Project
2D miR_HR1_12
Navigation
Downloads
ISMARA results 2D miR_HR1_12
ISMARA - Integrated System for Motif Actitivity Response Analysis is a free online tool that recognizes most important transcription factors that are changing their activity in a set of samples.
All motifs sorted by activity significance
| Motif name | Z-value | Associated genes | Profile | Logo |
|---|---|---|---|---|
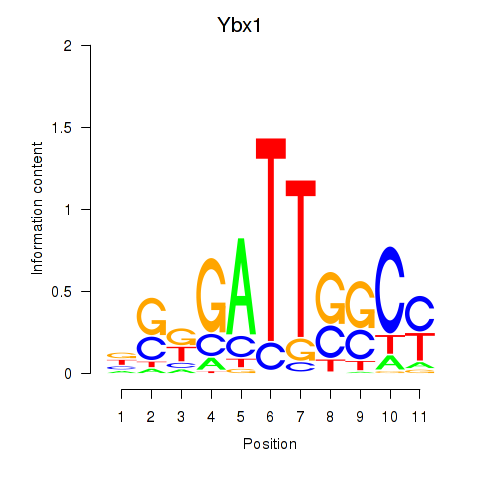
| Ybx1_Nfya_Nfyb_Nfyc_Cebpz | 9.07 |
|
|
|
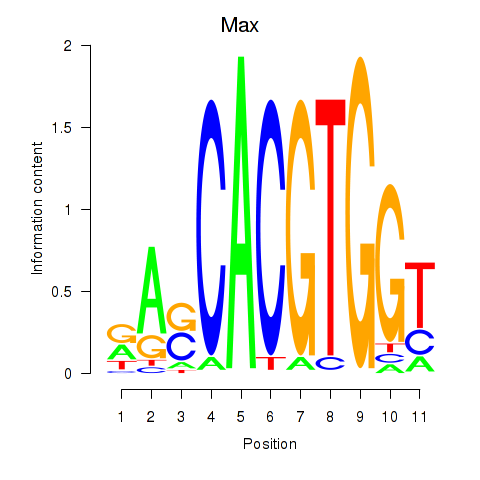
| Max_Mycn | 8.25 |
|

|
|
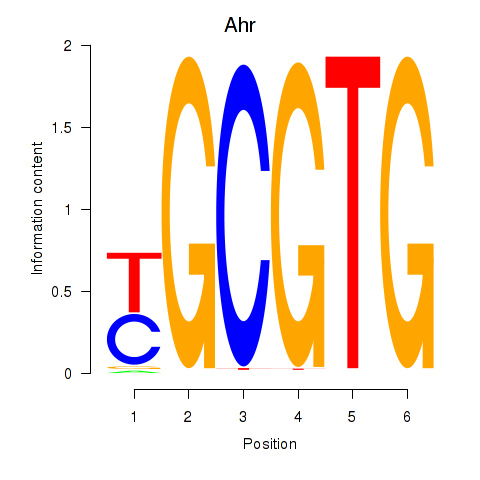
| Ahr | 7.60 |
|

|
|
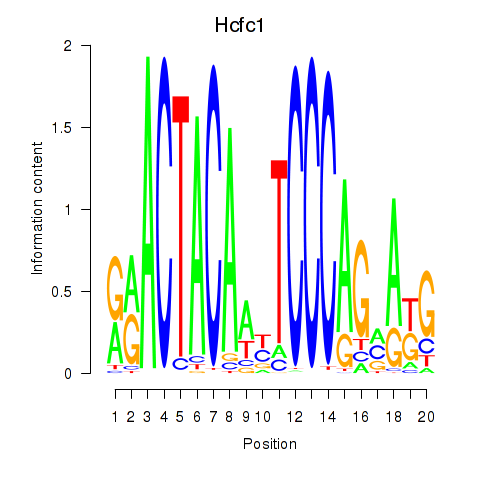
| Hcfc1_Six5_Smarcc2_Zfp143 | 6.42 |
|
|
|
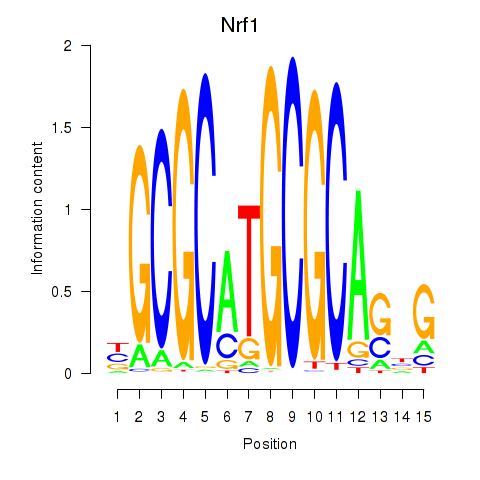
| Nrf1 | 6.16 |
|
|
|
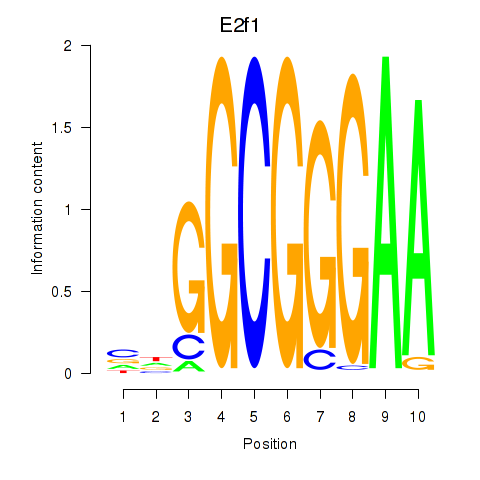
| E2f1 | 6.14 |
|
|
|
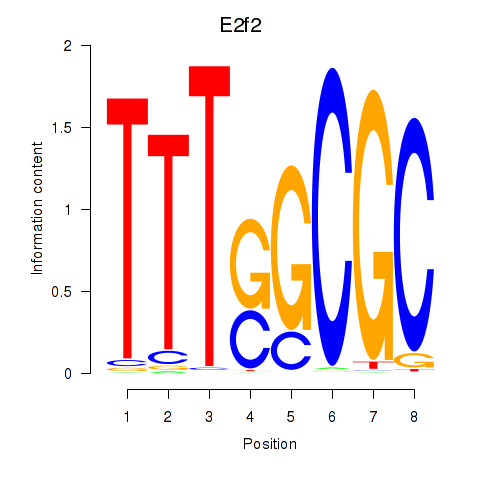
| E2f2_E2f5 | 6.10 |
|
|
|
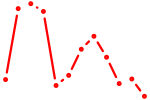
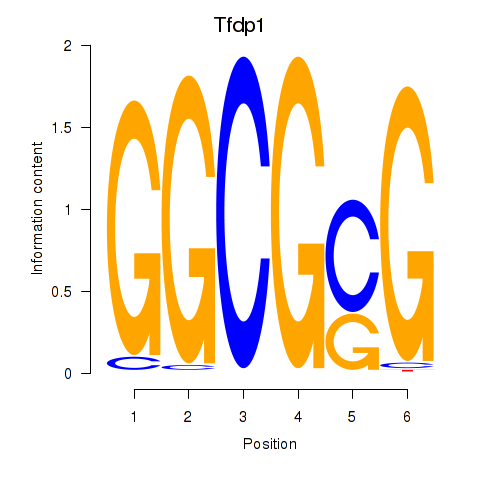
| Tfdp1_Wt1_Egr2 | 4.94 |
|
|
|
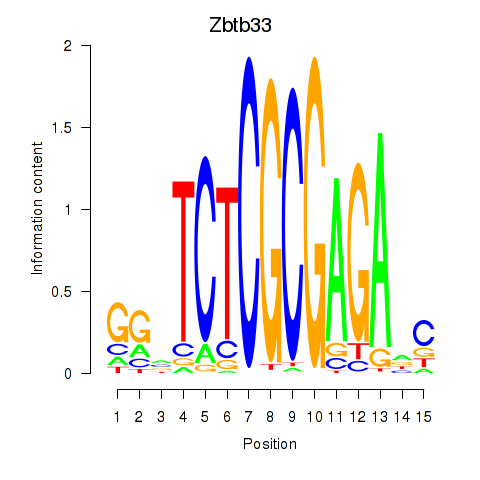
| Zbtb33_Chd2 | 4.43 |
|

|
|
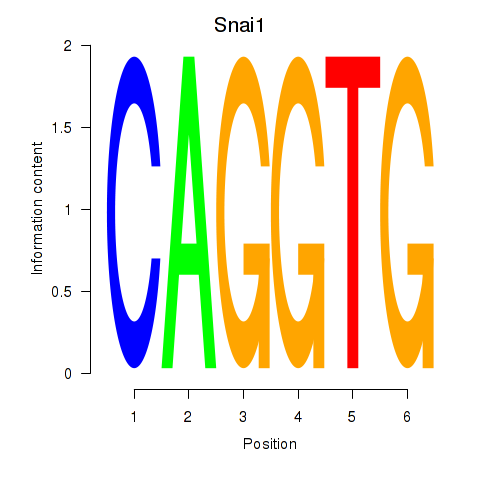
| Snai1_Zeb1_Snai2 | 4.03 |
|

|
|
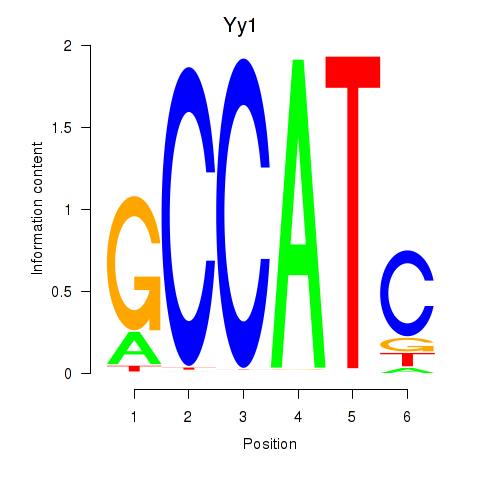
| Yy1_Yy2 | 3.73 |
|

|
|
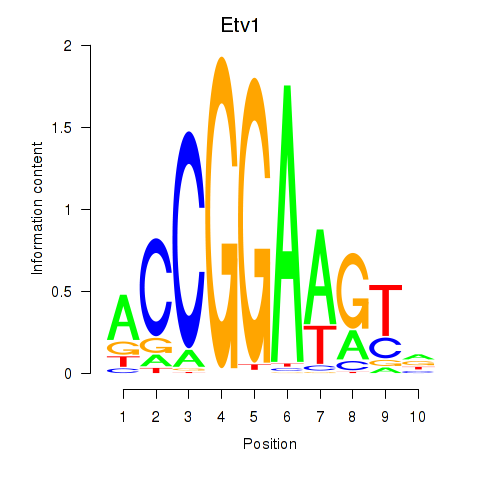
| Etv1_Etv5_Gabpa | 3.65 |
|
|
|
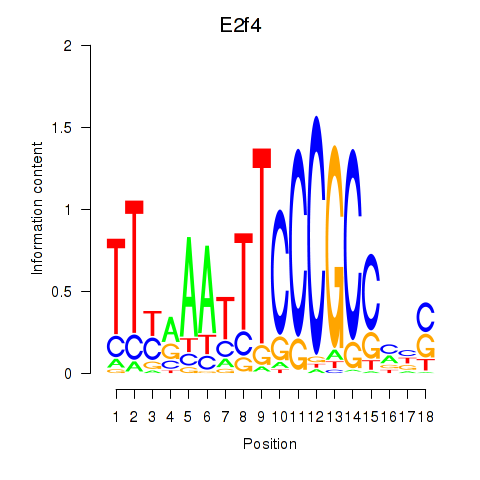
| E2f4 | 3.30 |
|
|
|
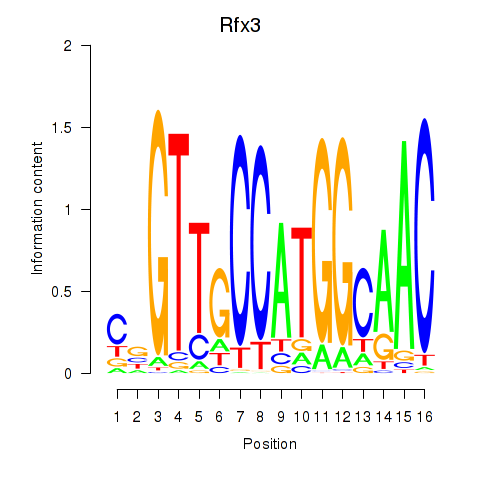
| Rfx3_Rfx1_Rfx4 | 3.24 |
|

|
|
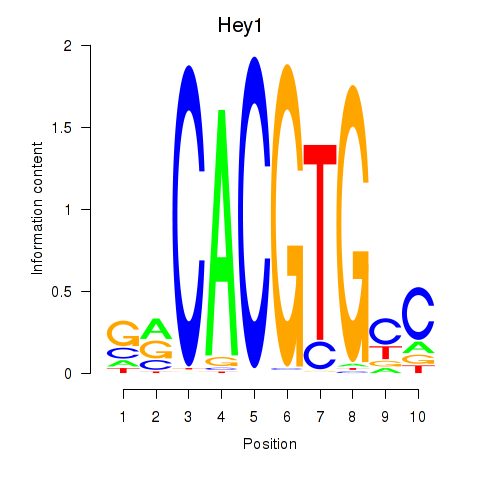
| Hey1_Myc_Mxi1 | 3.02 |
|
|
|
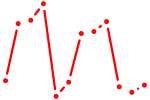
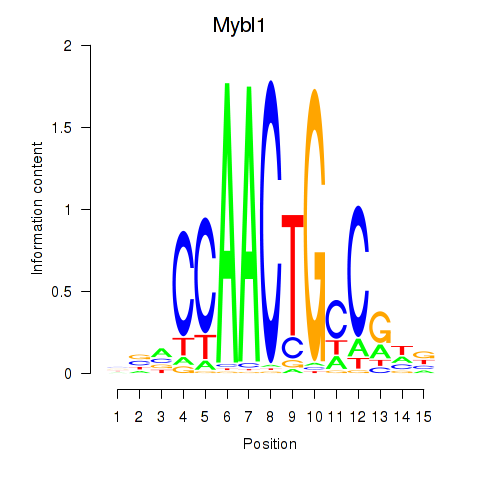
| Mybl1 | 2.99 |
|
|
|
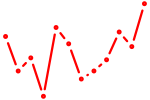
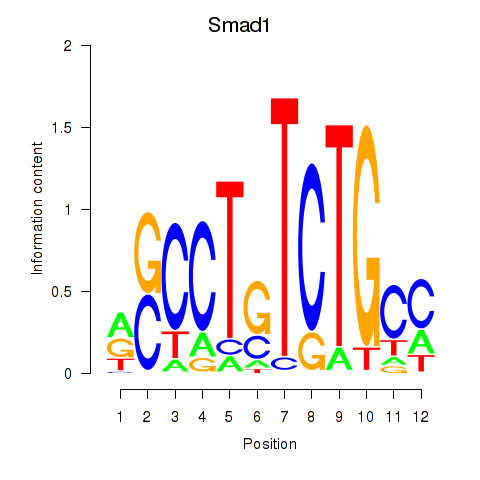
| Smad1 | 2.97 |
|
|
|
| Xbp1_Creb3l1 | 2.97 |
|

|

|
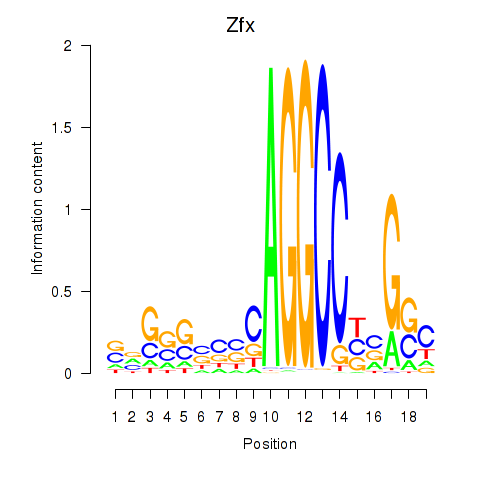
| Zfx_Zfp711 | 2.96 |
|

|
|
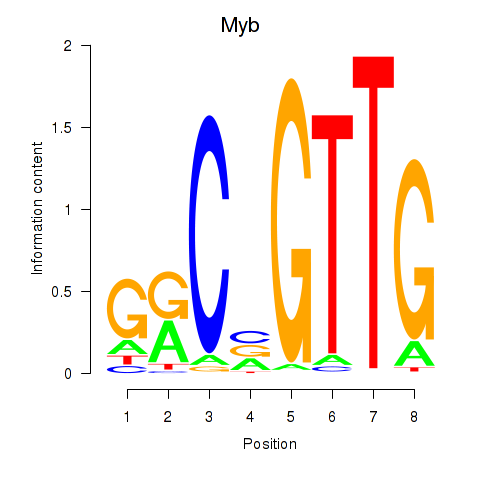
| Myb | 2.95 |
|
|
|
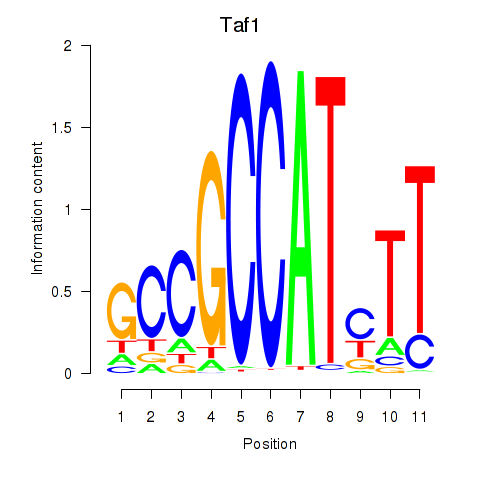
| Taf1 | 2.88 |
|
|
|
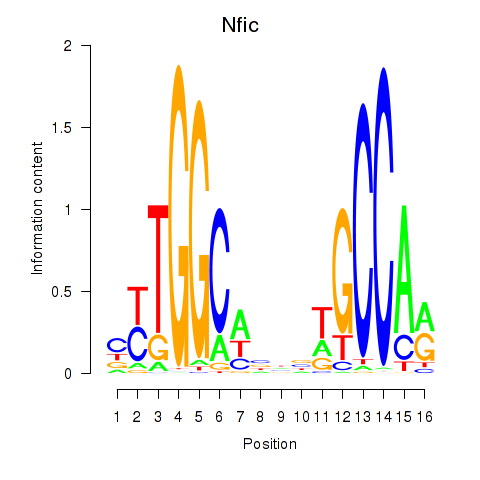
| Nfic_Nfib | 2.79 |
|
|
|
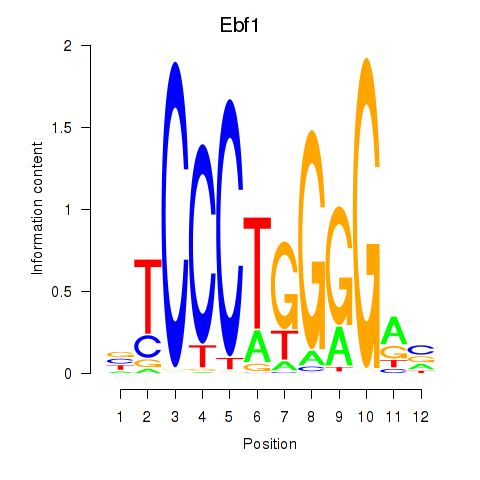
| Ebf1 | 2.68 |
|
|
|
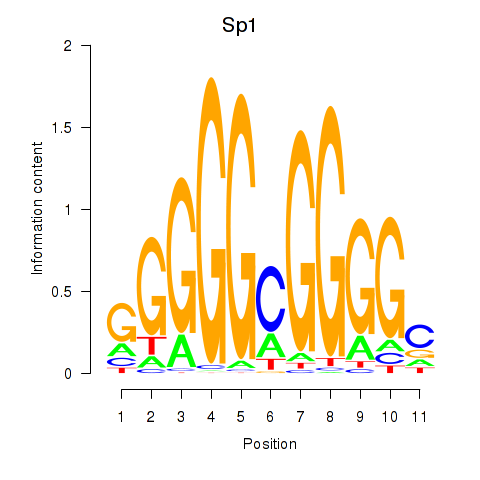
| Sp1 | 2.66 |
|
|
|
| Chd1_Pml | 2.59 |
|
|
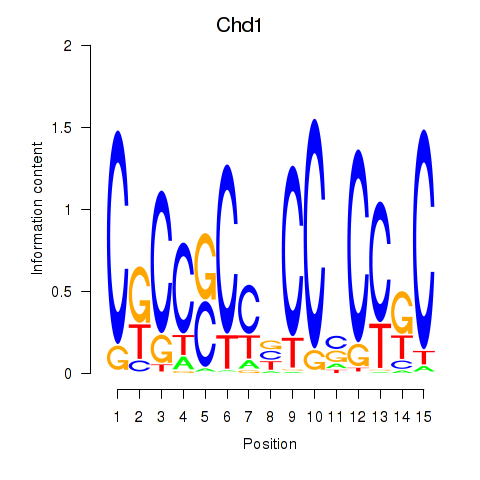
|
| Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2 | 2.50 |
|

|
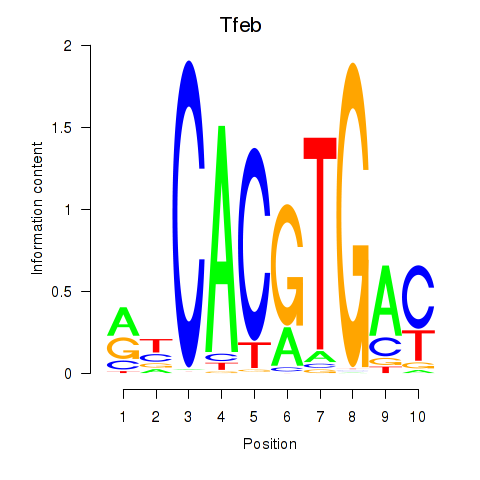
|
| Atf6 | 2.48 |
|
|
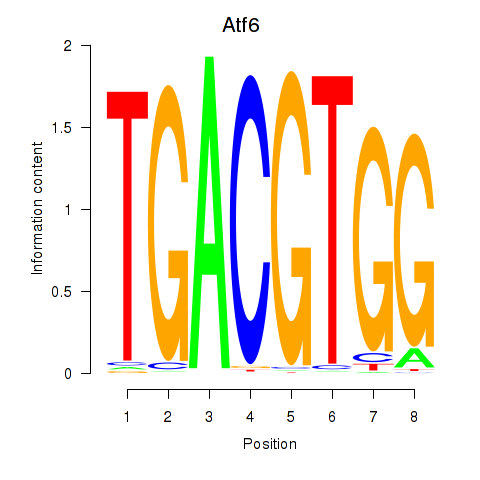
|
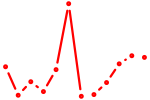
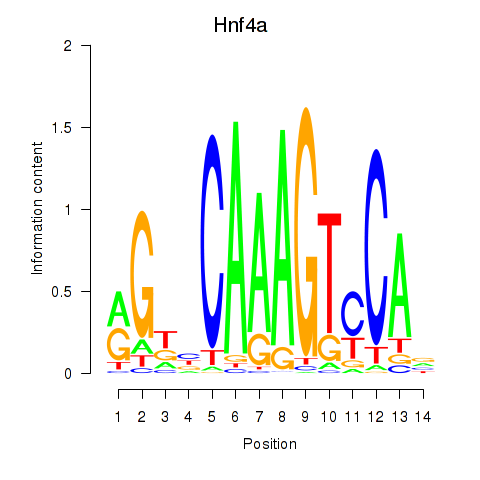
| Hnf4a | 2.48 |
|
|
|
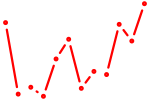
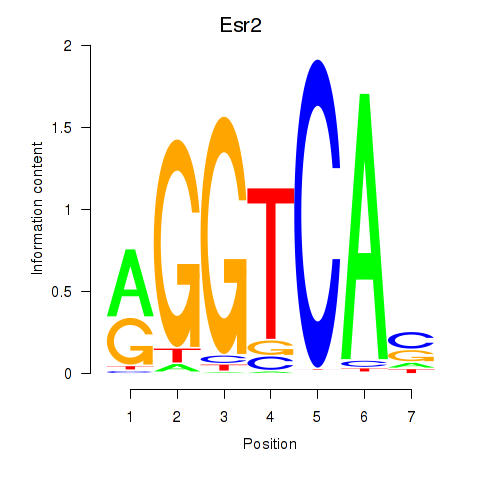
| Esr2 | 2.47 |
|
|
|
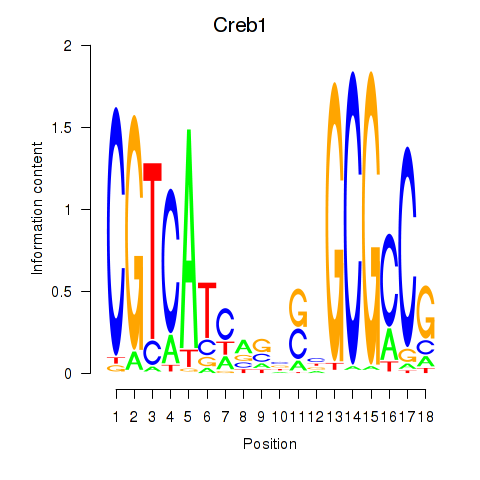
| Creb1 | 2.45 |
|

