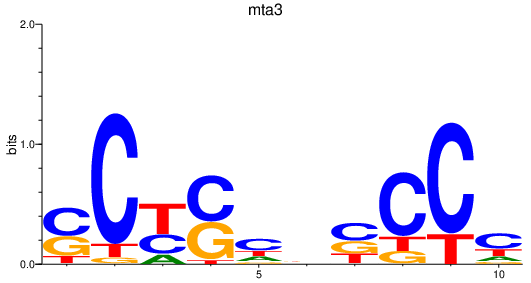
| mta3 | 0.40 |
|
|
|
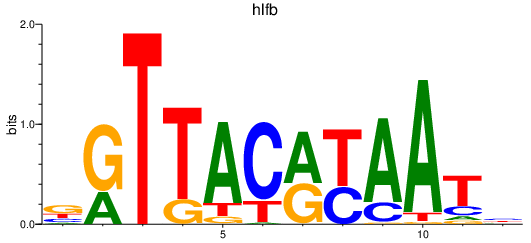
| hlfb | 0.40 |
|

|
|
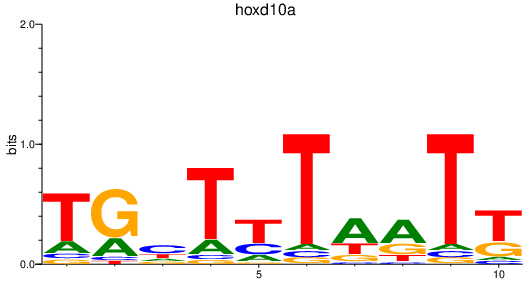
| hoxd10a | 0.39 |
|
|
|
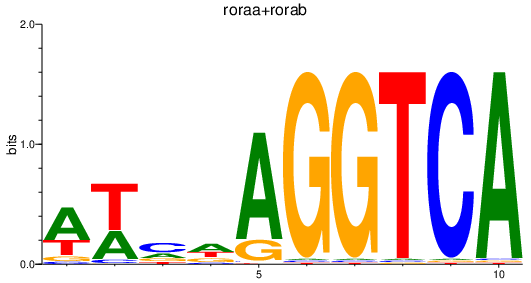
| roraa+rorab | 0.39 |
|
|
|
| cers4a+hoxb13a | 0.39 |
|
|

|
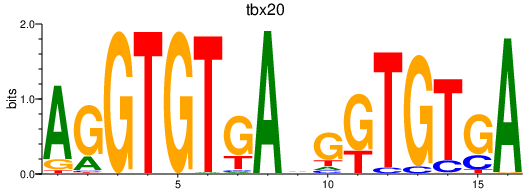
| tbx20 | 0.38 |
|
|
|
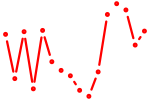
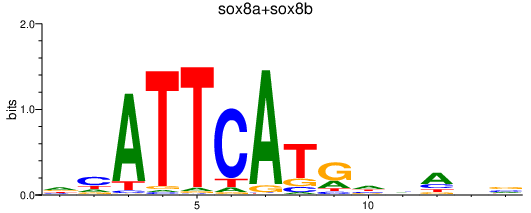
| sox8a+sox8b | 0.38 |
|
|
|
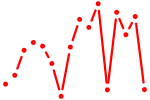
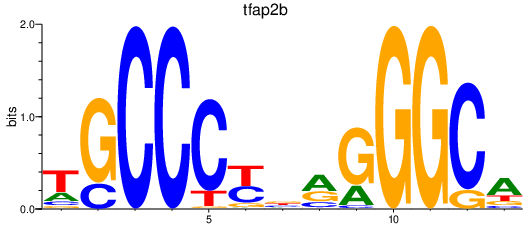
| tfap2b | 0.37 |
|
|
|
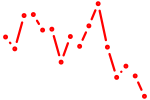
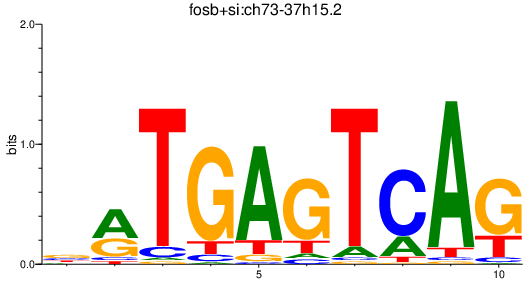
| fosb+si:ch73-37h15.2 | 0.37 |
