ISMARA results
ISMARA - Integrated System for Motif Actitivity Response Analysis is a free online tool that recognizes most important transcription factors that are changing their activity in a set of samples.
All motifs sorted by activity significance
| Motif name | Z-value | Associated genes | Profile | Logo |
|---|---|---|---|---|
| STB3 | 8.90 |
|

|

|
| RAP1 | 8.56 |
|

|

|
| SFP1 | 6.73 |
|

|

|
| XBP1 | 4.11 |
|

|

|
| SUM1 | 4.01 |
|

|

|
| RCS1 | 3.75 |
|

|
|
| RPH1 | 3.74 |
|

|

|
| AFT2 | 3.67 |
|

|

|
| HAP3 | 3.56 |
|

|

|
| UME6 | 3.50 |
|

|

|
| ABF1 | 3.42 |
|

|

|
| STP4 | 2.90 |
|

|

|
| PBF1 | 2.90 |
|

|

|
| STE12 | 2.89 |
|

|

|
| SIP4 | 2.84 |
|

|

|
| HAP4 | 2.74 |
|

|

|
| GIS1 | 2.69 |
|

|

|
| CIN5 | 2.66 |
|

|

|
| MBP1 | 2.64 |
|

|

|
| AZF1 | 2.59 |
|

|

|
| FHL1 | 2.57 |
|

|

|
| YDR026c | 2.57 |
|

|

|
| ACE2 | 2.52 |
|

|

|
| RIM101 | 2.50 |
|

|

|
| PBF2 | 2.50 |
|

|

|
| HCM1 | 2.45 |
|

|

|
| RGT1 | 2.31 |
|

|

|
| HSF1 | 2.17 |
|

|

|
| LEU3 | 2.15 |
|

|

|
| YPR022C | 2.04 |
|

|

|
| YPL230W | 2.04 |
|

|
|
| ADR1 | 1.99 |
|

|

|
| SWI4 | 1.97 |
|

|

|
| OAF1 | 1.94 |
|

|

|
| HAP5 | 1.89 |
|

|

|
| NDT80 | 1.88 |
|

|

|
| ABF2 | 1.87 |
|

|

|
| MCM1 | 1.79 |
|

|

|
| SPT15 | 1.76 |
|

|

|
| ECM22 | 1.73 |
|

|

|
| PHO4 | 1.70 |
|

|

|
| YLR278C | 1.69 |
|

|

|
| HMRA2 | 1.65 |
|

|

|
| SIG1 | 1.65 |
|

|

|
| RSC3 | 1.64 |
|

|

|
| CAD1 | 1.61 |
|

|

|
| NHP10 | 1.61 |
|

|

|
| SWI6 | 1.61 |
|

|

|
| MET32 | 1.60 |
|

|

|
| GCR1 | 1.58 |
|

|

|
| GSM1 | 1.58 |
|

|

|
| EDS1 | 1.56 |
|

|

|
| GAT1 | 1.55 |
|

|

|
| TEC1 | 1.52 |
|

|

|
| YKL222C | 1.46 |
|

|

|
| YAP7 | 1.46 |
|

|
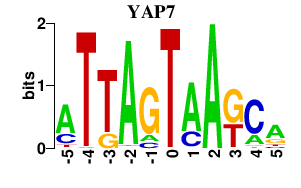
|
| MIG1 | 1.45 |
|

|

|
| DOT6 | 1.45 |
|

|
|
| STP3 | 1.44 |
|

|

|
| GAL4 | 1.44 |
|

|

|
| MIG3 | 1.44 |
|

|

|
| STB4 | 1.42 |
|

|

|
| REI1 | 1.34 |
|

|

|
| TOS8 | 1.34 |
|

|

|
| YPR013C | 1.32 |
|

|

|
| YBR239C | 1.31 |
|

|

|
| YDR520C | 1.30 |
|

|

|
| NRG1 | 1.26 |
|

|

|
| CAT8 | 1.24 |
|

|

|
| CUP2 | 1.23 |
|

|

|
| SFL1 | 1.23 |
|

|

|
| CRZ1 | 1.21 |
|

|

|
| FKH1 | 1.18 |
|

|

|
| ROX1 | 1.18 |
|

|