|
|
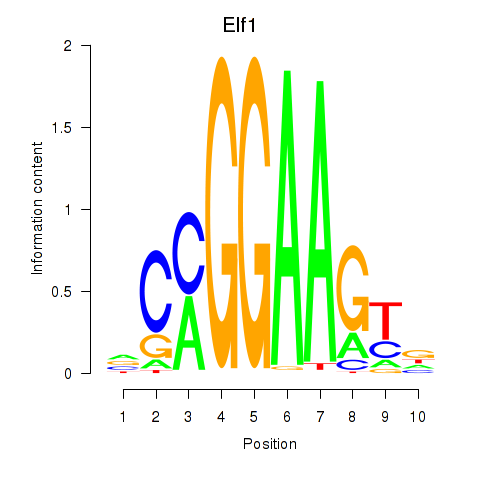
| Elf1_Elf2_Etv2_Elf4 | 2.41 |
|
|
|
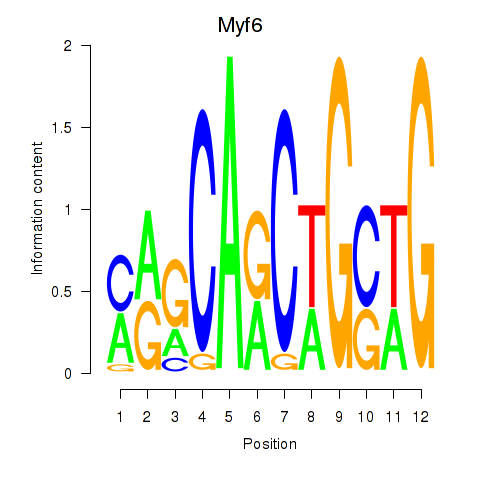
| Myf6 | 2.39 |
|

|
|
| Epas1_Bcl3 | 2.38 |
|

|
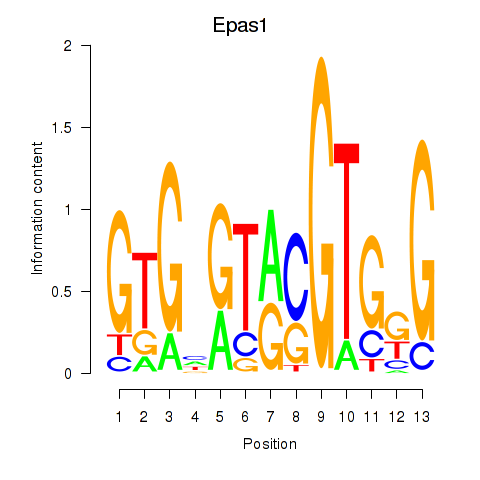
|
| Wrnip1_Mta3_Rcor1 | 2.29 |
|

|
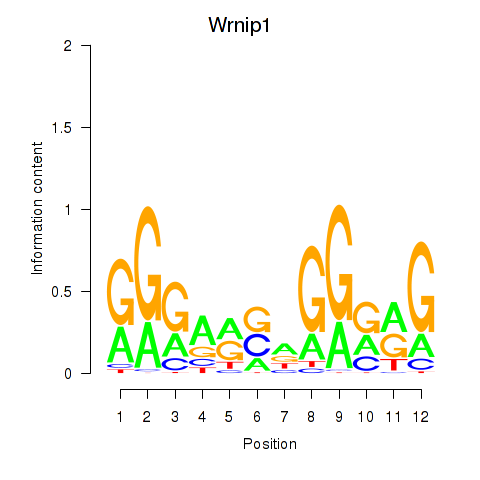
|
| Mecp2 | 2.08 |
|

|
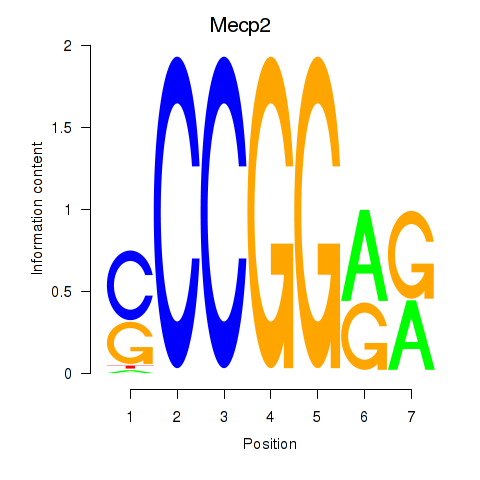
|
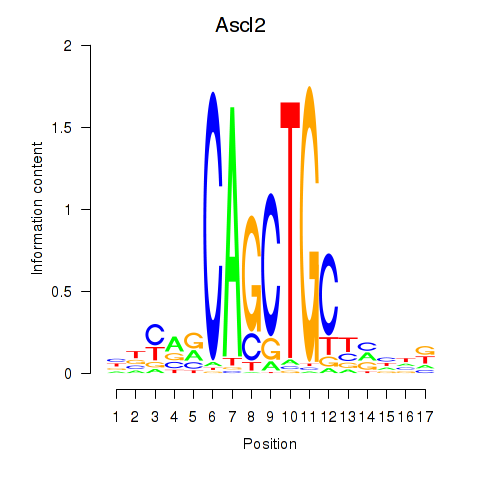
| Ascl2 | 2.04 |
|
|
|
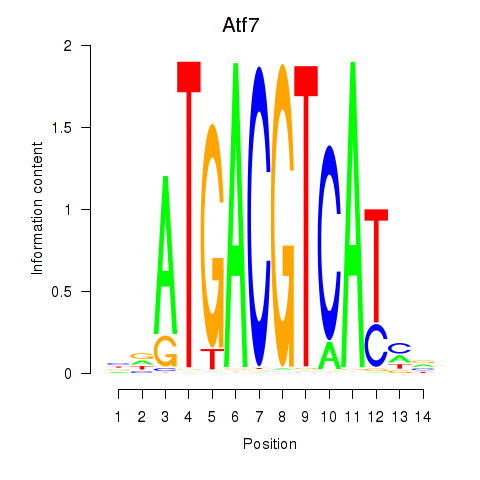
| Atf7_E4f1 | 2.00 |
|
|
|
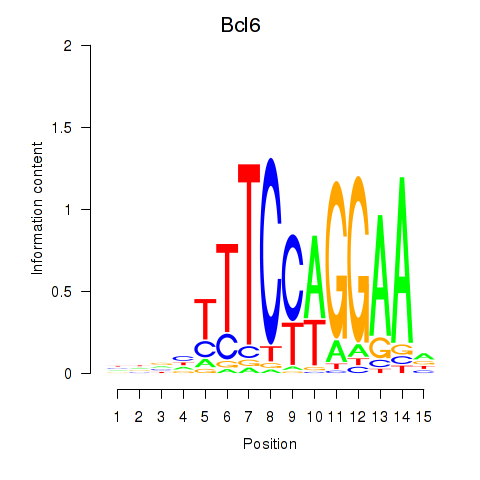
| Bcl6 | 1.99 |
|
|
|
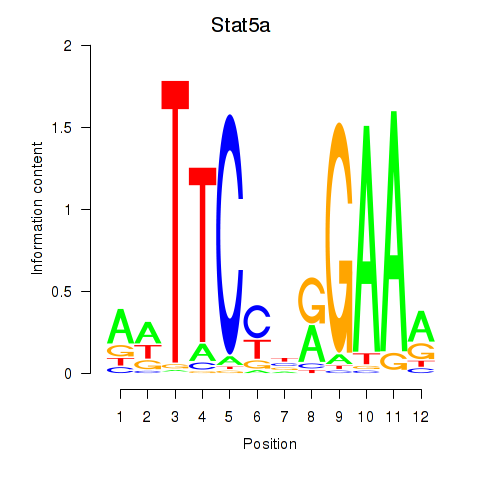
| Stat5a | 1.97 |
|
|
|
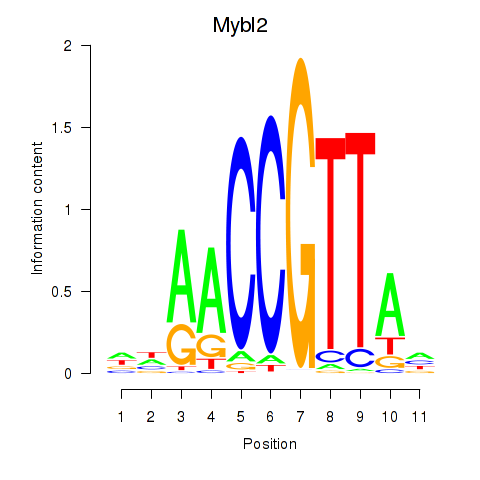
| Mybl2 | 1.95 |
|
|
|
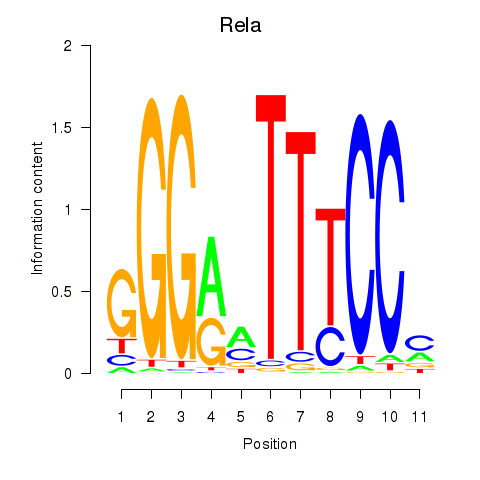
| Rela_Rel_Nfkb1 | 1.94 |
|
|
|
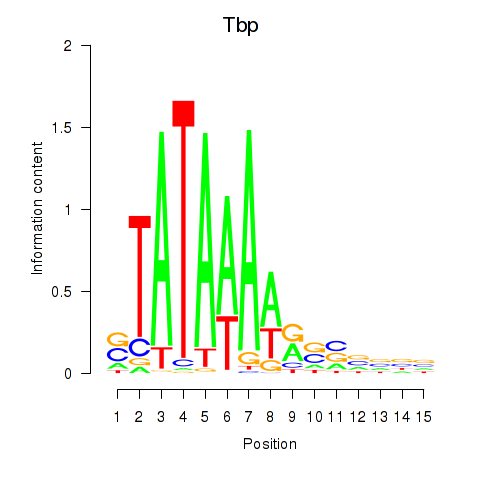
| Tbp | 1.89 |
|
|
|
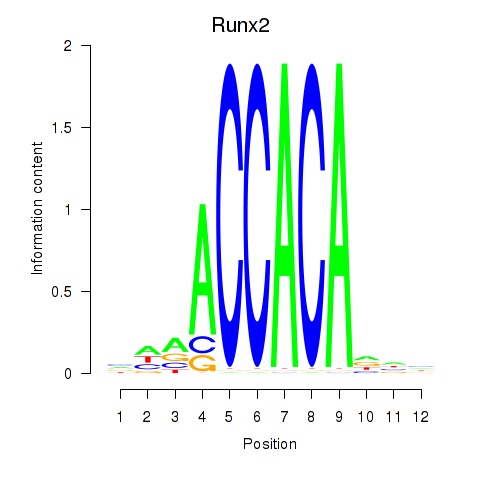
| Runx2_Bcl11a | 1.88 |
|

|
|
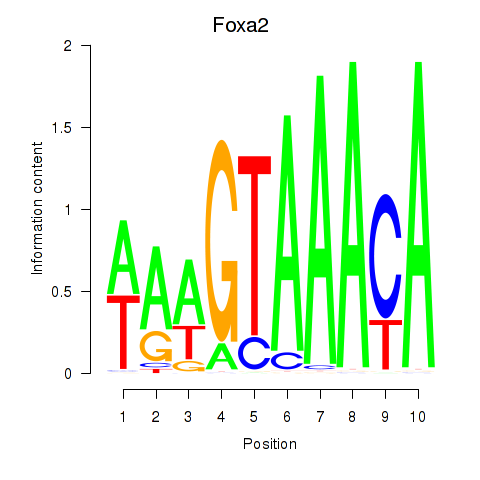
| Foxa2_Foxa1 | 1.84 |
|
|
|
| Klf6_Patz1 | 1.83 |
|

|
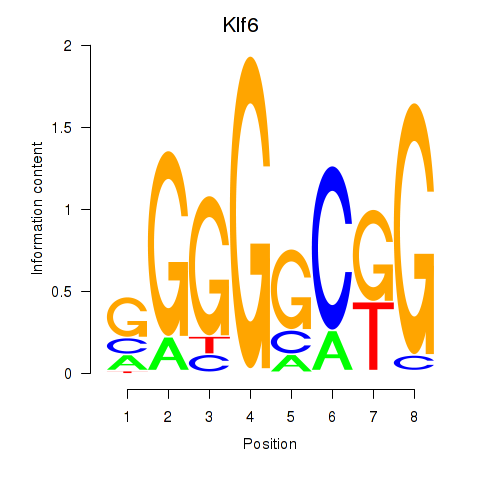
|
| Esrrb_Esrra | 1.79 |
|
|
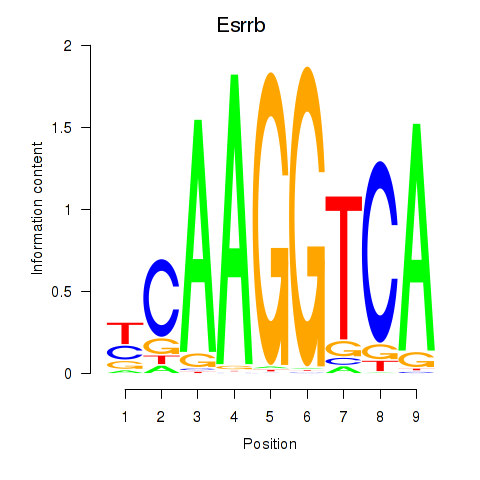
|
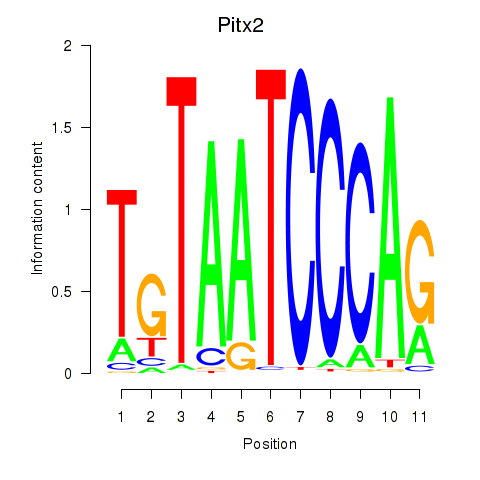
| Pitx2_Otx2 | 1.79 |
|
|
|
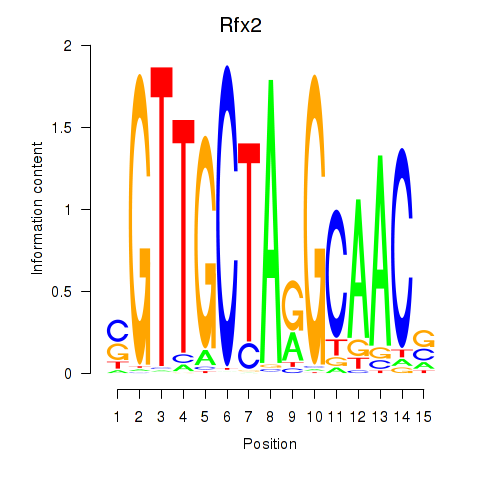
| Rfx2_Rfx7 | 1.79 |
|

|
|
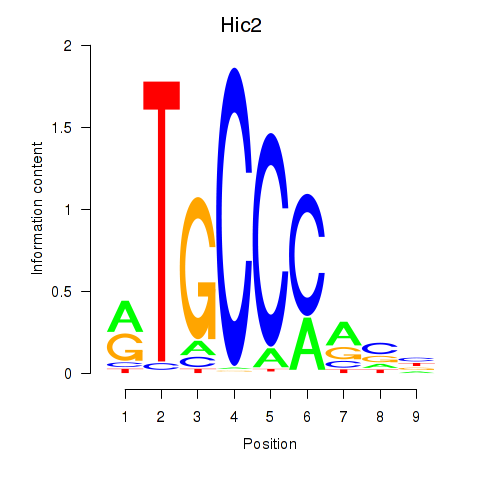
| Hic2 | 1.77 |
|
|
|
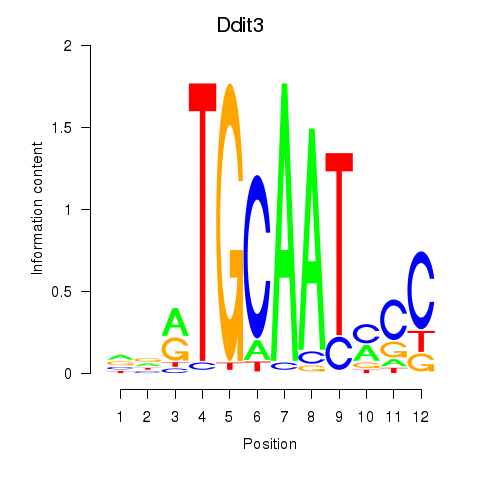
| Ddit3 | 1.76 |
|
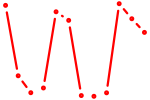
|
|
| AAGGCAC | 1.75 |
|
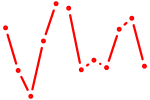
|
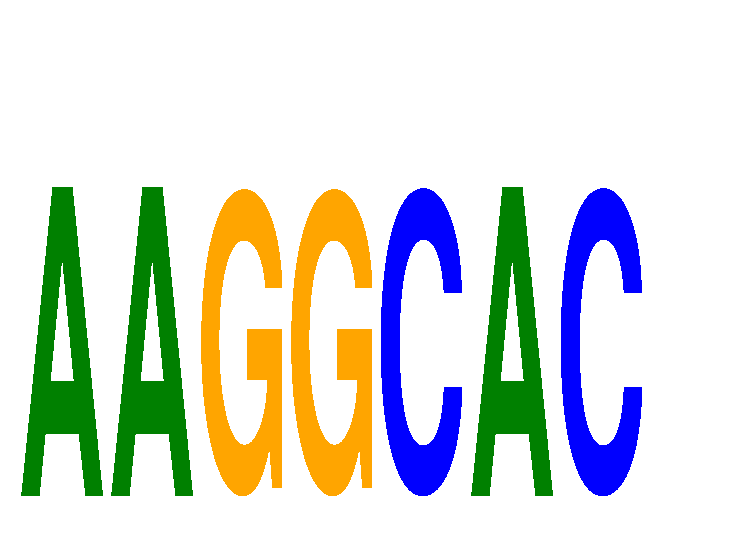
|
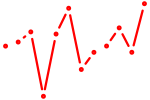
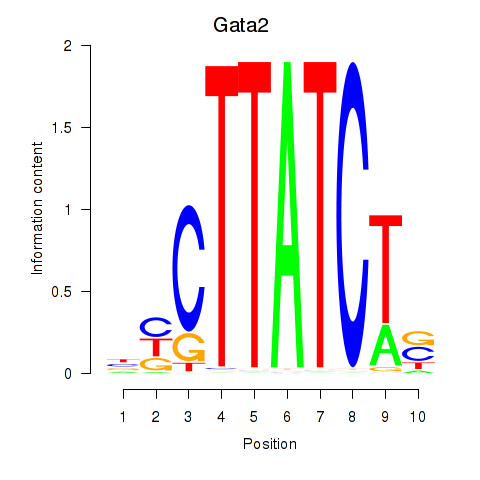
| Gata2_Gata1 | 1.75 |
|
|
|
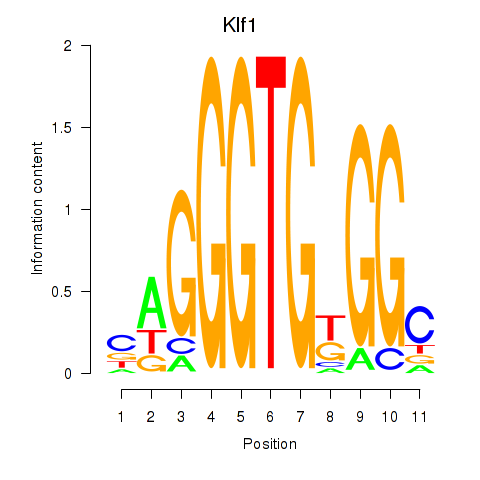
| Klf1 | 1.74 |
|
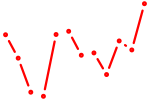
|
|
| Spib | 1.73 |
|