|
|
|
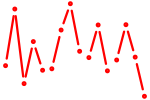
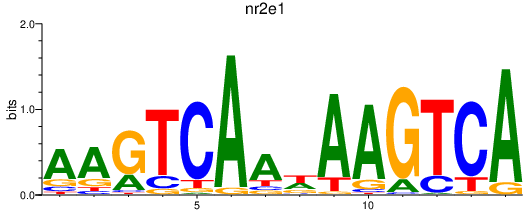
| nr2e1 | 0.37 |
|
|
|
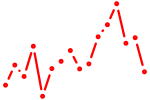
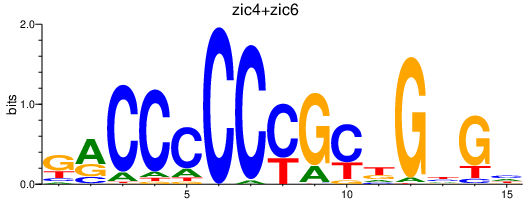
| zic4+zic6 | 0.36 |
|
|
|
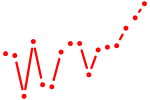
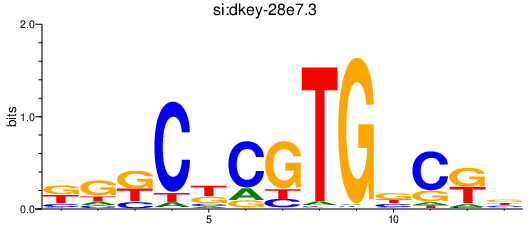
| si:dkey-28e7.3 | 0.36 |
|
|
|
| msx1b+msx3 | 0.36 |
|
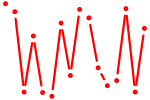
|
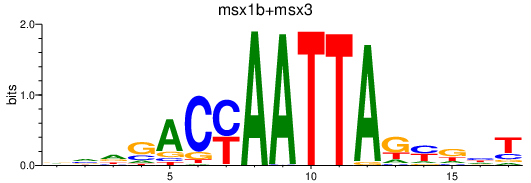
|
| nr1d2a | 0.36 |
|
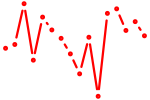
|
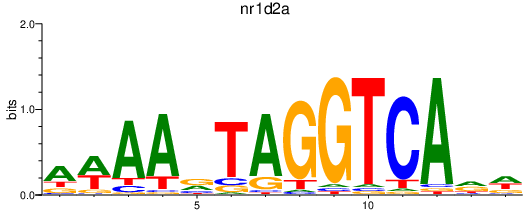
|
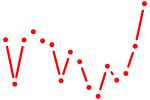
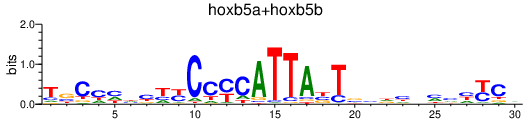
| hoxb5a+hoxb5b | 0.35 |
|
|
|
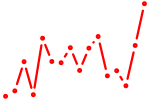