|
| THI2 | 1.17 |
|

|

|
| CBF1 | 1.17 |
|

|

|
| RLM1 | 1.13 |
|

|

|
| CST6 | 1.12 |
|

|

|
| FKH2 | 1.07 |
|

|

|
| GAL80 | 1.06 |
|

|

|
| YOX1 | 1.05 |
|

|

|
| HAP1 | 1.05 |
|

|

|
| GCN4 | 1.05 |
|

|

|
| ORC1 | 1.04 |
|

|

|
| CHA4 | 1.02 |
|

|

|
| HAP2 | 1.02 |
|

|

|
| MSN4 | 1.01 |
|

|

|
| TBS1 | 0.99 |
|

|

|
| UPC2 | 0.99 |
|

|

|
| CUP9 | 0.98 |
|

|

|
| PHD1 | 0.98 |
|

|

|
| REB1 | 0.92 |
|

|

|
| RGM1 | 0.91 |
|

|

|
| YBL054W | 0.90 |
|

|
|
| RPN4 | 0.90 |
|

|

|
| AFT1 | 0.88 |
|

|

|
| MSN2 | 0.88 |
|

|

|
| YER130C | 0.88 |
|

|

|
| YGR067C | 0.87 |
|

|

|
| HAC1 | 0.84 |
|

|

|
| YLL054C | 0.84 |
|

|

|
| SWI5 | 0.84 |
|

|

|
| MET31 | 0.83 |
|

|

|
| INO2 | 0.83 |
|

|

|
| INO4 | 0.83 |
|

|

|
| BAS1 | 0.83 |
|

|

|
| STB1 | 0.82 |
|

|

|
| MAC1 | 0.82 |
|

|

|
| GLN3 | 0.81 |
|

|

|
| RFX1 | 0.80 |
|

|

|
| RDR1 | 0.80 |
|

|

|
| YER184C | 0.80 |
|

|

|
| YRM1 | 0.78 |
|

|

|
| MIG2 | 0.78 |
|

|

|
| PDR8 | 0.77 |
|

|

|
| DAL82 | 0.77 |
|

|

|
| ARO80 | 0.76 |
|

|

|
| ZMS1 | 0.76 |
|

|

|
| YJL103C | 0.76 |
|

|
|
| STP2 | 0.75 |
|

|

|
| SKO1 | 0.75 |
|

|

|
| UGA3 | 0.75 |
|

|

|
| CEP3 | 0.74 |
|

|

|
| SNT2 | 0.73 |
|

|

|
| HAL9 | 0.73 |
|

|

|
| RDS1 | 0.73 |
|

|

|
| USV1 | 0.73 |
|

|

|
| GAT4 | 0.73 |
|

|

|
| YPR196W | 0.72 |
|

|

|
| ECM23 | 0.72 |
|

|

|
| RDS2 | 0.69 |
|

|

|
| YAP3 | 0.69 |
|

|

|
| PUT3 | 0.69 |
|

|

|
| LYS14 | 0.68 |
|

|

|
| RTG3 | 0.68 |
|

|

|
| TBF1 | 0.67 |
|

|

|
| RSC30 | 0.67 |
|

|

|
| GZF3 | 0.66 |
|

|

|
| NHP6B | 0.64 |
|

|

|
| FZF1 | 0.63 |
|

|

|
| YAP1 | 0.62 |
|

|

|
| MATALPHA2 | 0.60 |
|

|

|
| PDR1 | 0.59 |
|

|

|
| YAP6 | 0.59 |
|

|

|
| SMP1 | 0.58 |
|

|

|
| YNR063W | 0.56 |
|

|

|
| DAL80 | 0.54 |
|

|

|
| ASG1 | 0.54 |
|

|

|
| SUT2 | 0.52 |
|

|

|
| PHO2 | 0.51 |
|

|

|
| PDR3 | 0.49 |
|

|

|
| SOK2 | 0.48 |
|

|

|
| YML081W | 0.47 |
|

|

|
| SRD1 | 0.45 |
|

|

|
| NHP6A | 0.41 |
|

|

|
| MGA1 | 0.41 |
|

|

|
| TYE7 | 0.39 |
|

|

|
| STB5 | 0.38 |
|

|

|
| SKN7 | 0.38 |
|

|

|
| YRR1 | 0.36 |
|

|

|
| GAT3 | 0.33 |
|

|

|
| YPR015C | 0.29 |
|

|

|
| TEA1 | 0.28 |
|

|

|