|
|
| Irf2_Irf1_Irf8_Irf9_Irf7 | 1.73 |
|

|
|
| E2f8 | 1.72 |
|
|
|
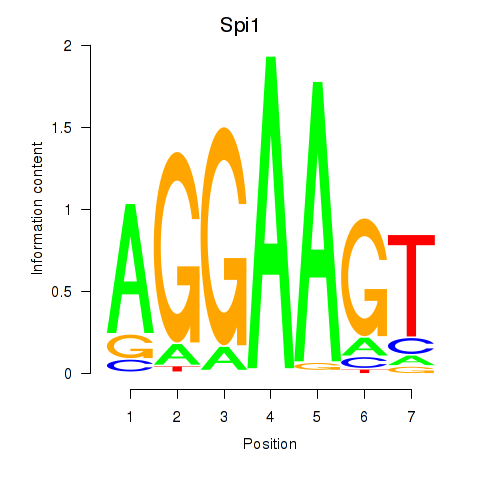
| Spi1 | 1.71 |
|
|
|
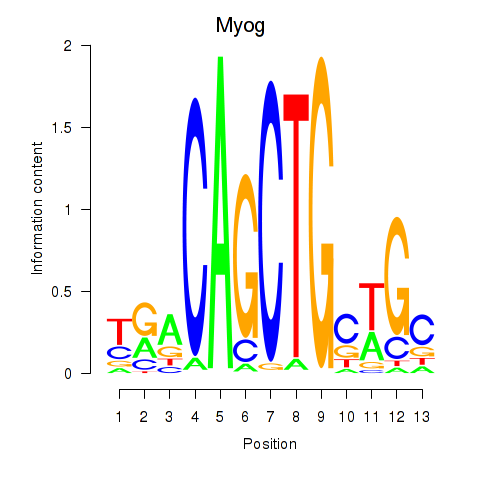
| Myog_Tcf12 | 1.70 |
|
|
|
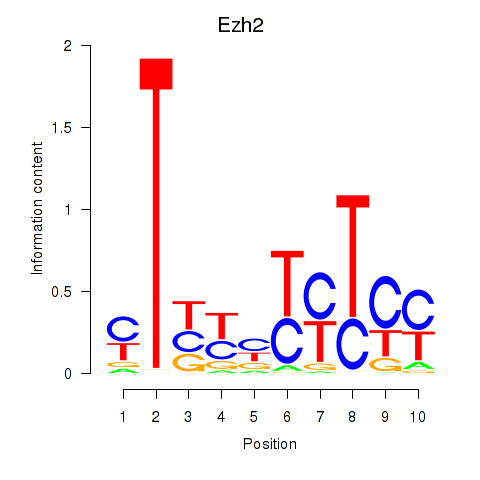
| Ezh2_Atf2_Ikzf1 | 1.69 |
|
|
|
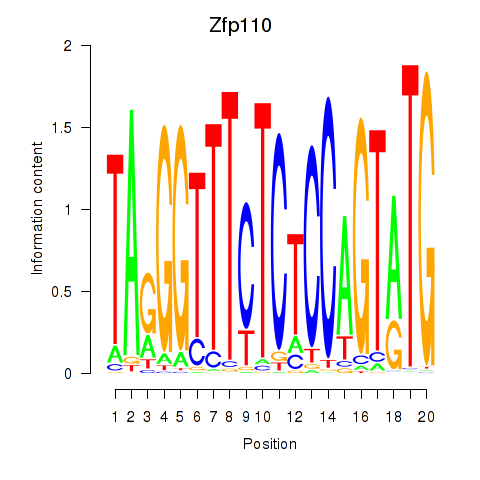
| Zfp110 | 1.68 |
|
|
|
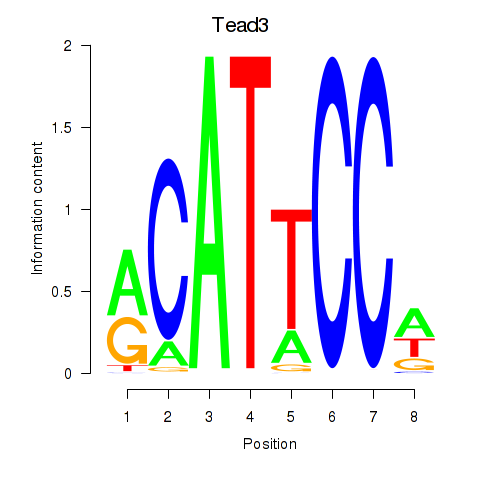
| Tead3_Tead4 | 1.67 |
|
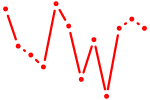
|
|
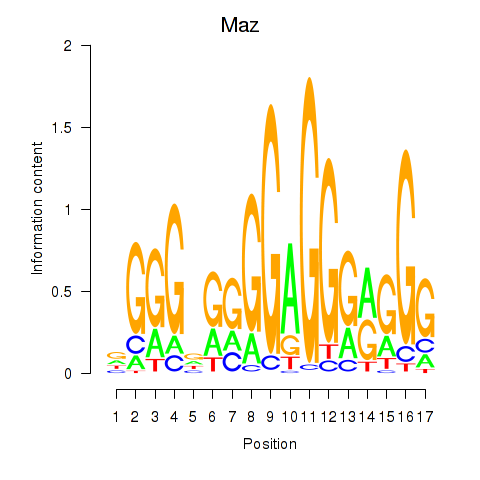
| Maz_Zfp281 | 1.65 |
|

|
|
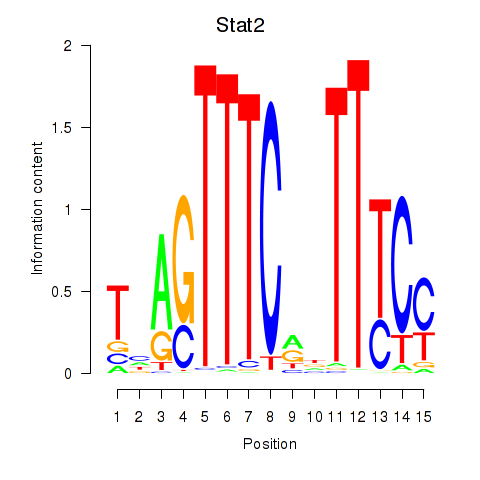
| Stat2 | 1.63 |
|
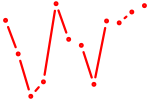
|
|
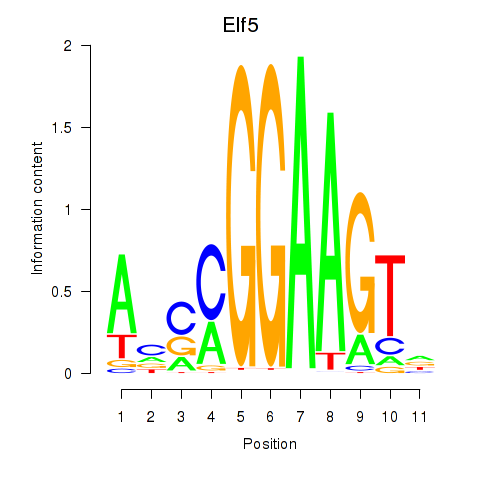
| Elf5 | 1.63 |
|

|
|
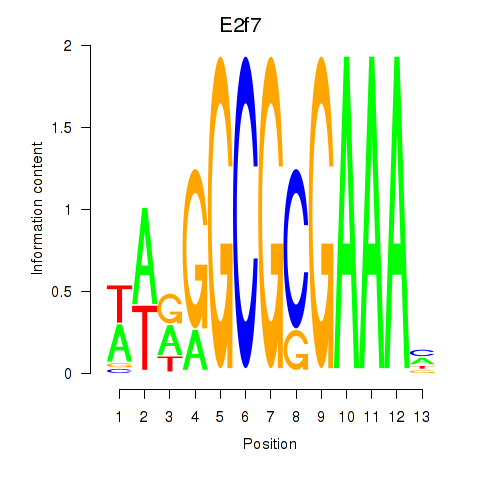
| E2f7 | 1.60 |
|

|
|
| Gtf2i_Gtf2f1 | 1.58 |
|
|
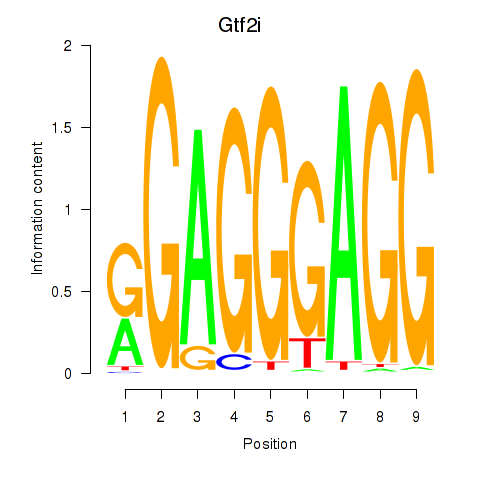
|
| Ets1 | 1.57 |
|

|
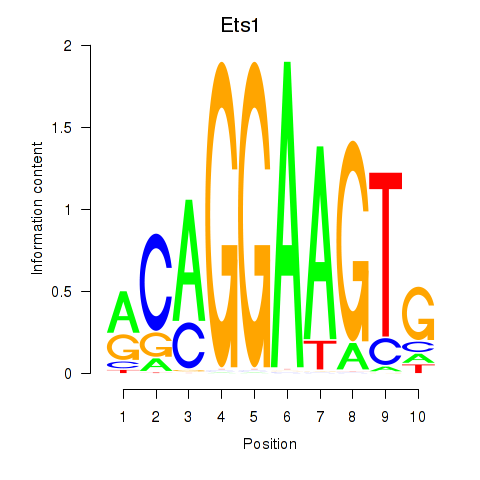
|
| AAAGUGC | 1.57 |
|
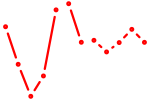
|

|
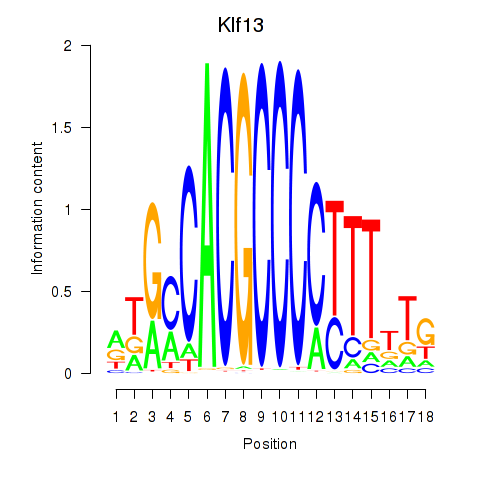
| Klf13 | 1.49 |
|
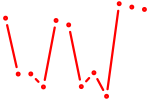
|
|
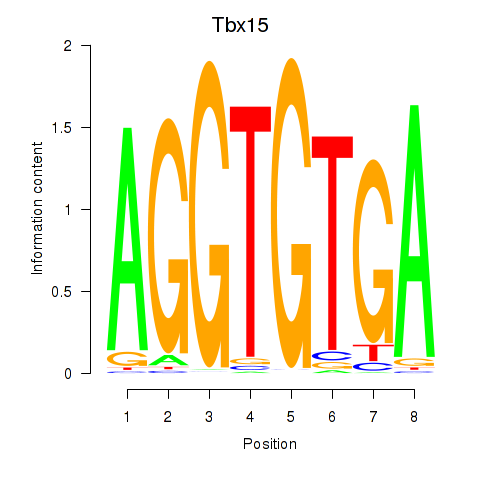
| Tbx15 | 1.47 |
|
|
|
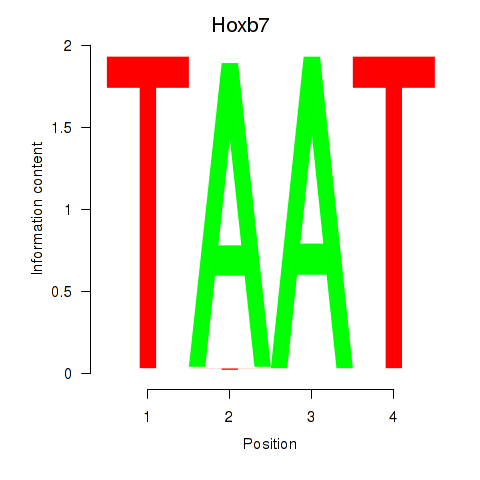
| Hoxb7 | 1.46 |
|
|
|
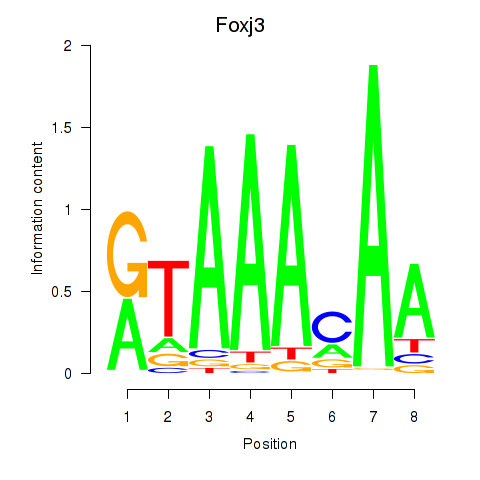
| Foxj3_Tbl1xr1 | 1.46 |
|
|
|
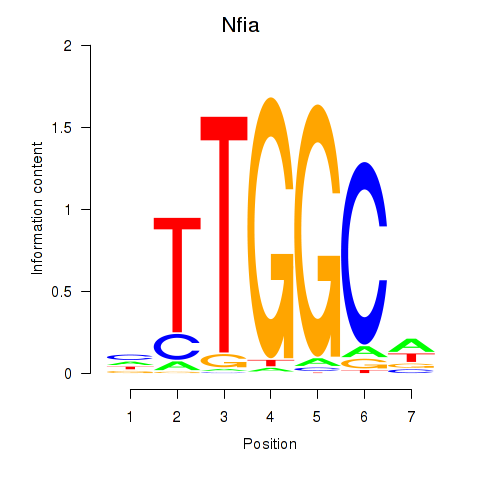
| Nfia | 1.43 |
|
|
|
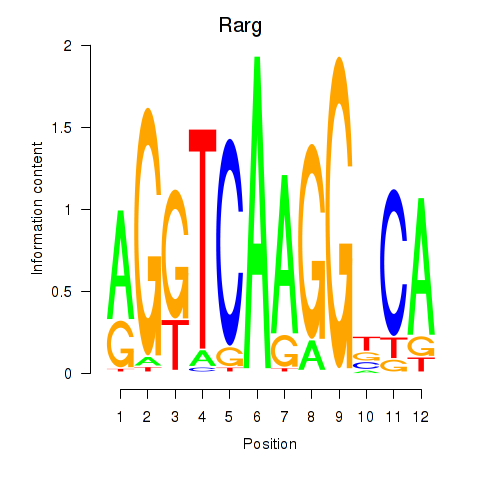
| Rarg | 1.43 |
|
|
|
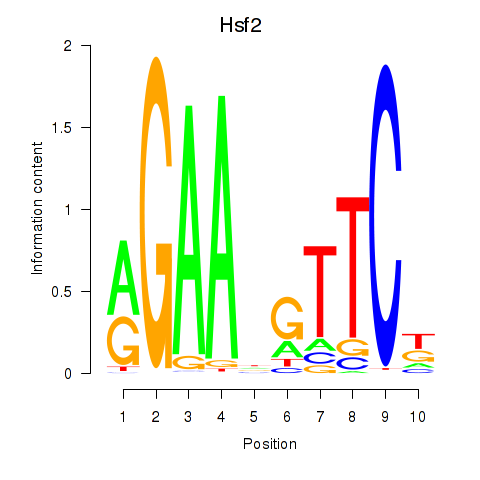
| Hsf2 | 1.42 |
|

|
|
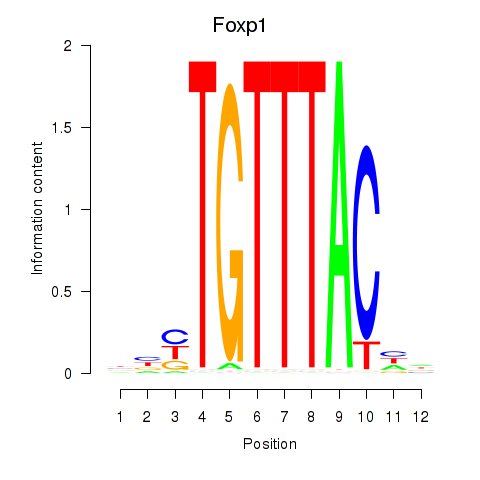
| Foxp1_Foxj2 | 1.41 |
|

|
|
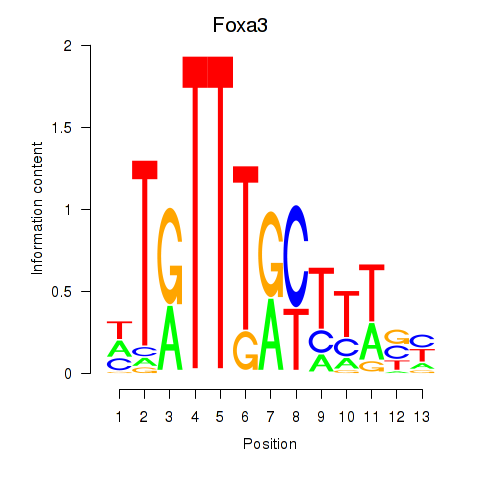
| Foxa3 | 1.41 |
|
|
|
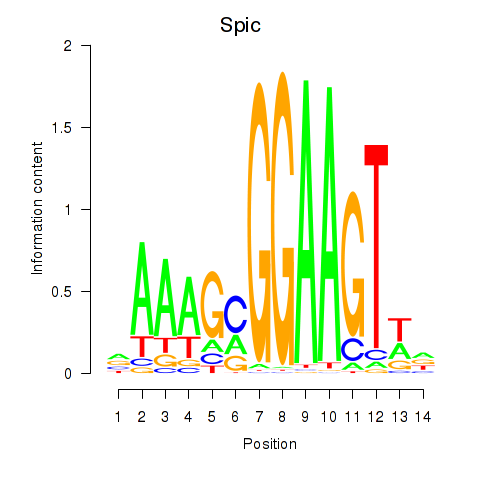
| Spic | 1.39 |
|
|
|
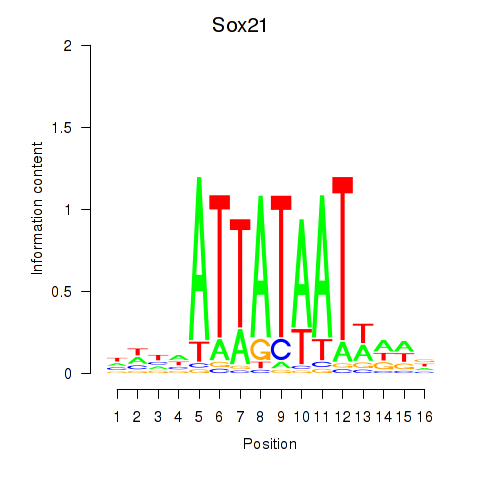
| Sox21 | 1.36 |
|
|
|
| CCCUGAG | 1.36 |
|
|
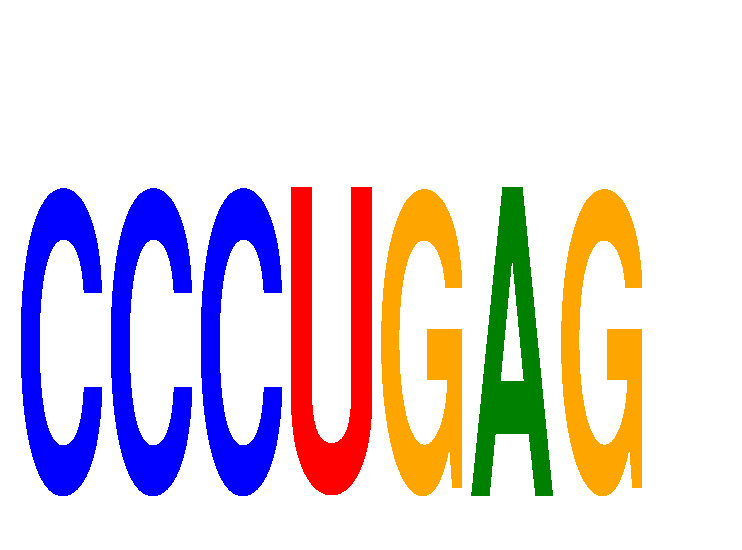
|
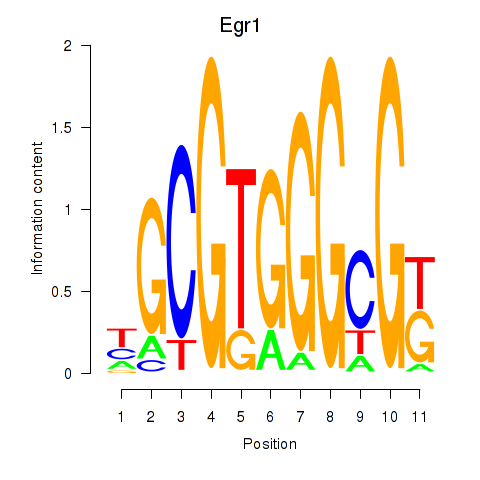
| Egr1 | 1.36 |
|

|
|
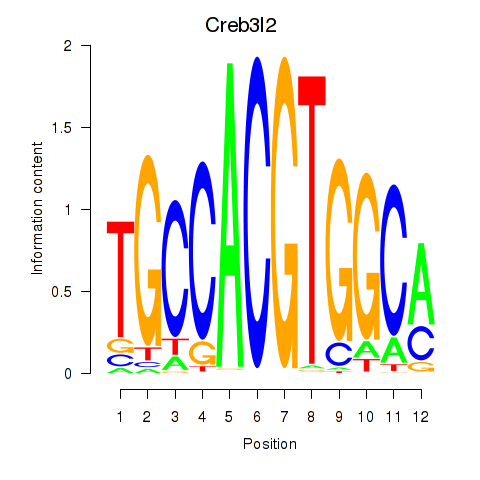
| Creb3l2 | 1.35 |
|

|
|
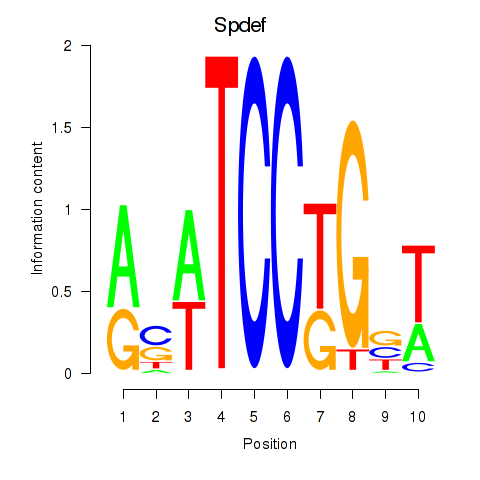
| Spdef | 1.34 |
|

|
|
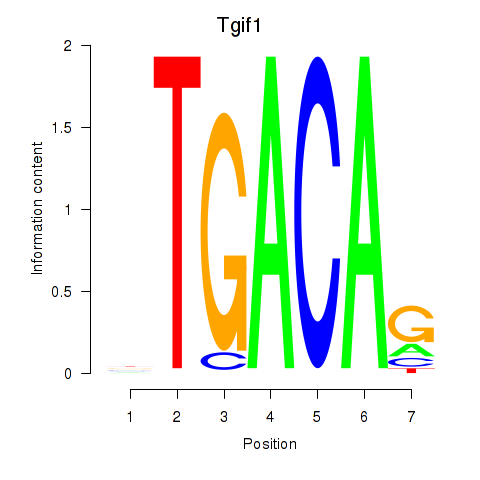
| Tgif1_Meis3 | 1.34 |
|
|
|
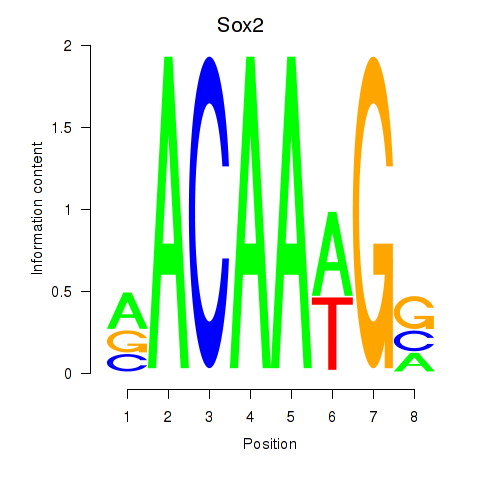
| Sox2 | 1.34 |
|