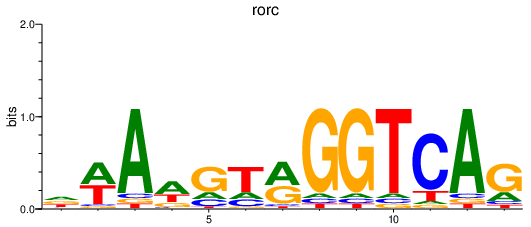
| rorc | 0.34 |
|
|
|
| si:dkey-68o6.8 | 0.34 |
|
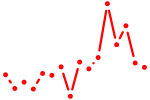
|
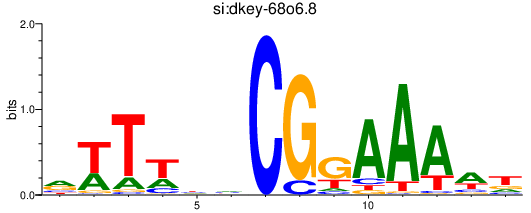
|
| msx2a | 0.33 |
|
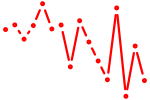
|
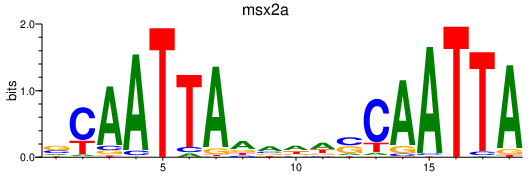
|
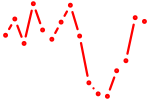
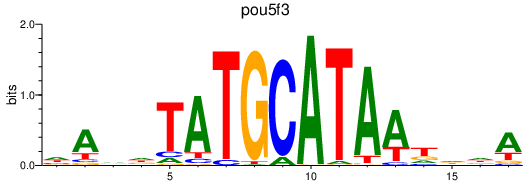
| pou5f3 | 0.33 |
|
|
|
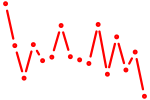
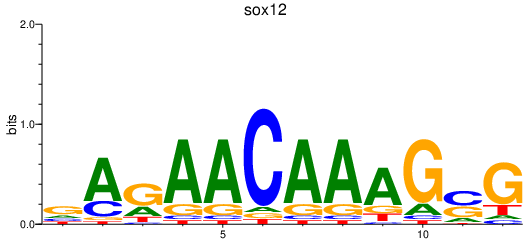
| sox12 | 0.33 |
|
|
|
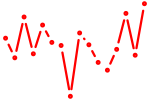
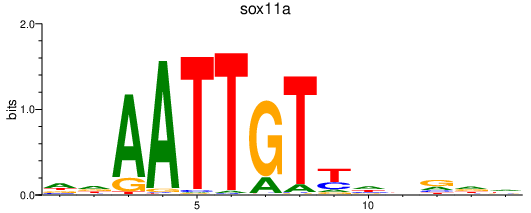
| sox11a | 0.32 |
|
|
|
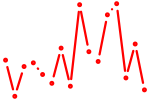
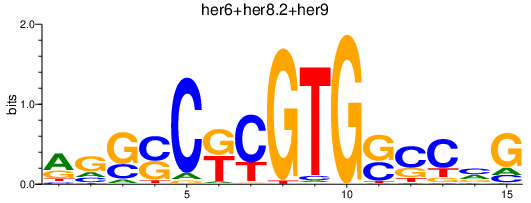