|
|
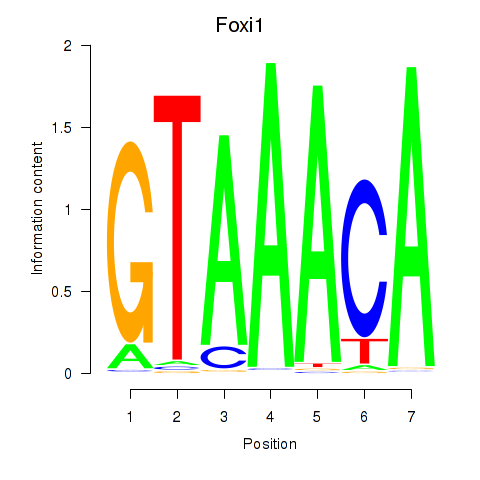
| Foxi1_Foxo1 | 1.32 |
|

|
|
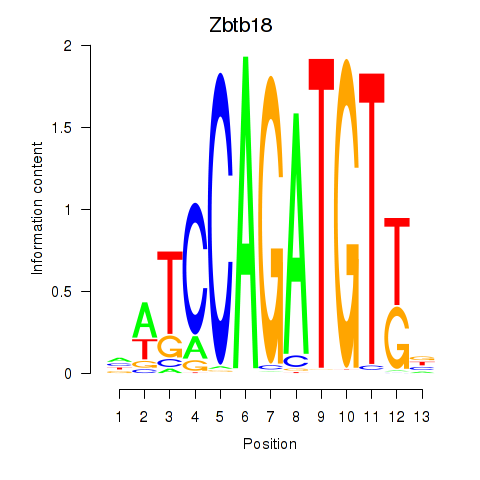
| Zbtb18 | 1.31 |
|
|
|
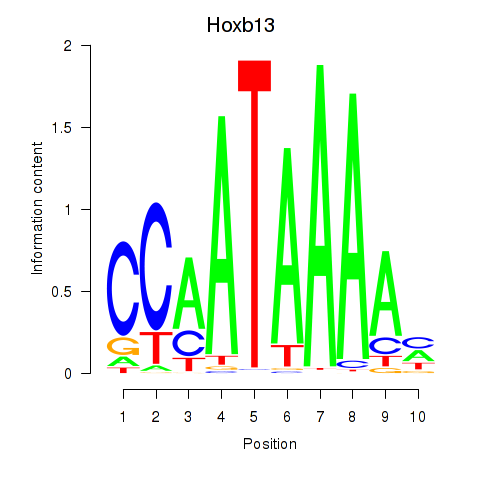
| Hoxb13 | 1.31 |
|
|
|
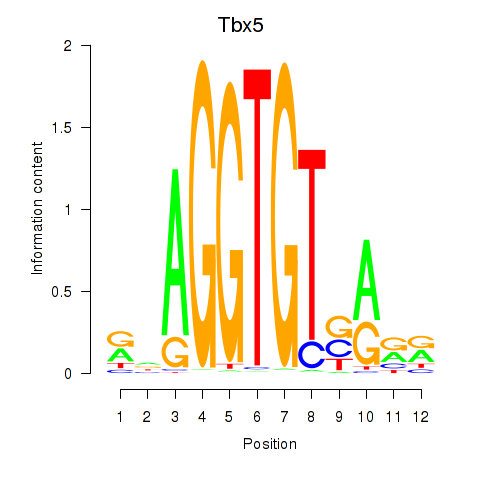
| Tbx5 | 1.30 |
|

|
|
| AAUACUG | 1.30 |
|

|

|
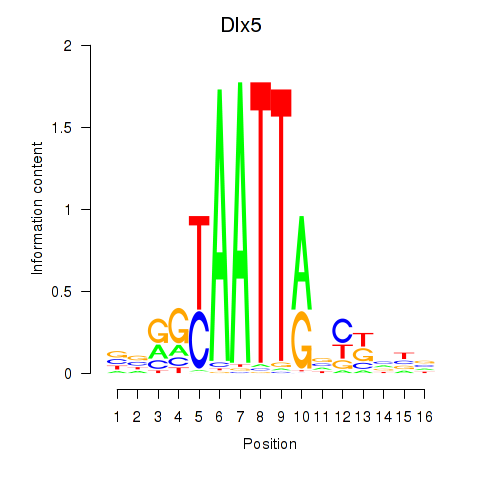
| Dlx5_Dlx4 | 1.29 |
|
|
|
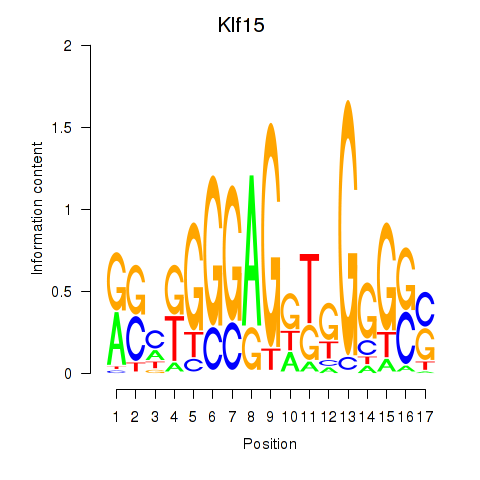
| Klf15 | 1.29 |
|

|
|
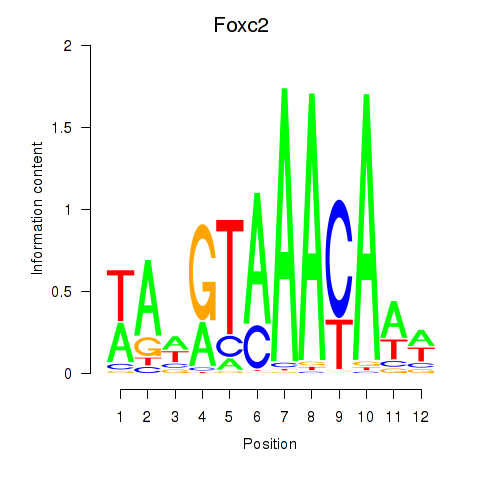
| Foxc2 | 1.29 |
|
|
|
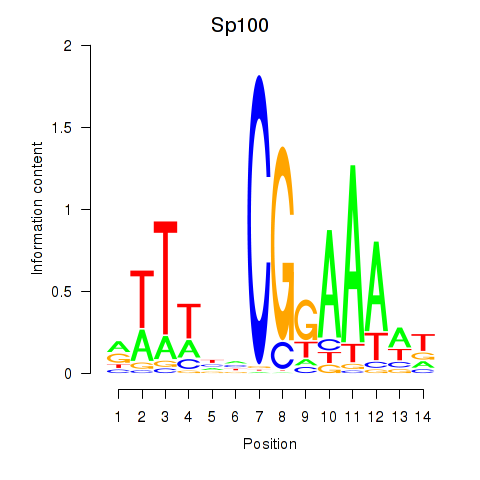
| Sp100 | 1.29 |
|
|
|
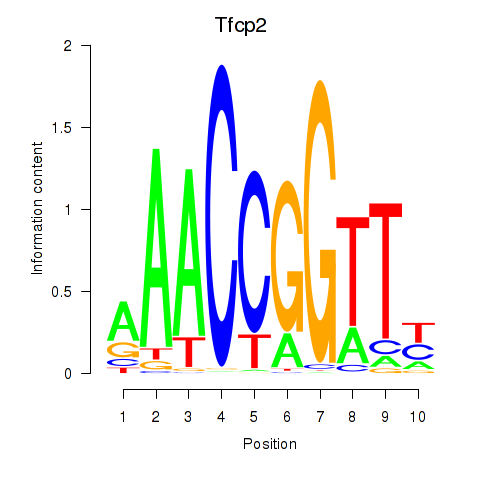
| Tfcp2 | 1.28 |
|

|
|
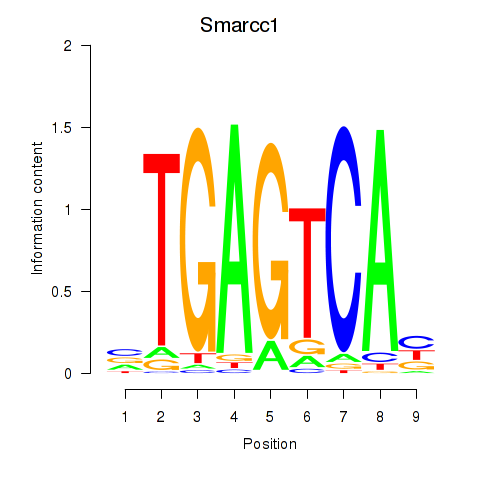
| Smarcc1_Fosl1 | 1.28 |
|
|
|
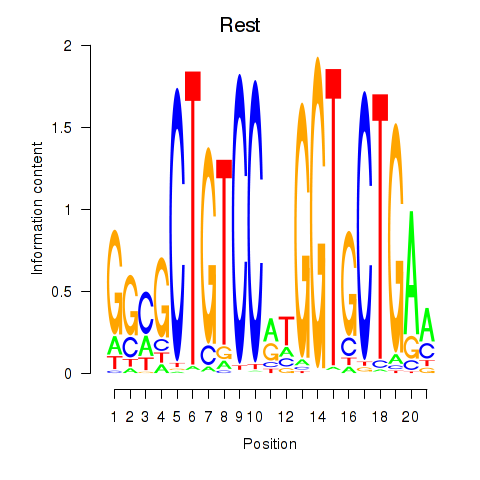
| Rest | 1.28 |
|
|
|
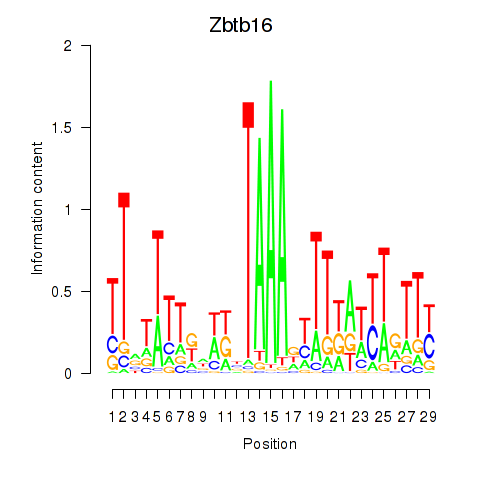
| Zbtb16 | 1.27 |
|
|
|
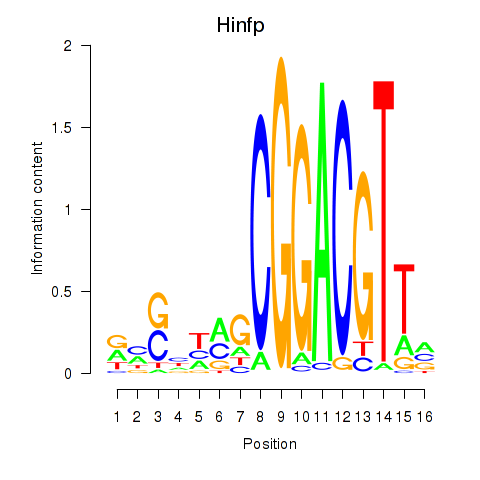
| Hinfp | 1.27 |
|
|
|
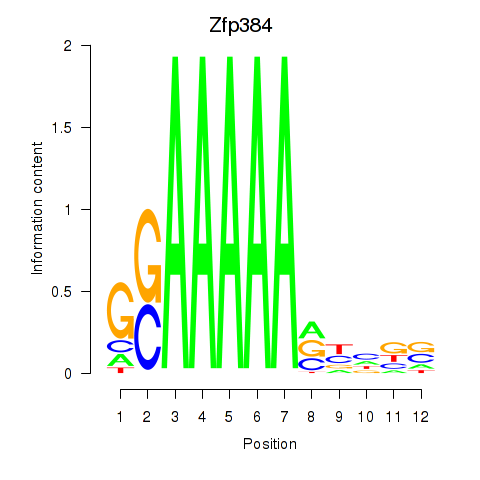
| Zfp384 | 1.27 |
|
|
|
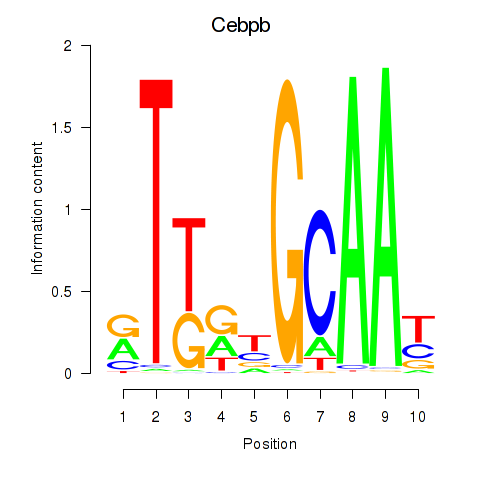
| Cebpb | 1.27 |
|
|
|
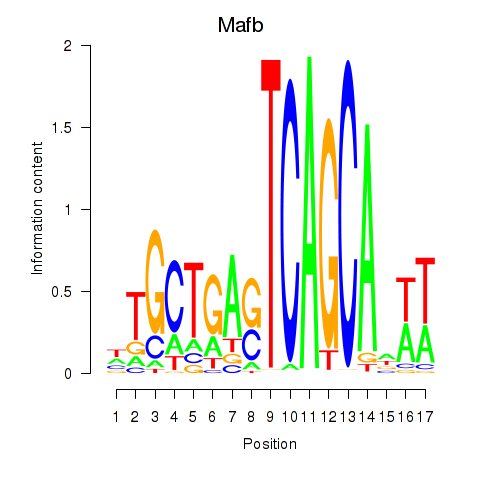
| Mafb | 1.27 |
|
|
|
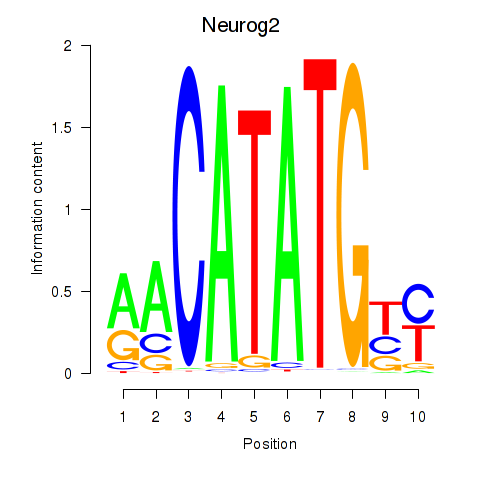
| Neurog2 | 1.27 |
|
|
|
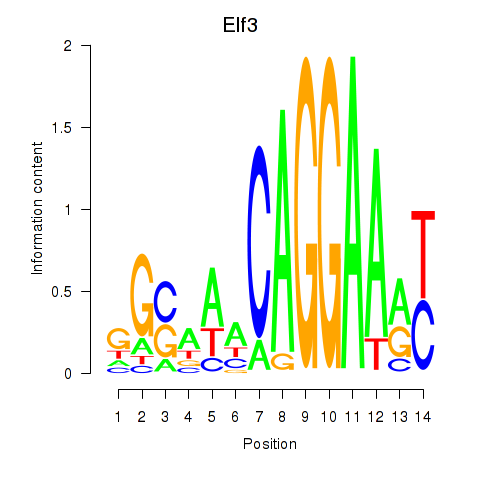
| Elf3 | 1.26 |
|

|
|
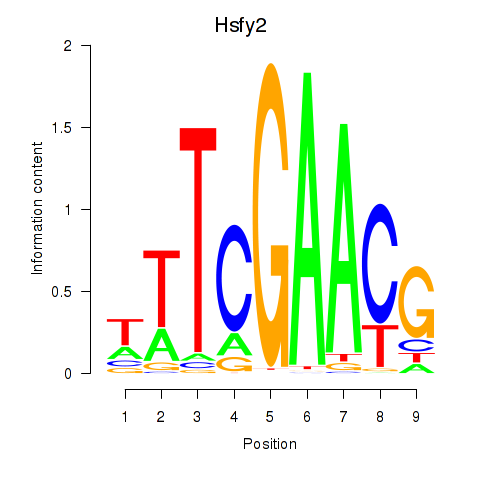
| Hsfy2 | 1.26 |
|
|
|
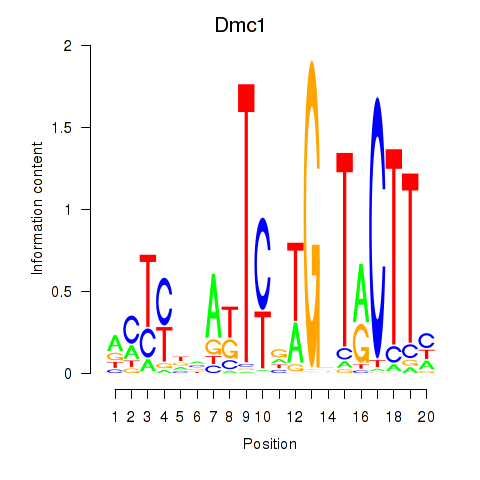
| Dmc1 | 1.25 |
|
|
|
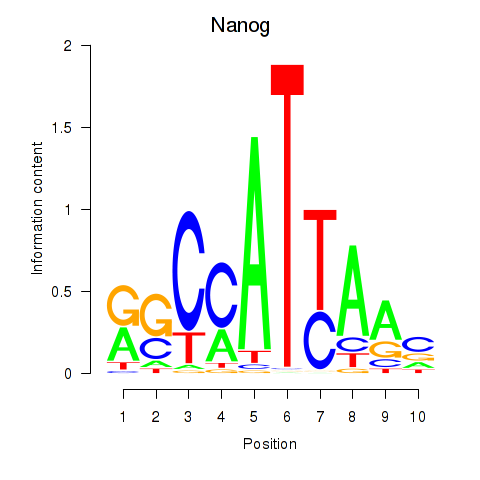
| Nanog | 1.24 |
|
|
|
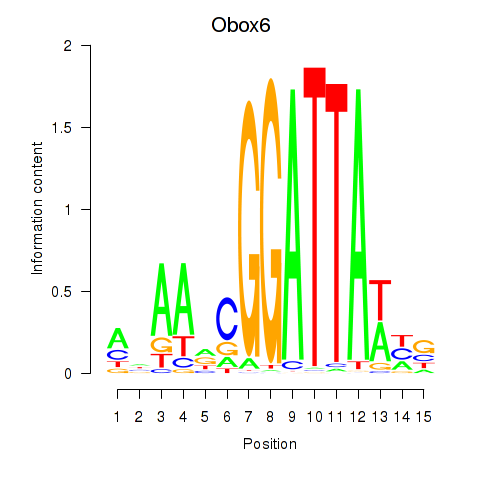
| Obox6_Obox5 | 1.24 |
|

|
|
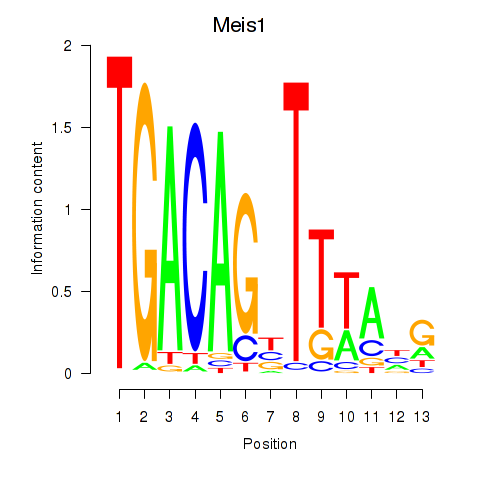
| Meis1 | 1.23 |
|
|
|
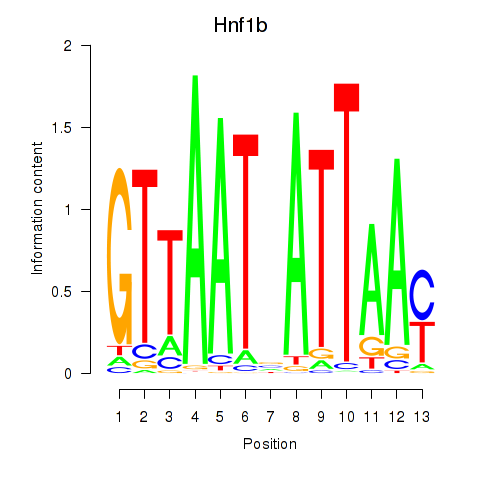
| Hnf1b | 1.23 |
|
|
|
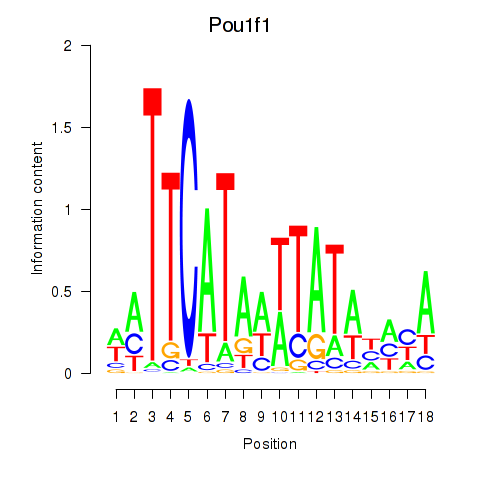
| Pou1f1 | 1.23 |
|

|
|
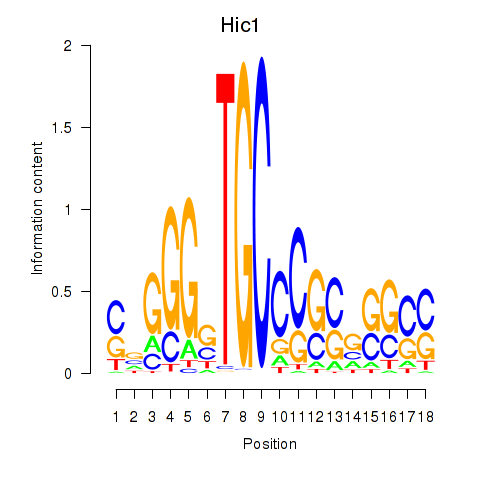
| Hic1 | 1.23 |
|
|
|
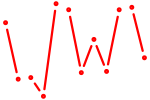
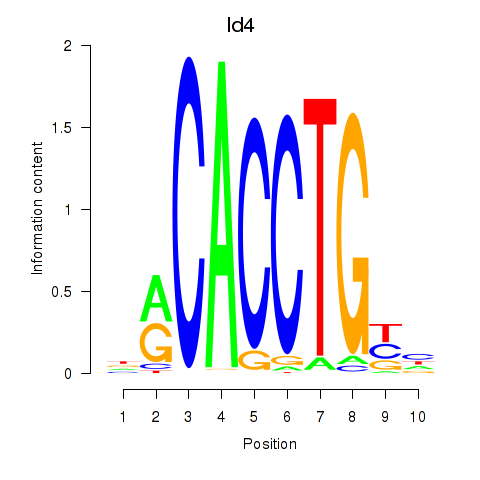
| Id4 | 1.21 |
|
|
|
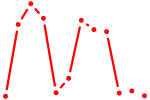
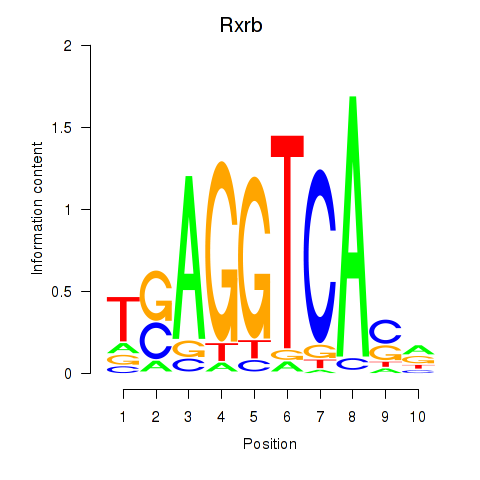
| Rxrb | 1.21 |
|
|
|
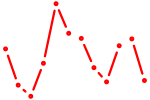
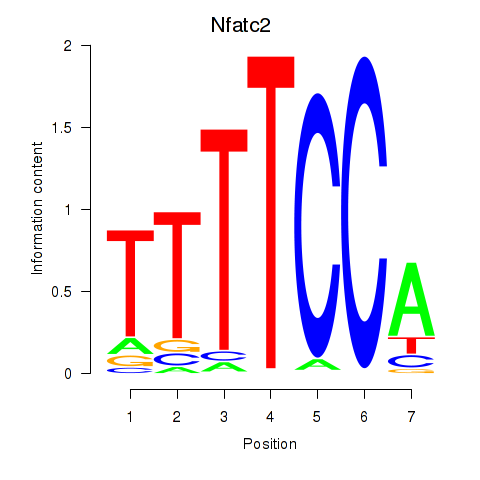
| Nfatc2 | 1.18 |
|
|
|
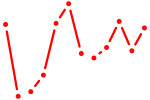
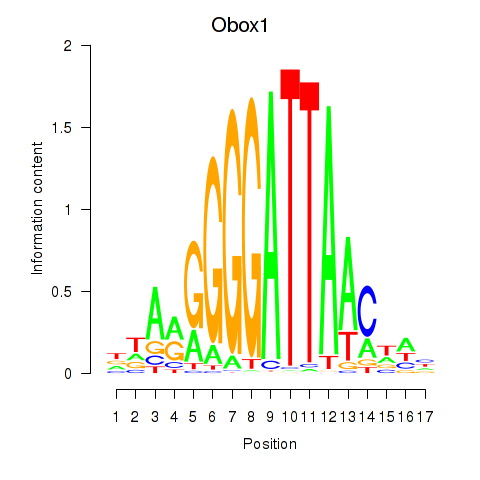
| Obox1 | 1.17 |
|
|
|
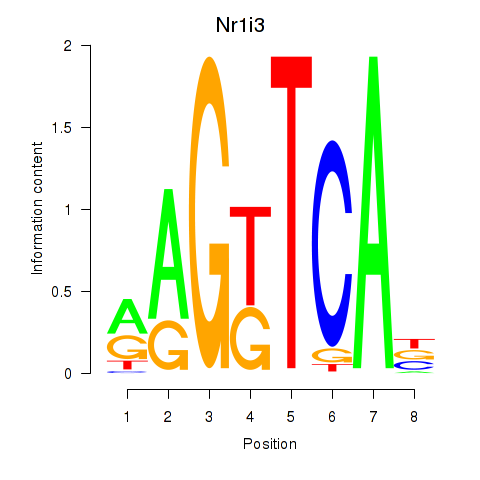
| Nr1i3 | 1.17 |
|

|
|
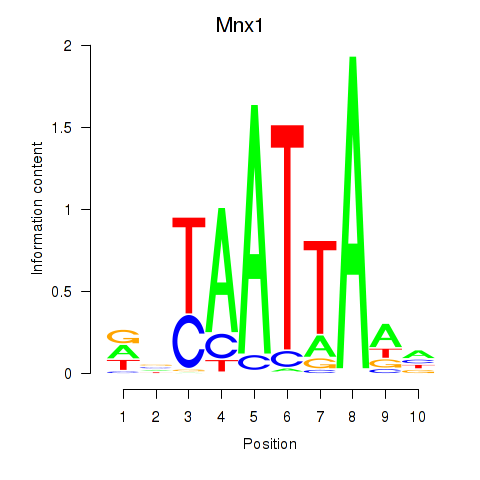
| Mnx1_Lhx6_Lmx1a | 1.17 |
|
|
|
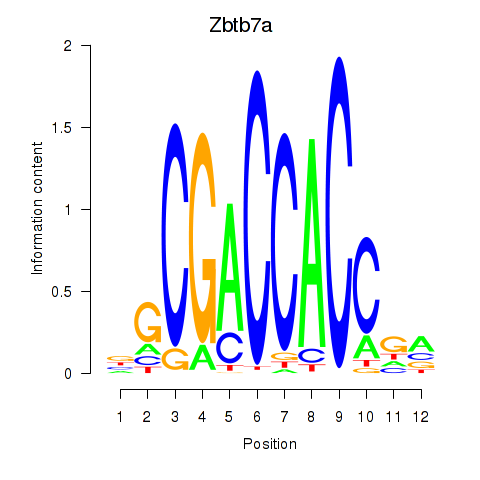
| Zbtb7a | 1.16 |
|
|
|
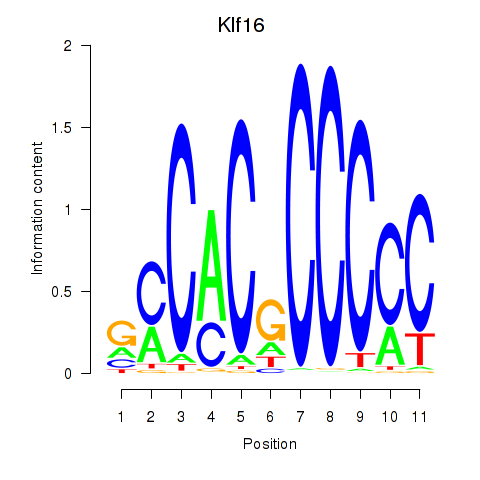
| Klf16_Sp8 | 1.16 |
|

|
|
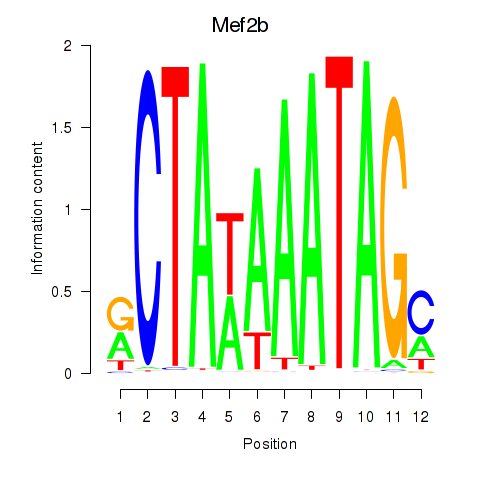
| Mef2b | 1.16 |
|
|
|
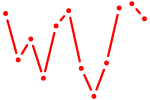
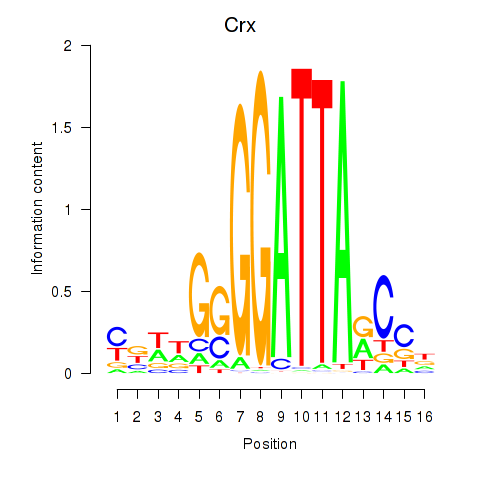
| Crx_Gsc | 1.15 |
|
|
|
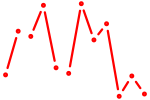
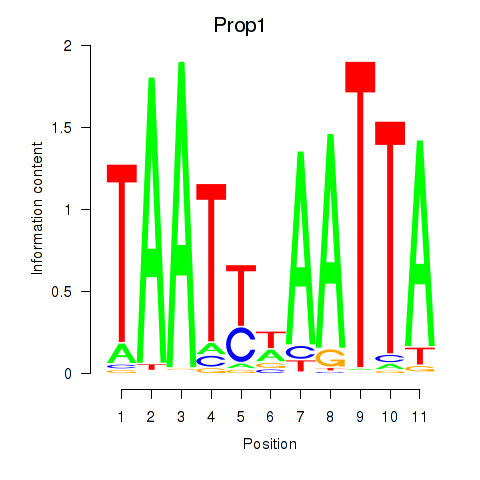
| Prop1 | 1.15 |
|
|
|
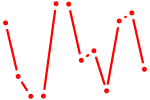
| GUGCAAA | 1.15 |
|
|

|
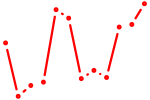
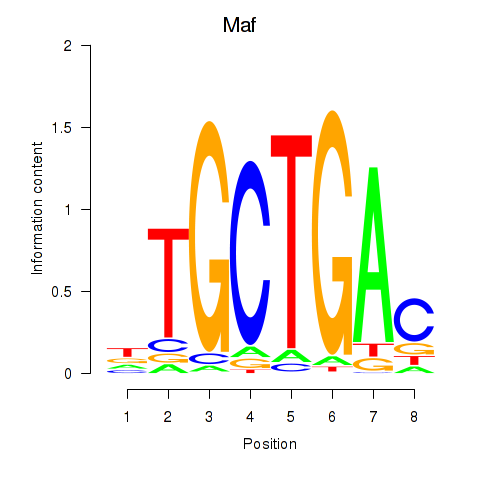
| Maf_Nrl | 1.15 |
|
|
|
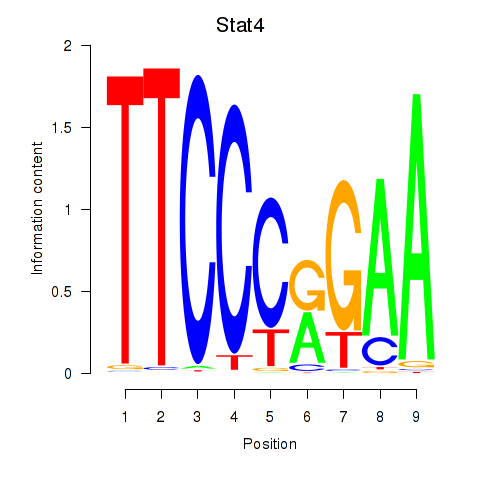
| Stat4_Stat3_Stat5b | 1.15 |
|

|
|
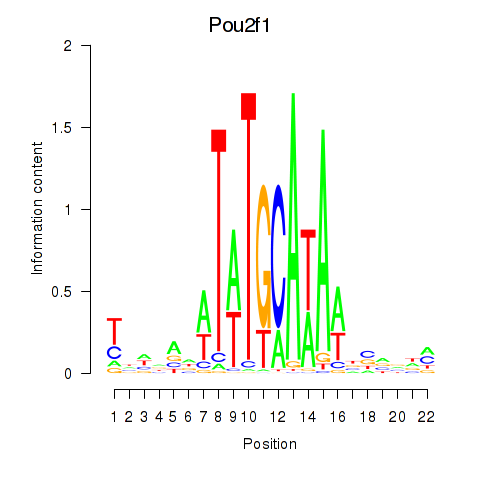
| Pou2f1 | 1.14 |
|
|
|
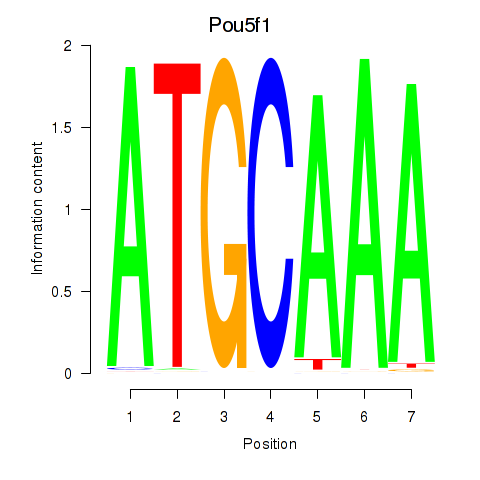
| Pou5f1 | 1.13 |
|
|
|
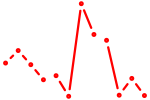
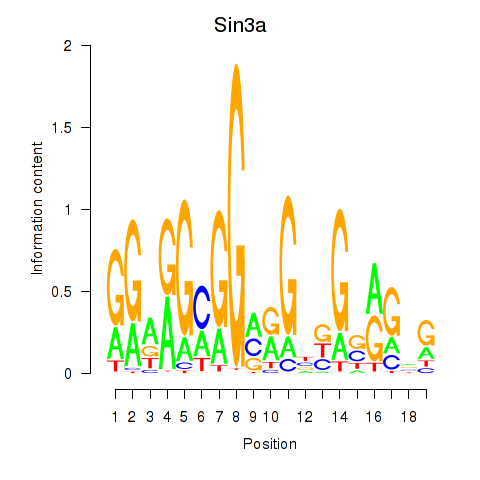
| Sin3a | 1.11 |
|
|
|
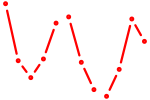
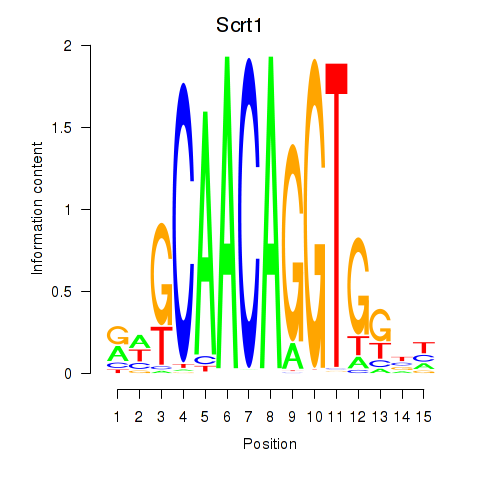
| Scrt1 | 1.09 |
|
|
|
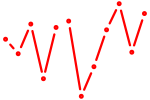
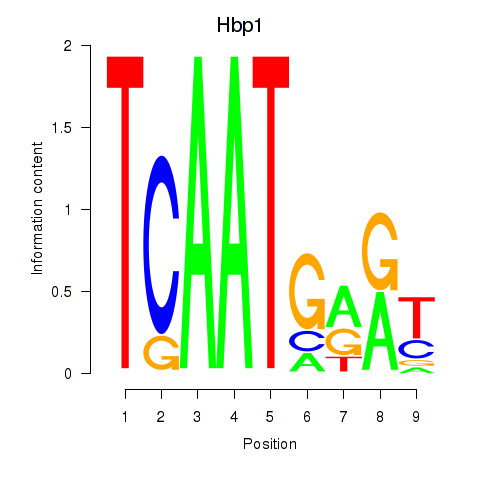
| Hbp1 | 1.08 |
|
|
|
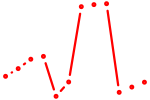
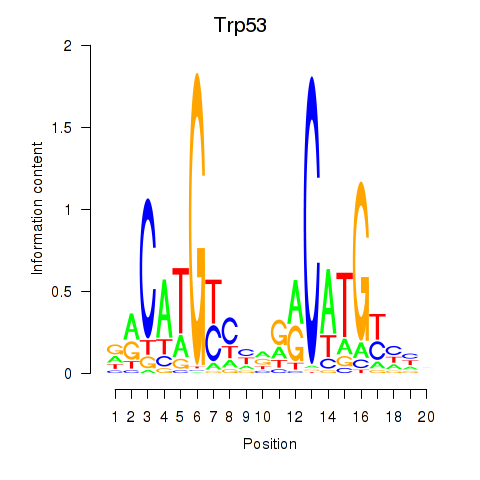
| Trp53 | 1.08 |
|
|
|
| UGUGCUU | 1.08 |
|
|

|
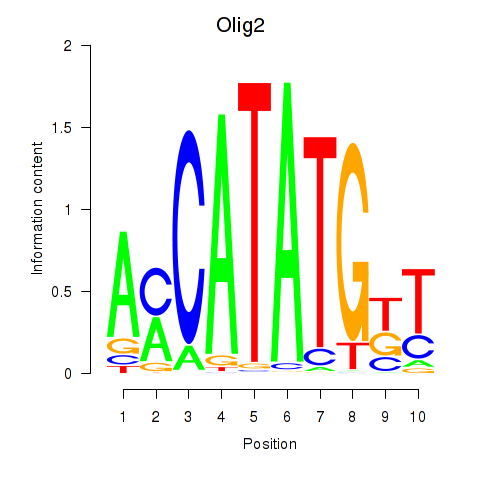
| Olig2_Olig3 | 1.07 |
|
|
|
| AGUGCAA | 1.07 |
|
|

|
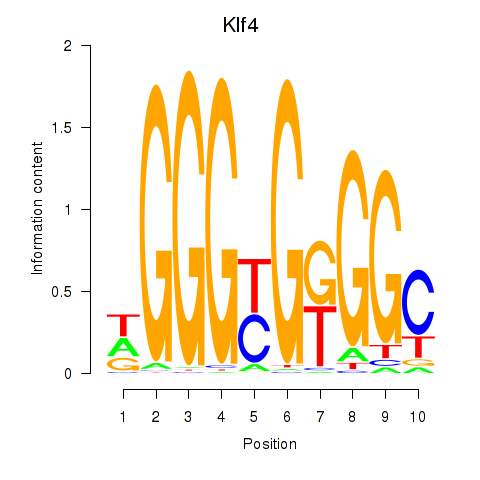
| Klf4_Sp3 | 1.07 |
|
|
|
| UAUUGCU | 1.06 |
|

|

|
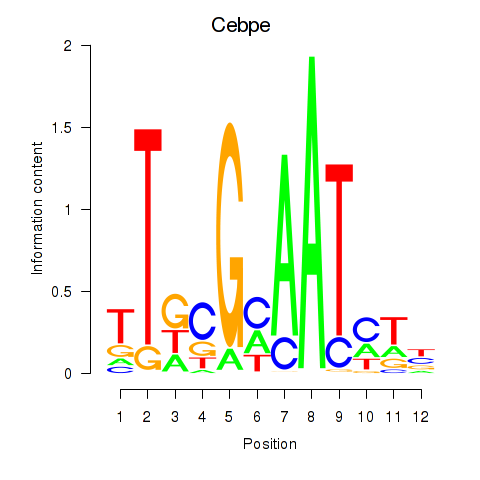
| Cebpe | 1.06 |
|
|
|
| AGCUGCC | 1.06 |
|

|

|
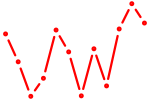
| Smad4 | 1.06 |
|
|
|
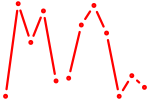
| Sox5_Sry | 1.06 |
|
|
|
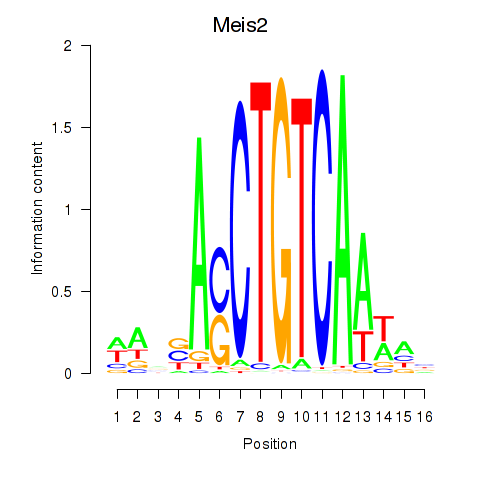
| Meis2 | 1.06 |
|
|
|
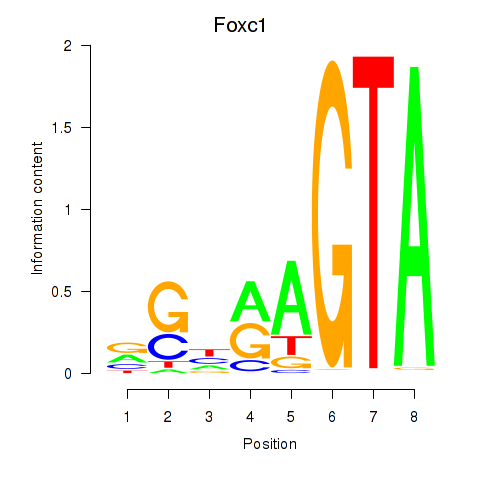
| Foxc1 | 1.05 |
|
|
|
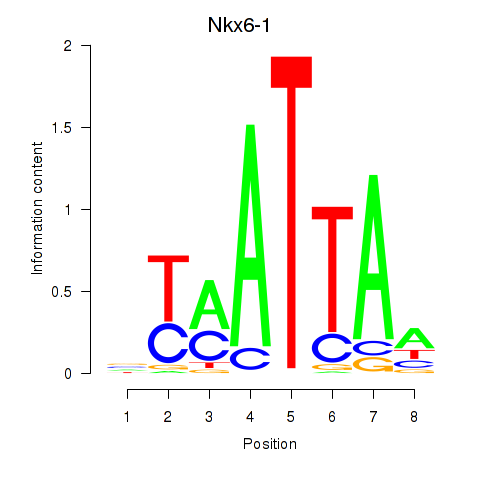
| Nkx6-1_Evx1_Hesx1 | 1.05 |
|

|
|
| CUUUGGU | 1.04 |
|
|

|
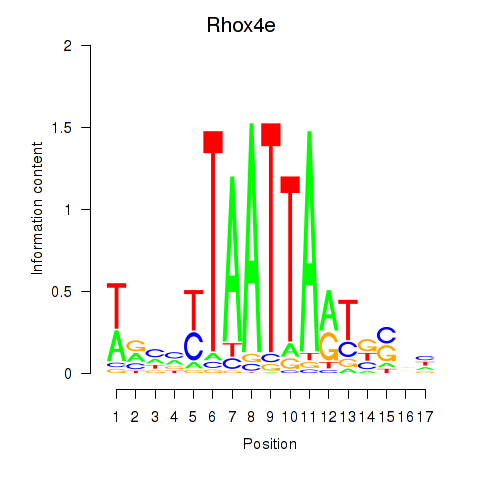
| Rhox4e_Rhox6_Vax2 | 1.04 |
|
|
|
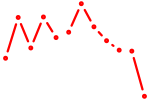
| ACAUUCA | 1.04 |
|
|