| her6+her8.2+her9 | 0.32 |
|
|
|
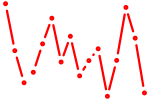
| isl1+isl1l | 0.32 |
|
|
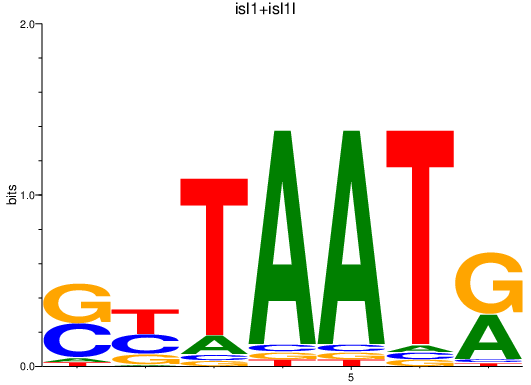
|
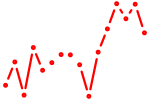
| phox2bb | 0.31 |
|
|
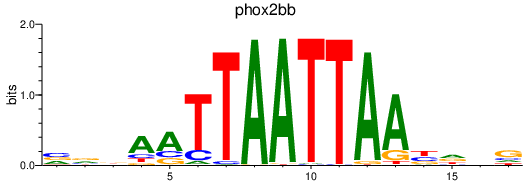
|
| lhx1a+lhx1b | 0.31 |
|
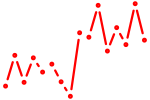
|
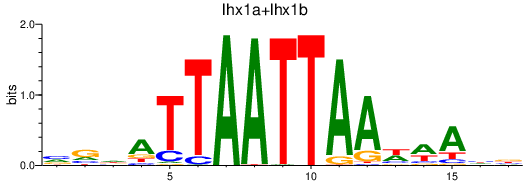
|
| nkx1.2lb | 0.30 |
|
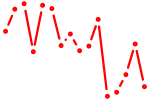
|
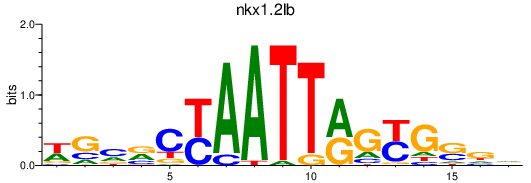
|
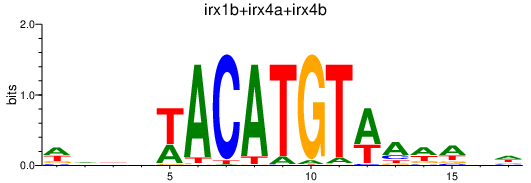
| irx1b+irx4a+irx4b | 0.30 |
|

|
|
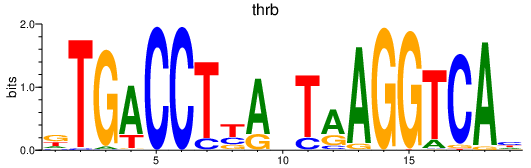
| thrb | 0.29 |
|

|
|
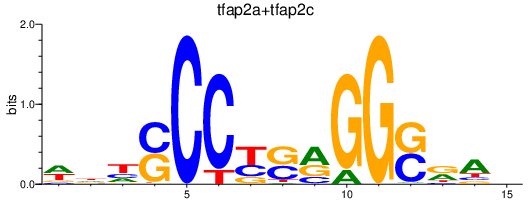
| tfap2a+tfap2c | 0.29 |
|

|
|