|
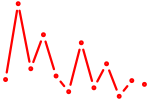
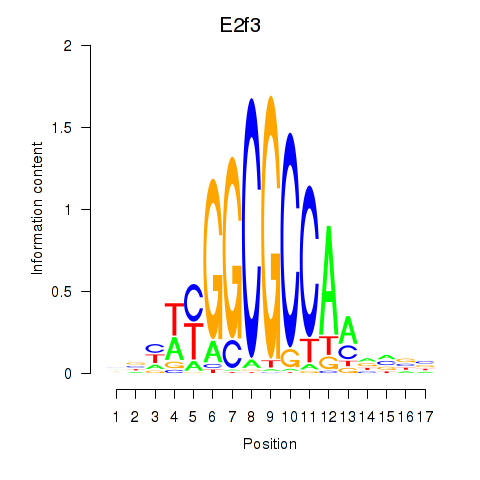
| E2f3 | 1.02 |
|
|
|
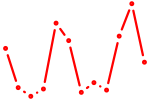
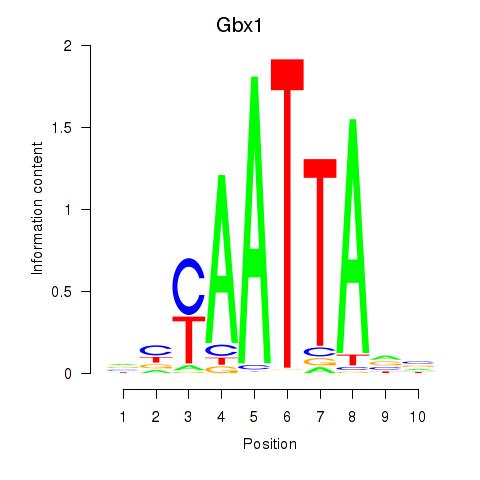
| Gbx1_Nobox_Alx3 | 1.02 |
|
|
|
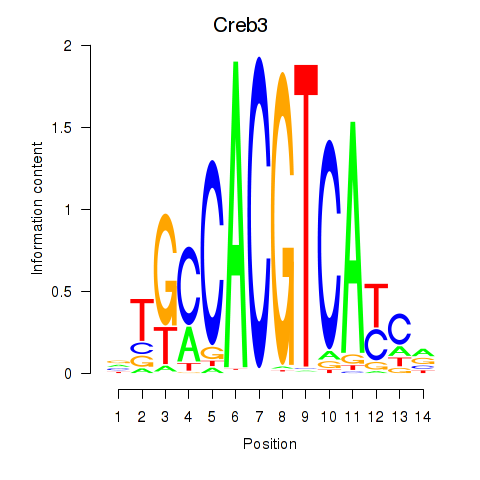
| Creb3 | 1.02 |
|
|
|
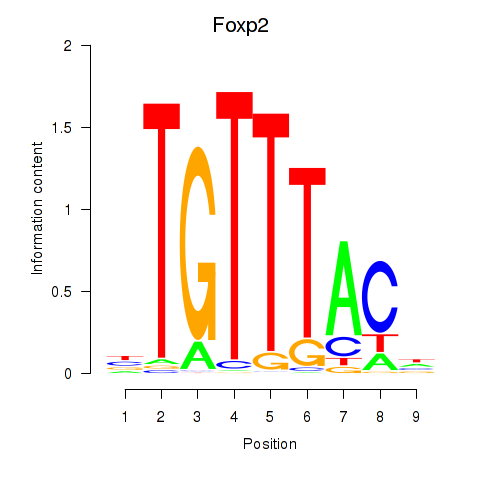
| Foxp2_Foxp3 | 1.02 |
|

|
|
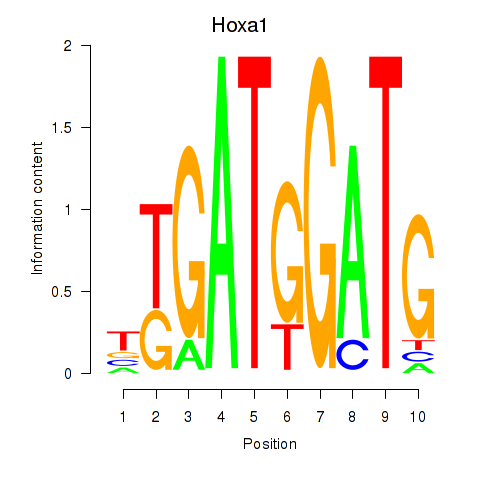
| Hoxa1 | 1.01 |
|
|
|
| GGCAGUG | 1.01 |
|

|

|
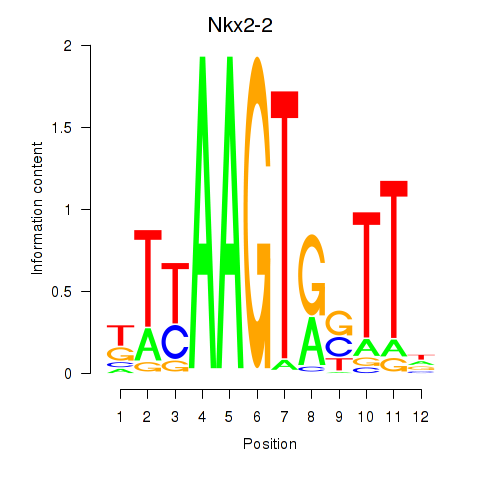
| Nkx2-2 | 1.01 |
|

|
|
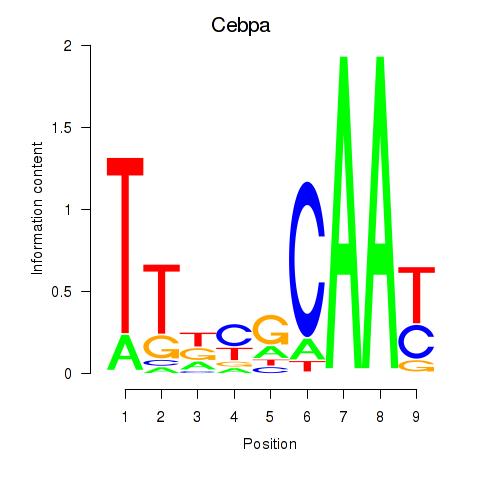
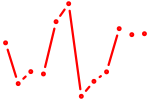
| Cebpa_Cebpg | 1.01 |
|
|
|
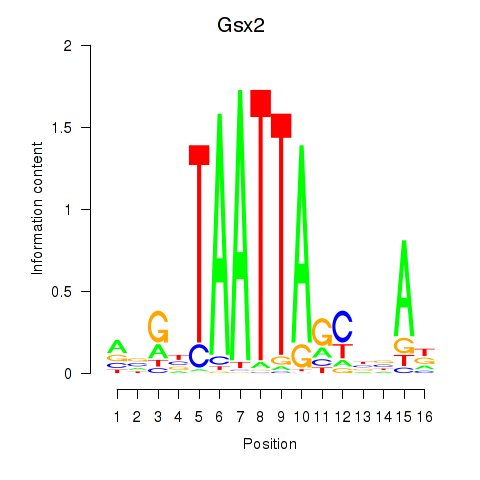
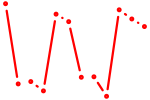
| Gsx2_Hoxd3_Vax1 | 1.00 |
|
|
|
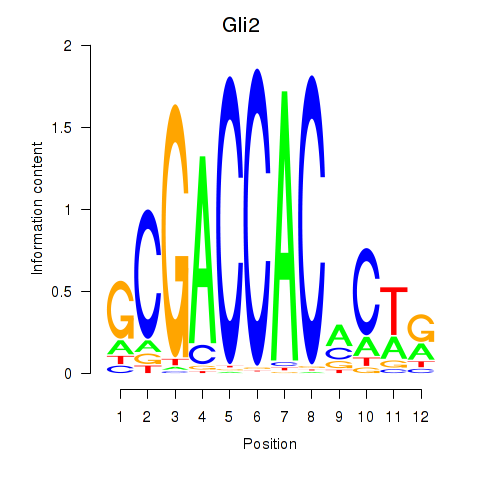
| Gli2 | 0.99 |
|
|
|
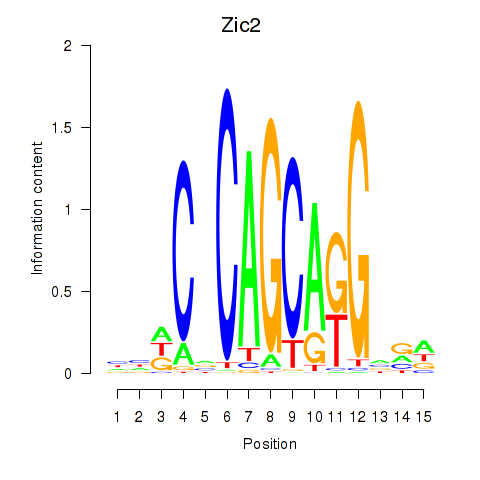
| Zic2 | 0.98 |
|
|
|
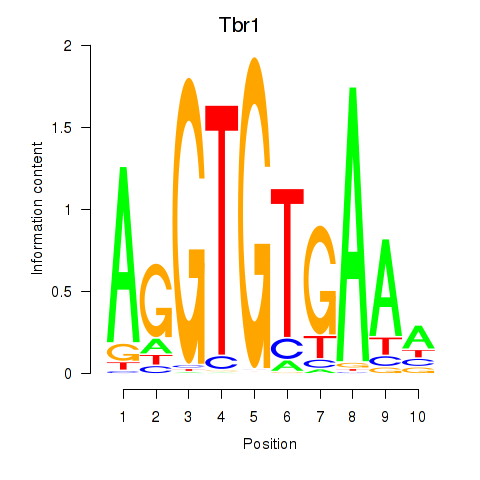
| Tbr1 | 0.98 |
|
|
|
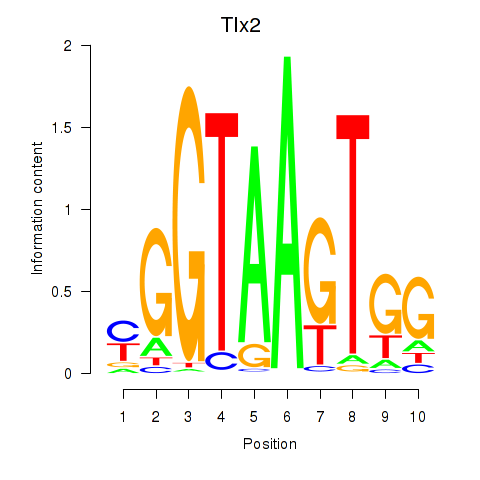
| Tlx2 | 0.98 |
|

|
|
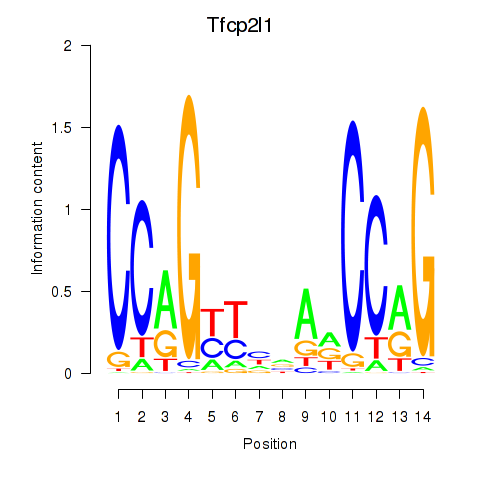
| Tfcp2l1 | 0.98 |
|

|
|
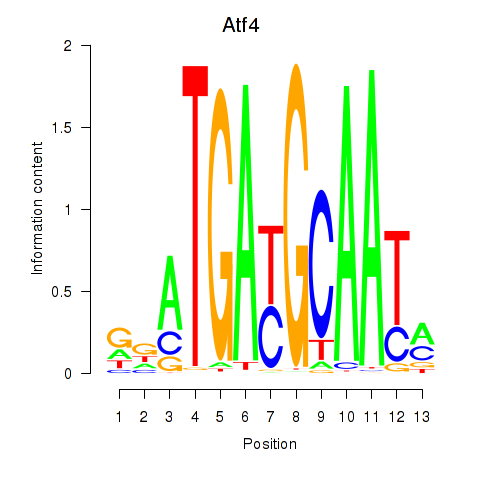
| Atf4 | 0.97 |
|
|
|
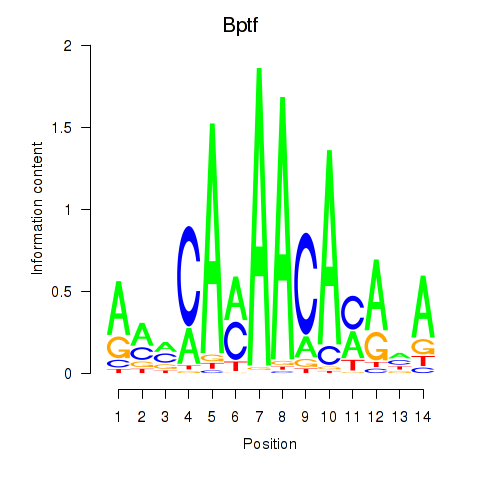
| Bptf | 0.97 |
|
|
|
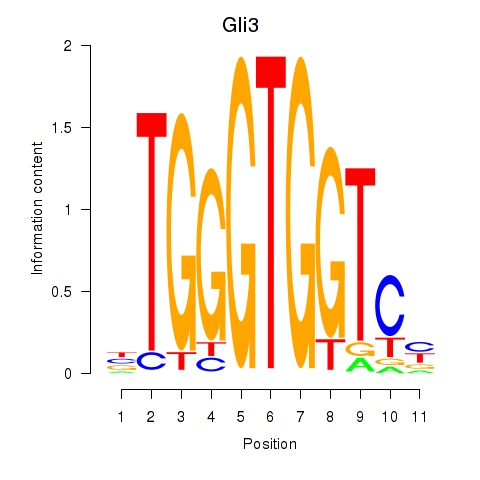
| Gli3_Zic1 | 0.97 |
|
|
|
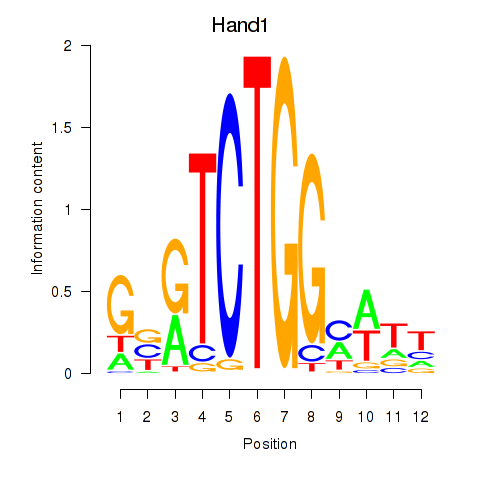
| Hand1 | 0.97 |
|
|
|
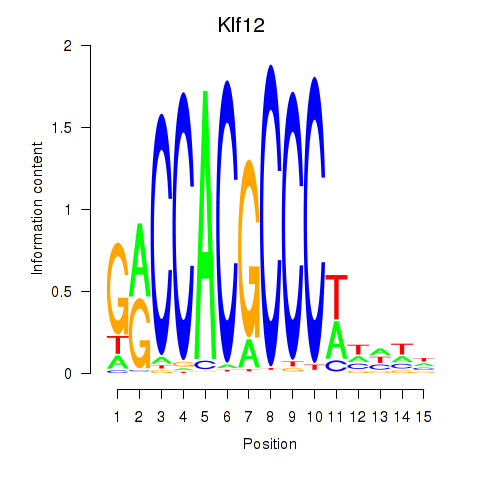
| Klf12_Klf14_Sp4 | 0.97 |
|
|
|
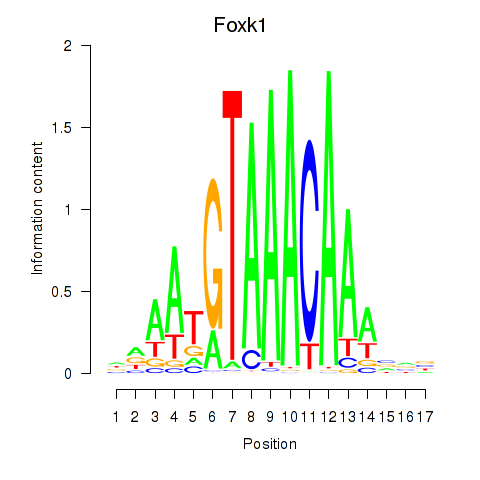
| Foxk1_Foxj1 | 0.97 |
|
|
|
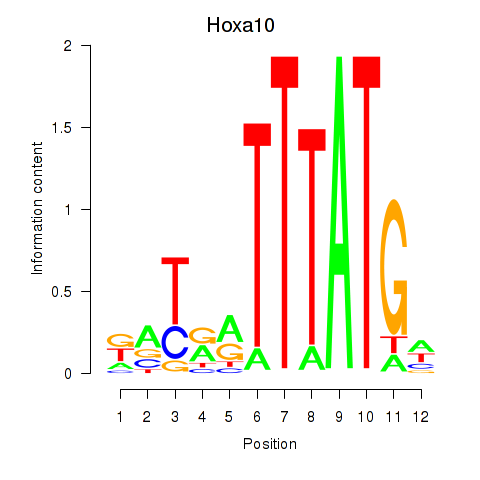
| Hoxa10 | 0.96 |
|
|
|
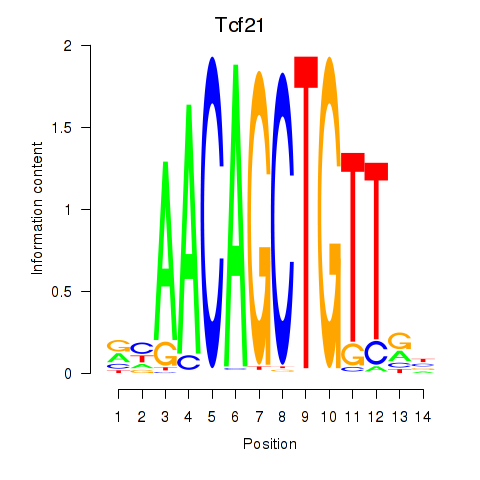
| Tcf21_Msc | 0.96 |
|
|
|
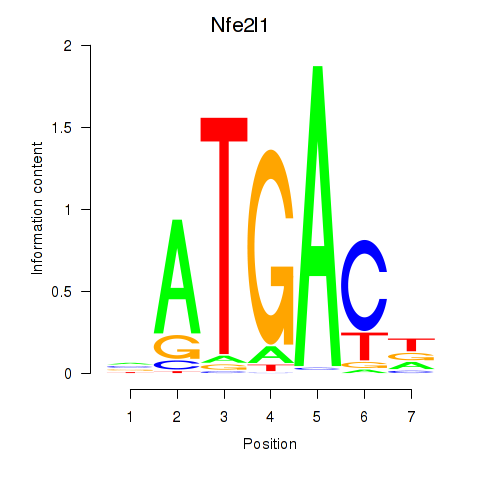
| Nfe2l1_Mafg | 0.96 |
|
|
|
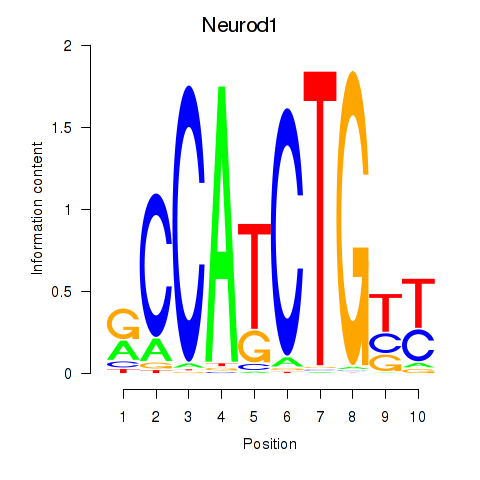
| Neurod1 | 0.95 |
|

|
|
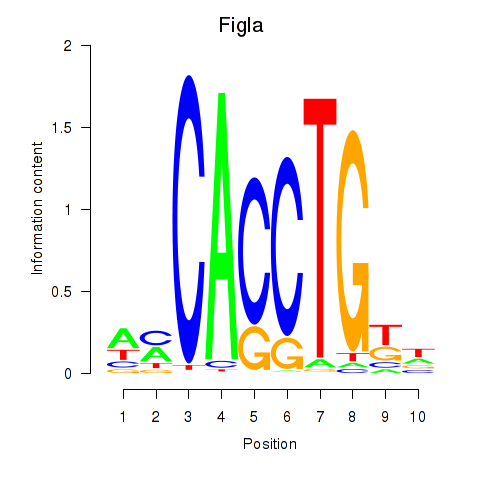
| Figla | 0.95 |
|
|
|
| Hoxb3 | 0.95 |
|
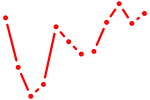
|
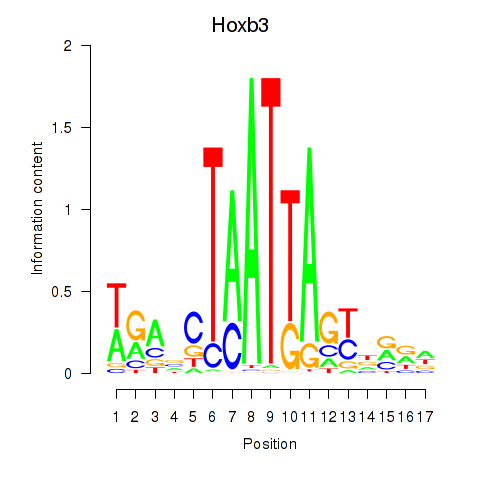
|
| Zbtb4 | 0.94 |
|
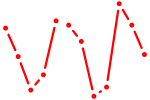
|
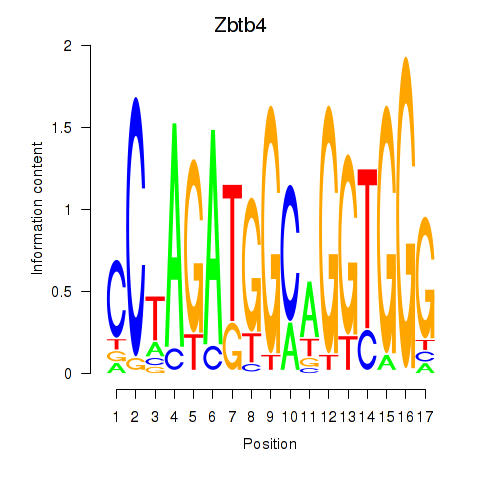
|
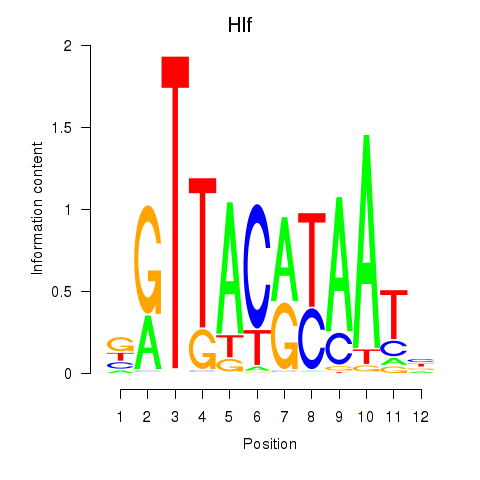
| Hlf | 0.94 |
|
|
|
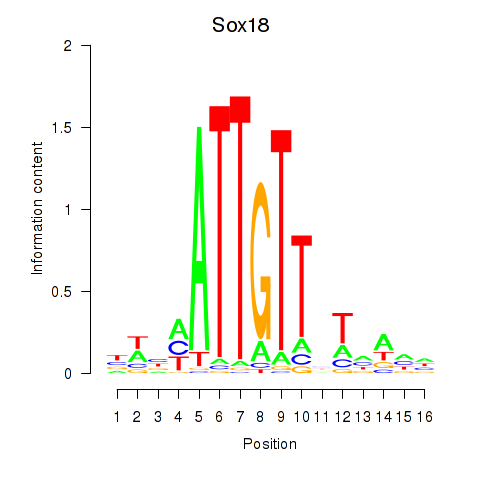
| Sox18_Sox12 | 0.94 |
|
|
|
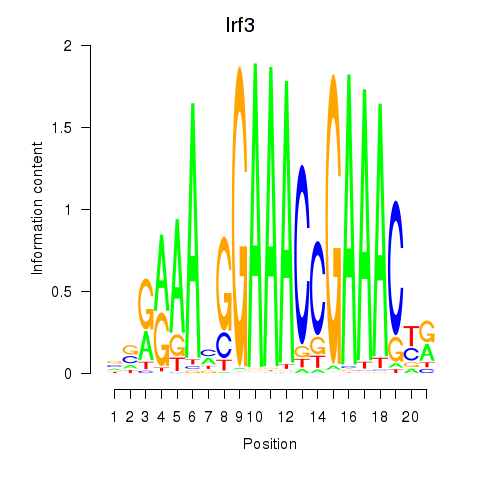
| Irf3 | 0.93 |
|

|
|
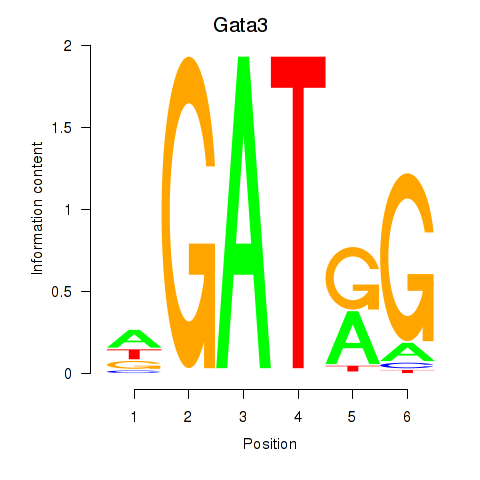
| Gata3 | 0.93 |
|
|
|
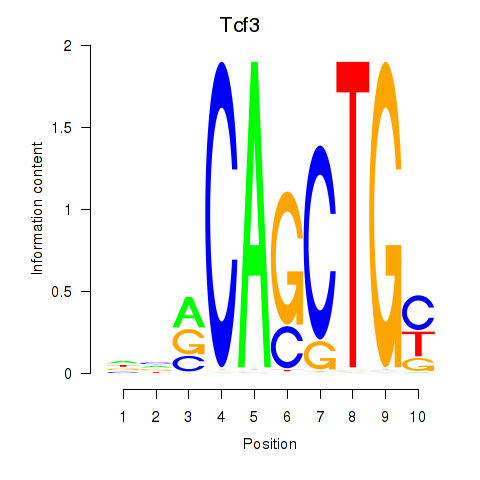
| Tcf3 | 0.93 |
|
|
|
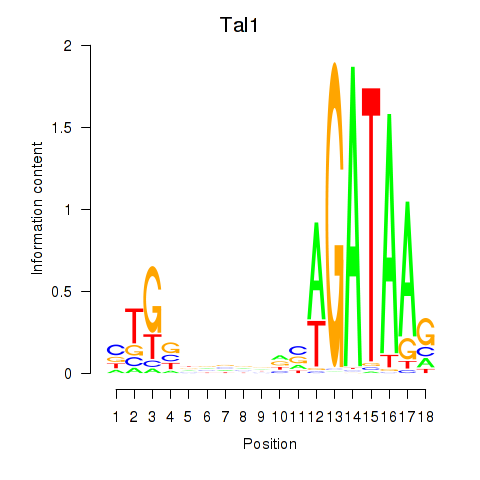
| Tal1 | 0.92 |
|
|
|
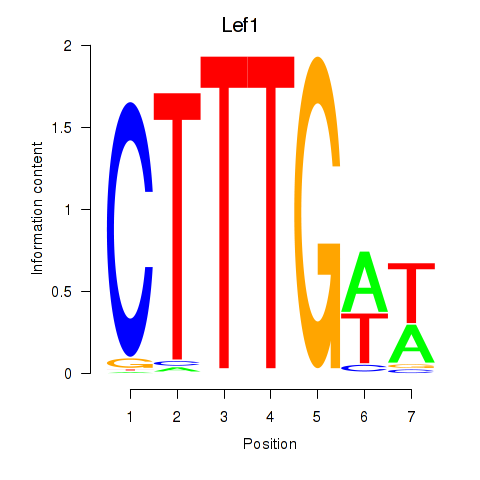
| Lef1 | 0.92 |
|
|
|
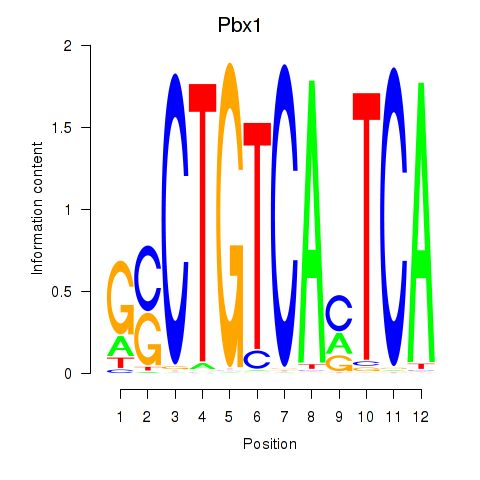
| Pbx1_Pbx3 | 0.92 |
|
|
|
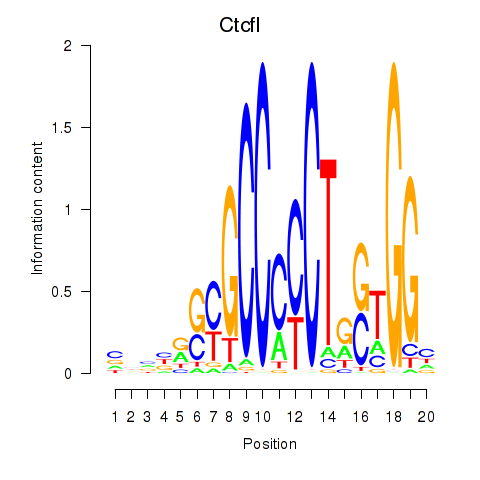
| Ctcfl_Ctcf | 0.92 |
|

|
|
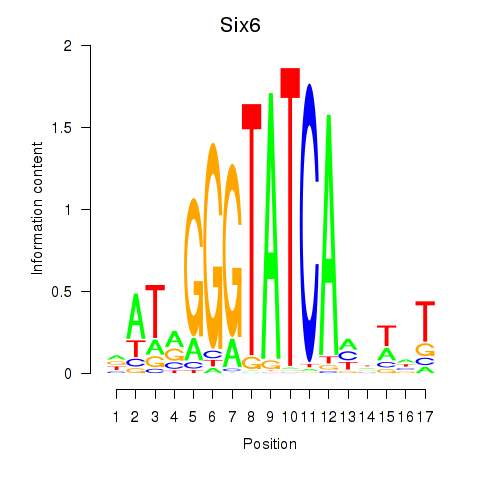
| Six6 | 0.92 |
|
|
|
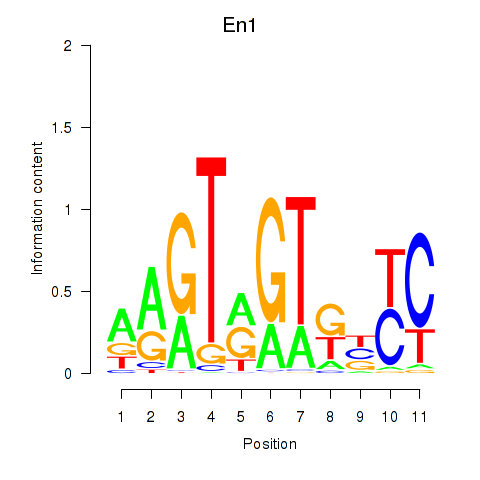
| En1 | 0.92 |
|
|
|
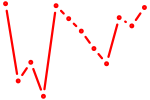
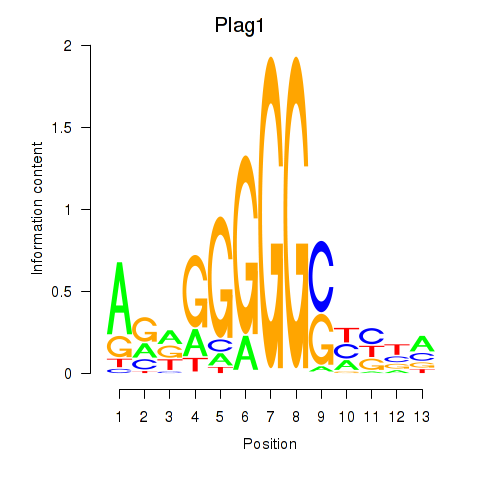
| Plag1 | 0.91 |
|
|
|
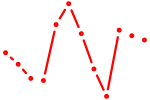
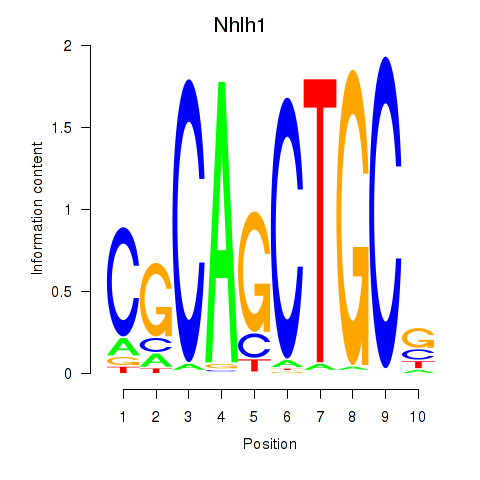
| Nhlh1 | 0.91 |
|
|
|
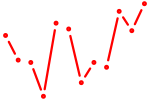
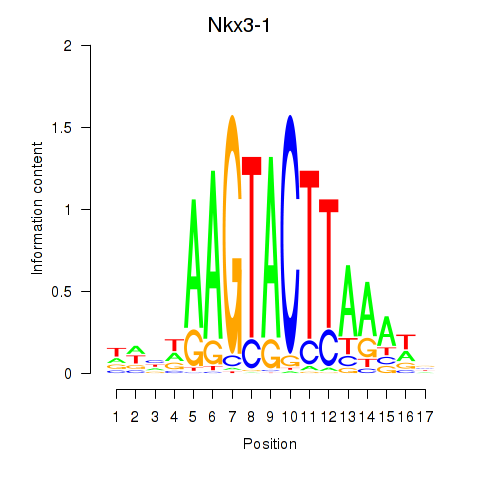
| Nkx3-1 | 0.90 |
|
|
|
| AGCACCA | 0.90 |
|
|

|
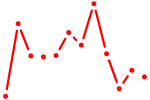
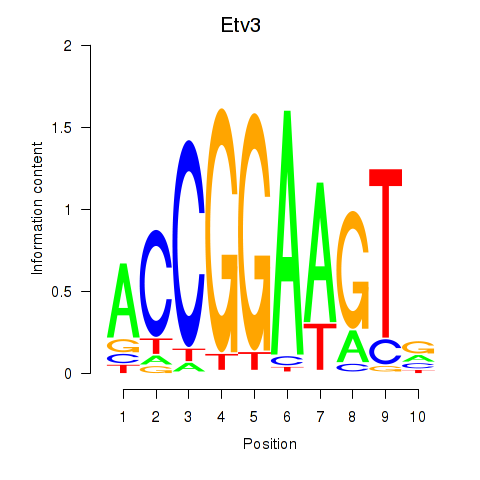
| Etv3_Erf_Fev_Elk4_Elk1_Elk3 | 0.89 |
|

|
|
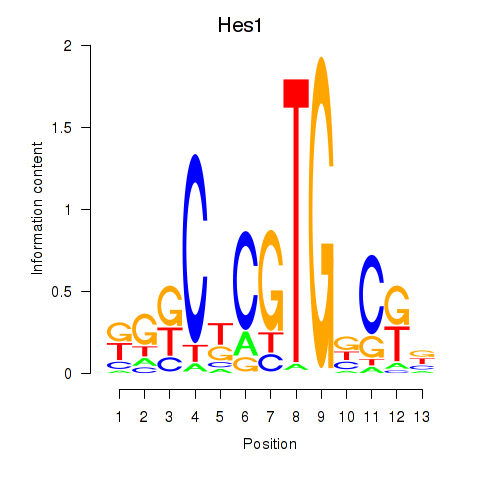
| Hes1 | 0.88 |
|
|
|
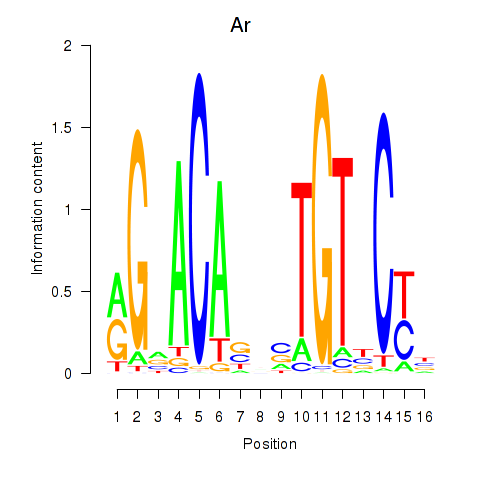
| Ar | 0.88 |
|
|
|
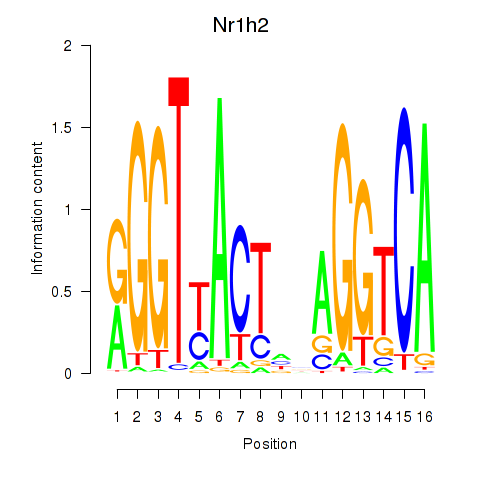
| Nr1h2 | 0.87 |
|
|
|
| GGCUCAG | 0.87 |
|

|

|
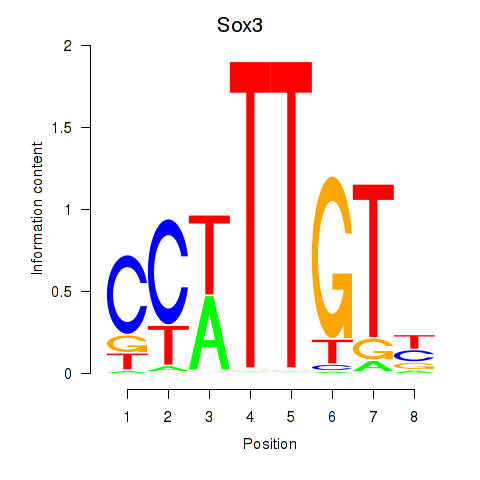
| Sox3_Sox10 | 0.87 |
|
|
|
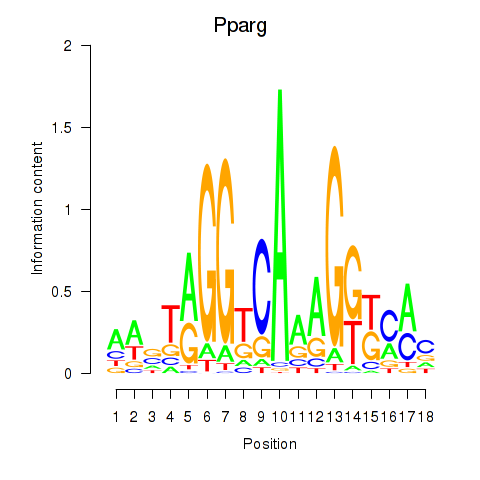
| Pparg_Rxrg | 0.87 |
|
|
|
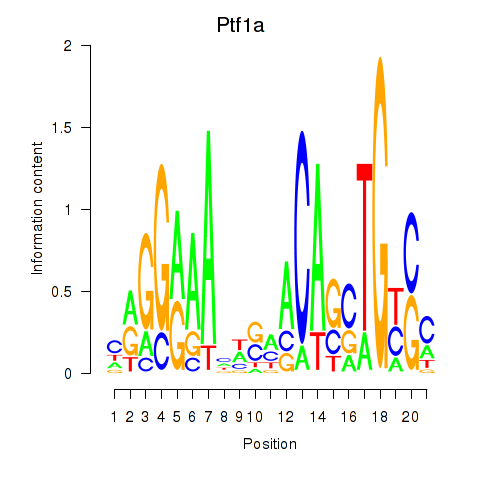
| Ptf1a | 0.87 |
|

|
|
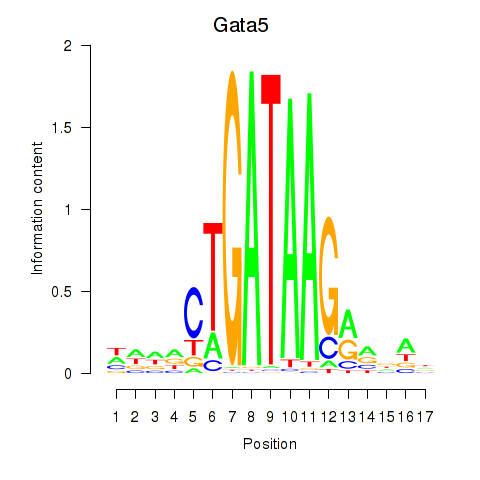
| Gata5 | 0.86 |
|
|
|
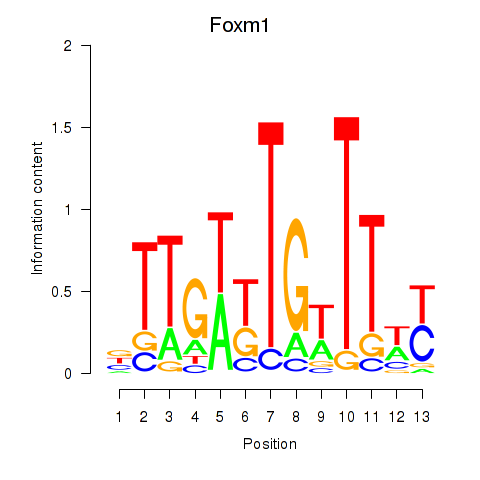
| Foxm1 | 0.86 |
|

|
|
| GAGGUAG | 0.85 |
|
|

|
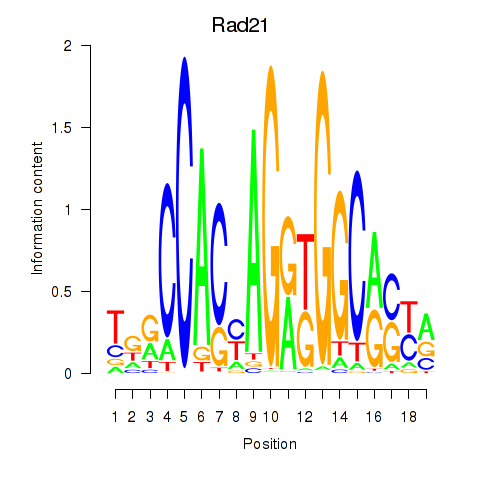
| Rad21_Smc3 | 0.85 |
|
|
|
| UUGGUCC | 0.85 |
|
|

|
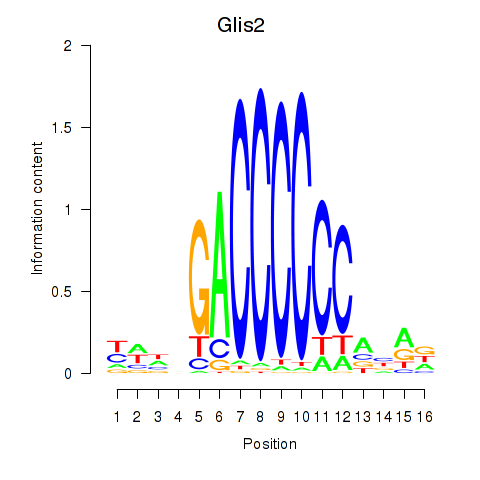
| Glis2 | 0.85 |
|
|
|
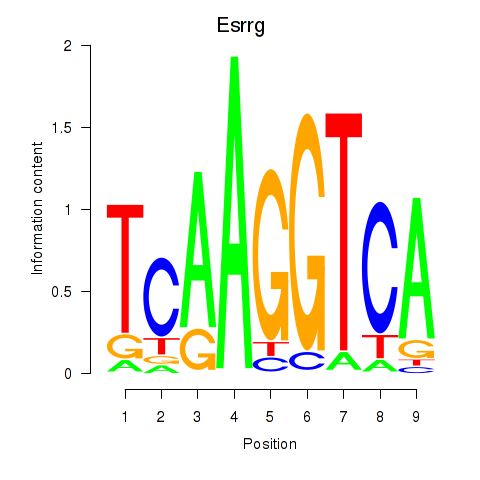
| Esrrg | 0.84 |
|
|
|
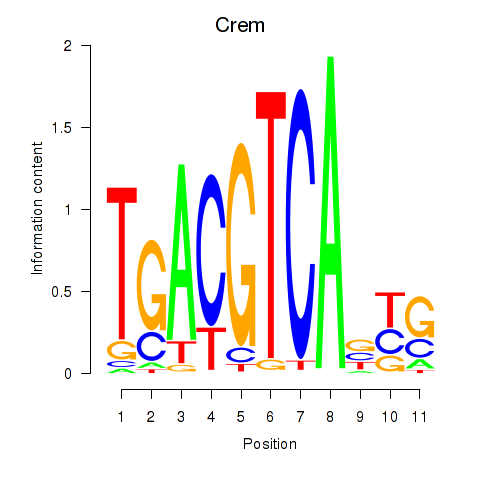
| Crem_Jdp2 | 0.84 |
|
|
|
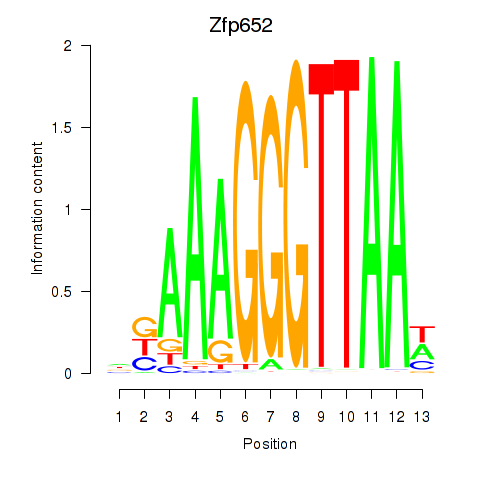
| Zfp652 | 0.84 |
|

|
|
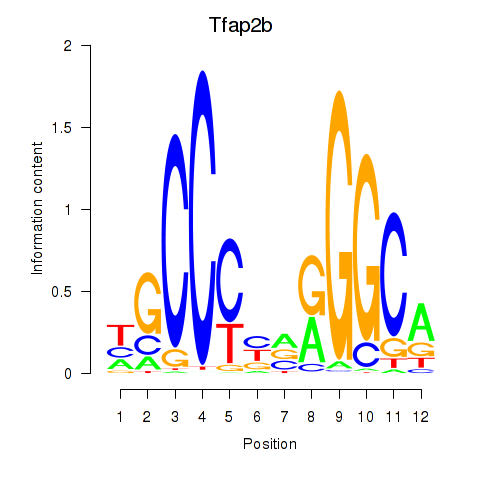
| Tfap2b | 0.84 |
|
|
|
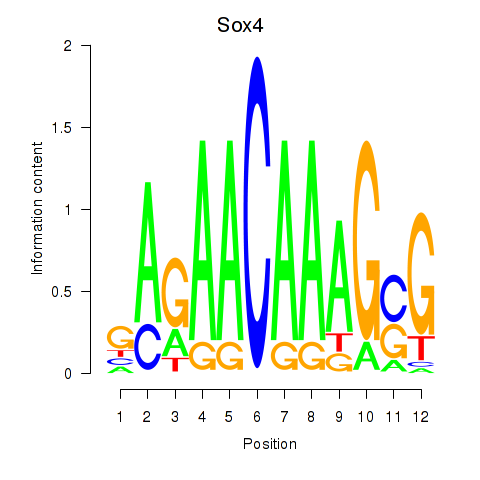
| Sox4 | 0.83 |
|
|
|
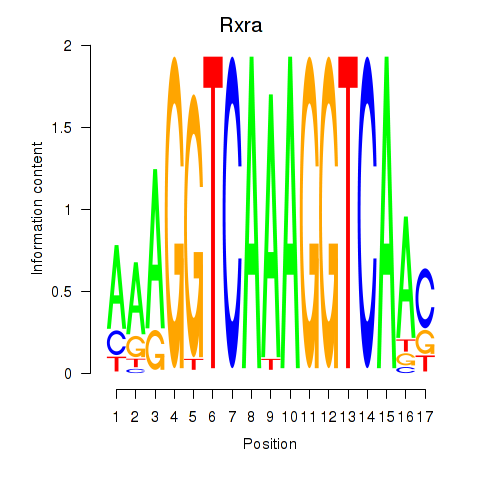
| Rxra | 0.83 |
|