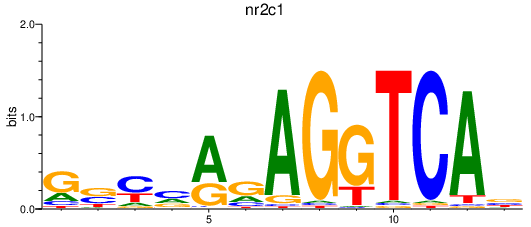
| nr2c1 | 0.28 |
|
|
|
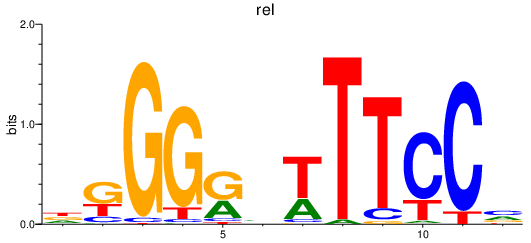
| rel | 0.28 |
|
|
|
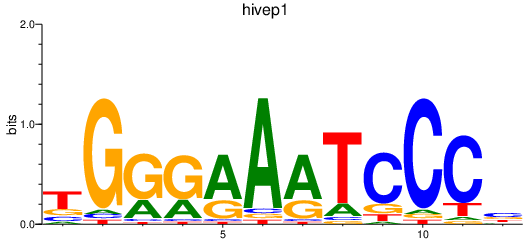
| hivep1 | 0.28 |
|
|
|
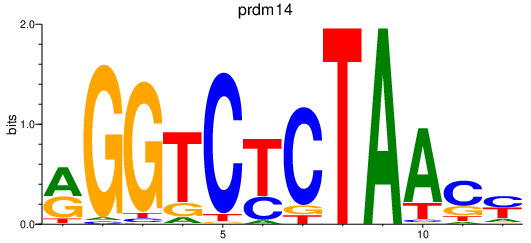
| prdm14 | 0.27 |
|
|
|
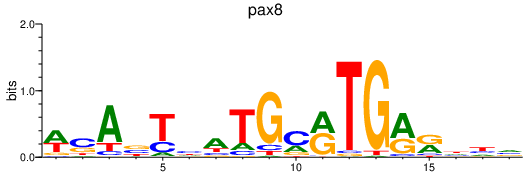
| pax8 | 0.27 |
|
|
|
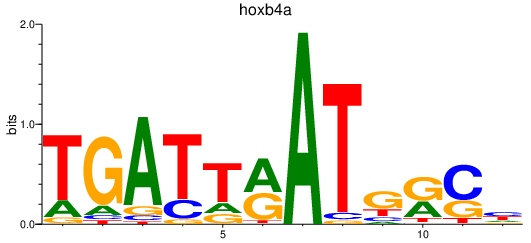
| hoxb4a | 0.26 |
|
|
|
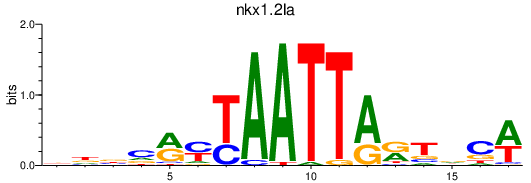
| nkx1.2la_rx1+rx2 | 0.26 |
|
|
|
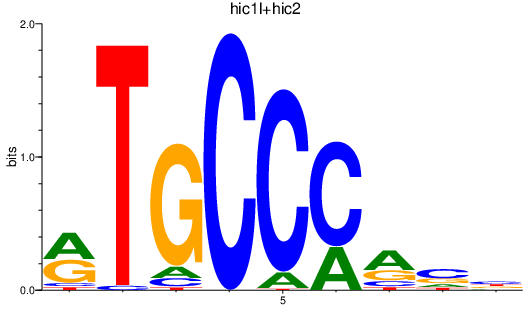
| hic1l+hic2 | 0.25 |
|
|
|
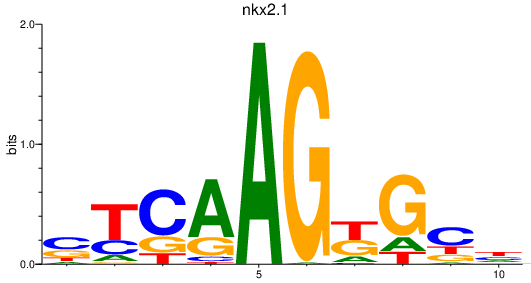
| nkx2.1 | 0.24 |
|
|
|
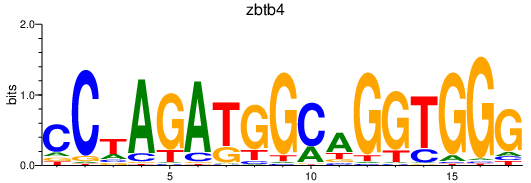
| zbtb4 | 0.24 |
|
|
|
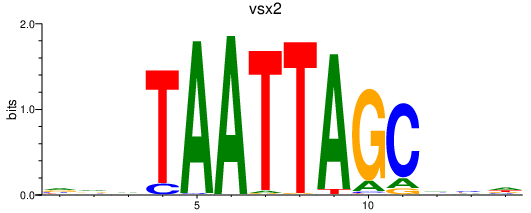
| vsx2 | 0.24 |
|
|
|
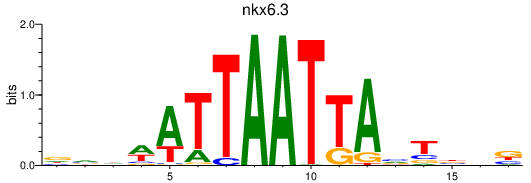
| nkx6.3 | 0.23 |
|

|
|
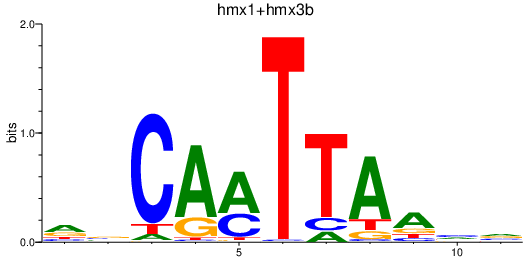
| hmx1+hmx3b | 0.22 |
|
|
|
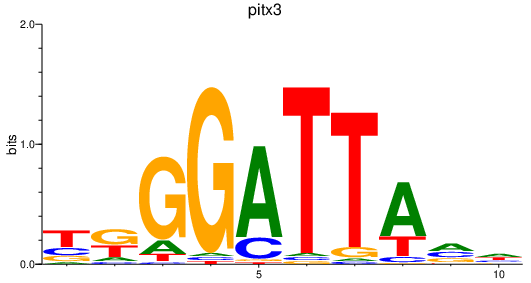
| pitx3 | 0.22 |
|
|
|
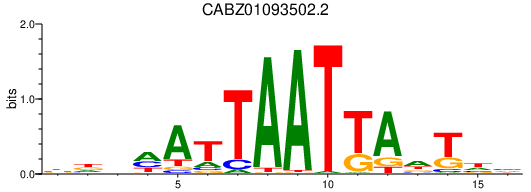
| CABZ01093502.2 | 0.21 |
|
|
|
| gata1a+gata2b | 0.21 |
|

|
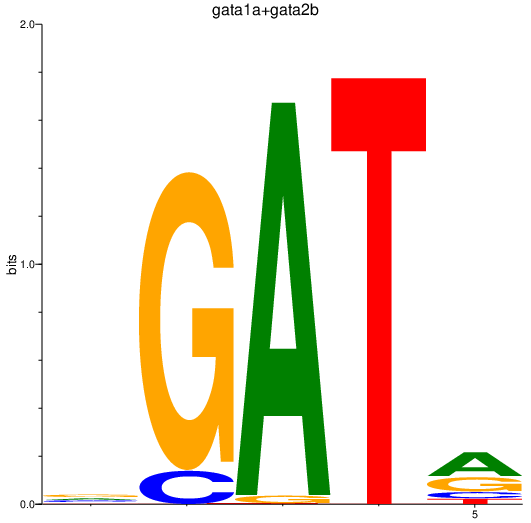
|
| cxxc1a+cxxc1b+mbd1a | 0.20 |
|

|
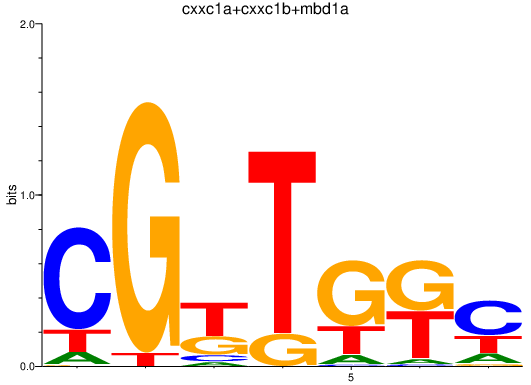
|
| six6a+six6b | 0.19 |
|
|
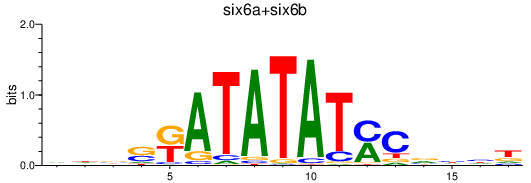
|
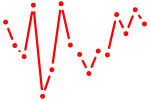
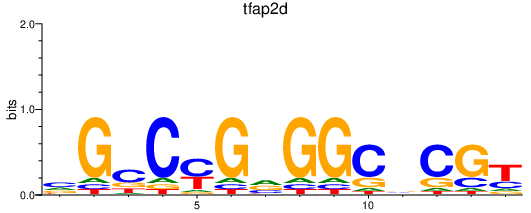
| tfap2d | 0.16 |
|
|
|
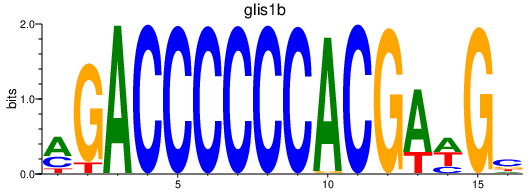
| glis1b | 0.12 |
|

|
|