|
|
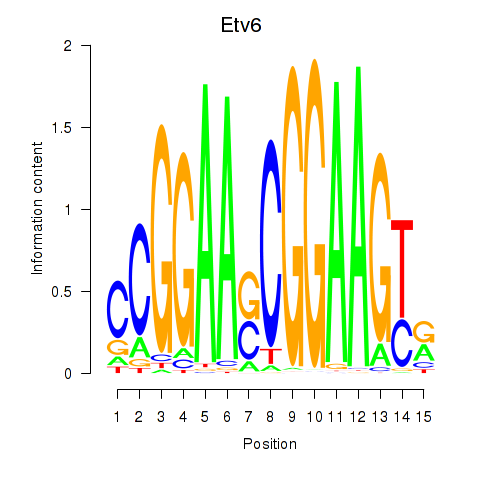
| Etv6 | 0.83 |
|

|
|
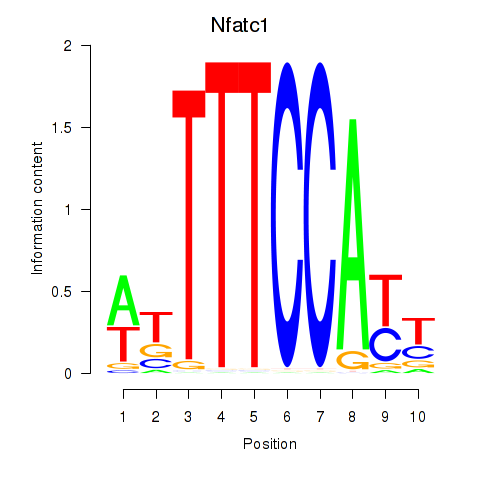
| Nfatc1 | 0.82 |
|
|
|
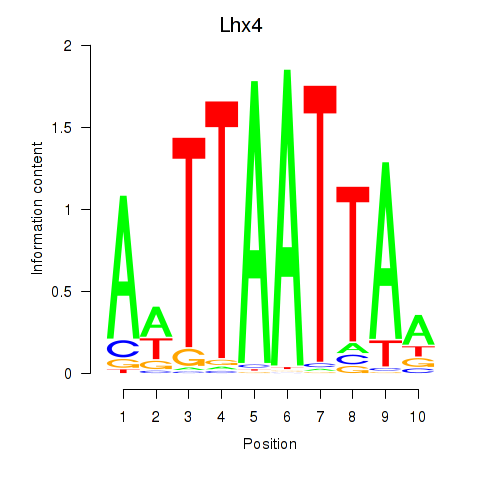
| Lhx4 | 0.82 |
|
|
|
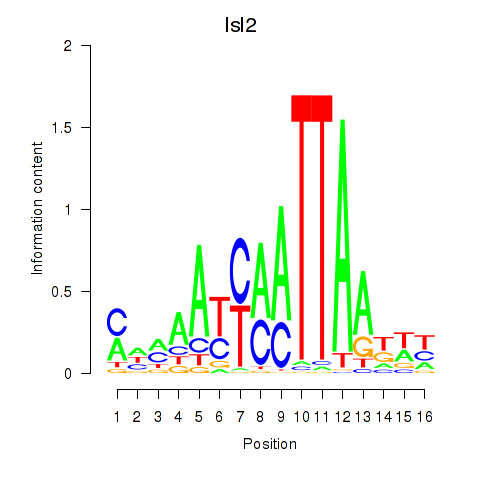
| Isl2 | 0.82 |
|
|
|
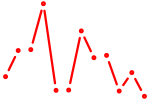
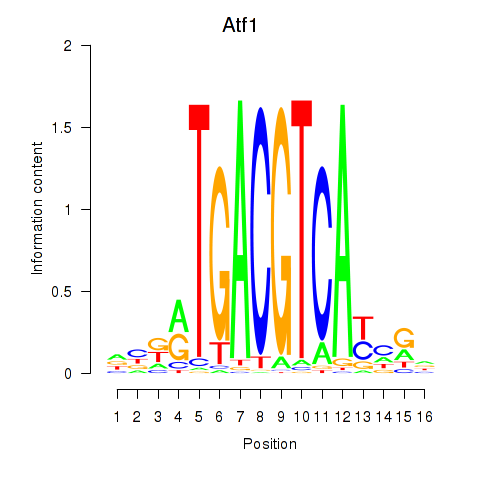
| Atf1_Creb5 | 0.82 |
|
|
|
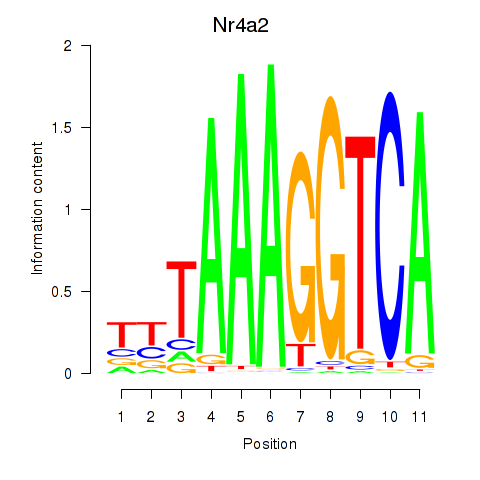
| Nr4a2 | 0.82 |
|

|
|
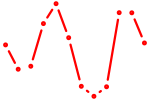
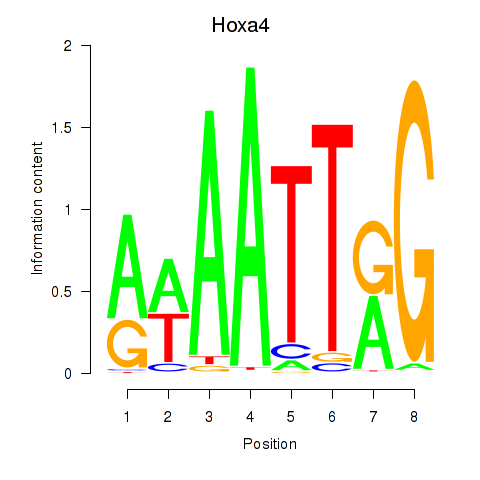
| Hoxa4 | 0.82 |
|
|
|
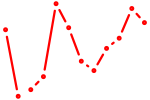
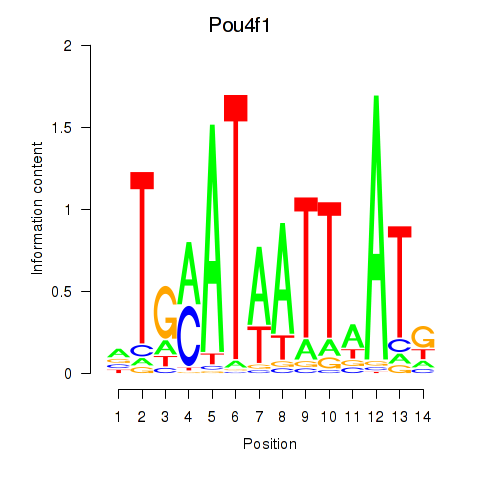
| Pou4f1_Pou6f1 | 0.81 |
|
|
|
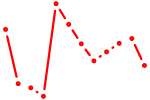
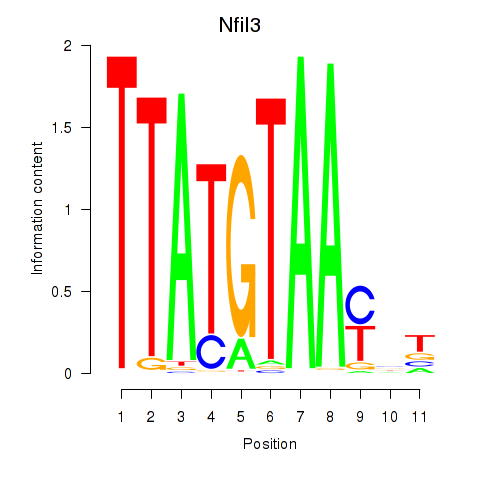
| Nfil3_Tef | 0.81 |
|
|
|
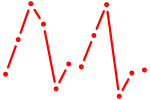
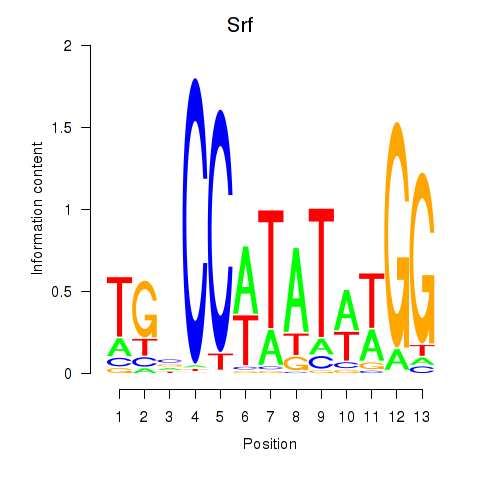
| Srf | 0.80 |
|
|
|
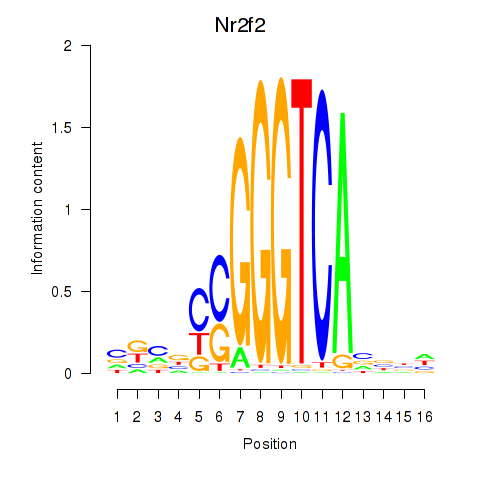
| Nr2f2 | 0.80 |
|
|
|
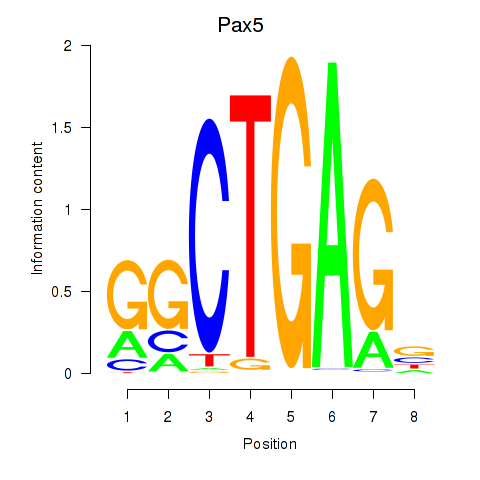
| Pax5 | 0.79 |
|
|
|
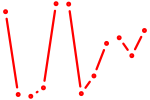
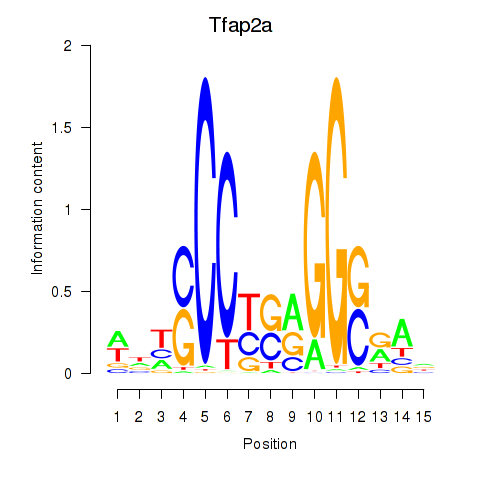
| Tfap2a | 0.79 |
|
|
|
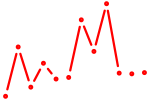
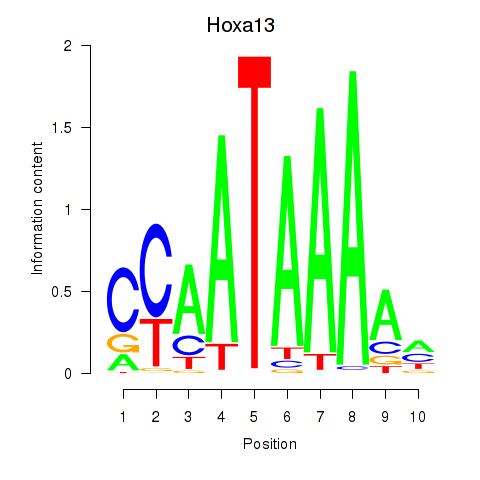
| Hoxa13 | 0.79 |
|
|
|
| Homez | 0.79 |
|
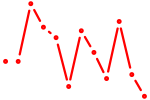
|
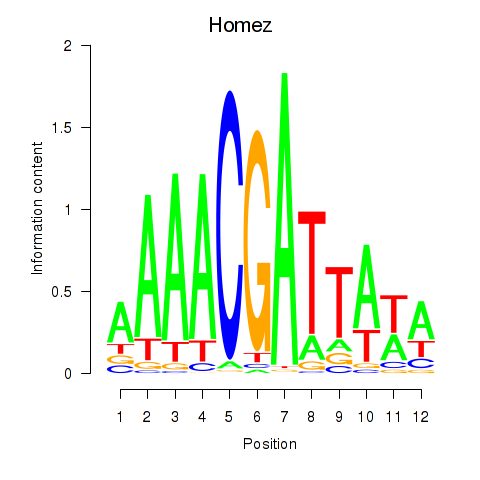
|
| Hoxb8_Pdx1 | 0.79 |
|
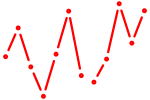
|
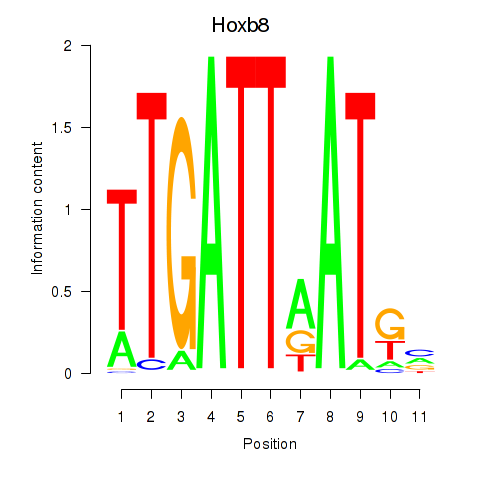
|
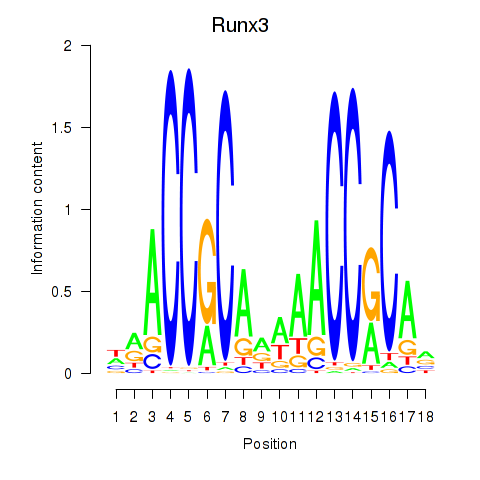
| Runx3 | 0.79 |
|

|
|
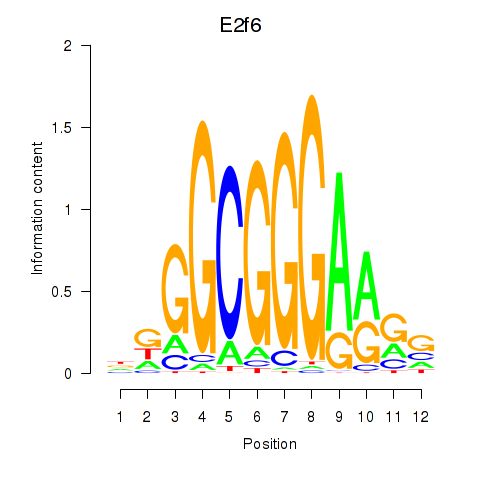
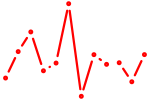
| E2f6 | 0.78 |
|

|
|
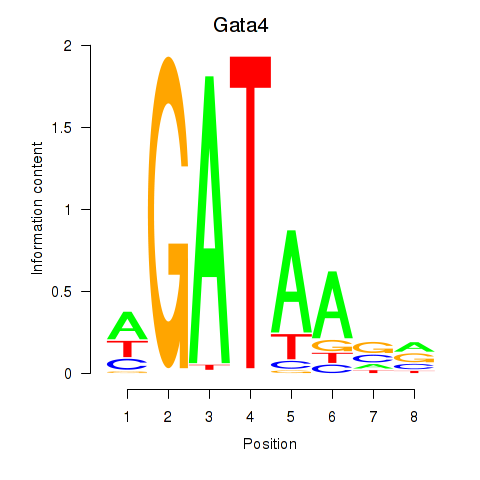
| Gata4 | 0.78 |
|
|
|
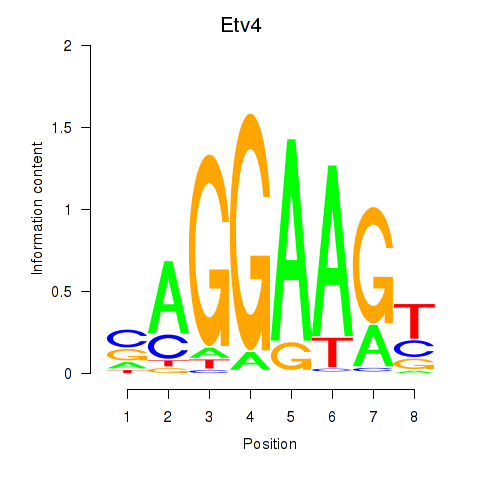
| Etv4 | 0.78 |
|
|
|
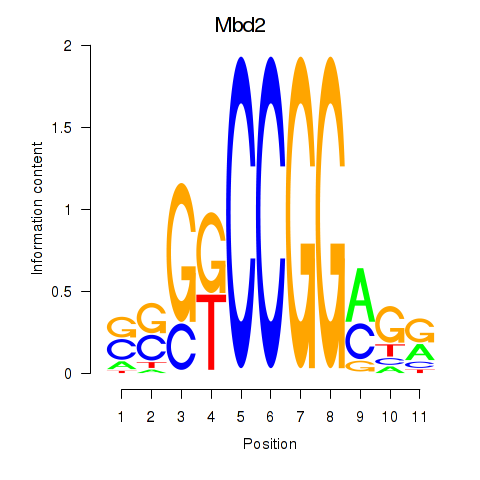
| Mbd2 | 0.77 |
|
|
|
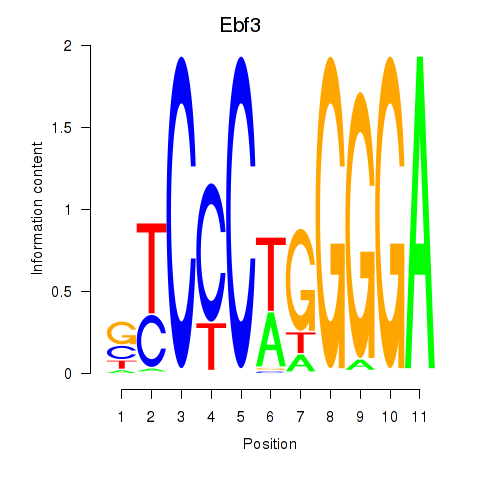
| Ebf3 | 0.77 |
|
|
|
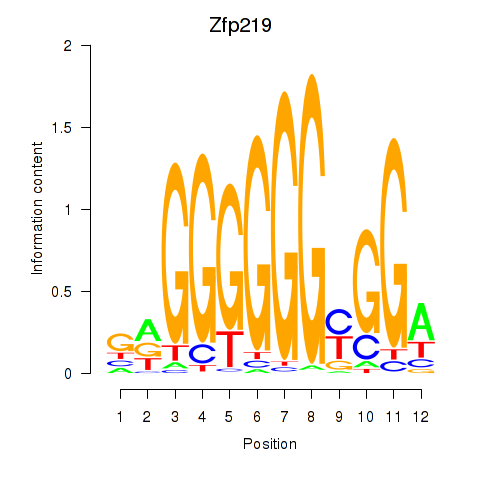
| Zfp219_Zfp740 | 0.77 |
|

|
|
| GUAAACA | 0.77 |
|
|

|
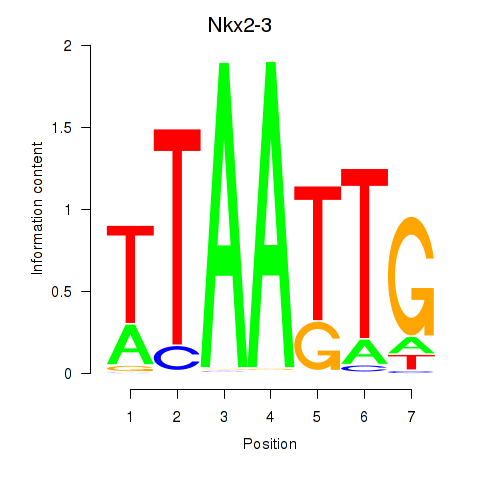
| Nkx2-3 | 0.77 |
|
|
|
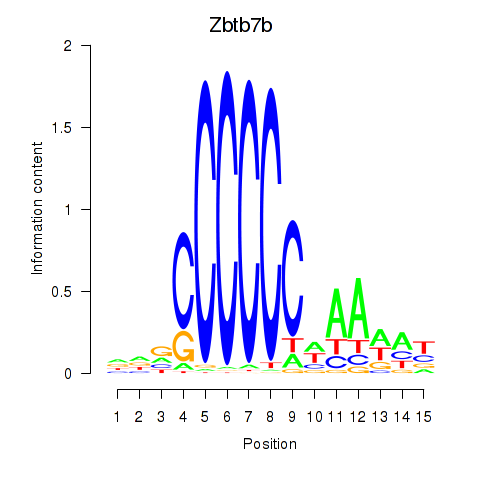
| Zbtb7b | 0.77 |
|
|
|
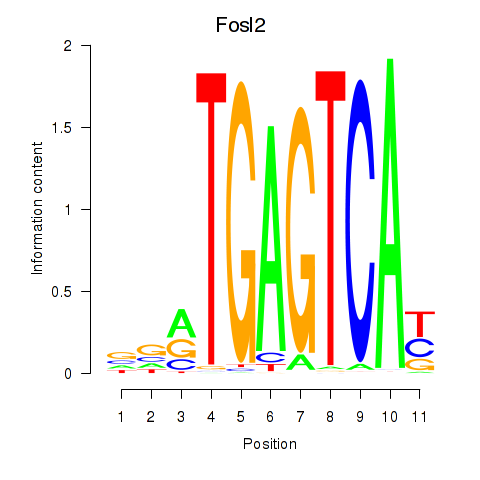
| Fosl2_Bach2 | 0.77 |
|
|
|
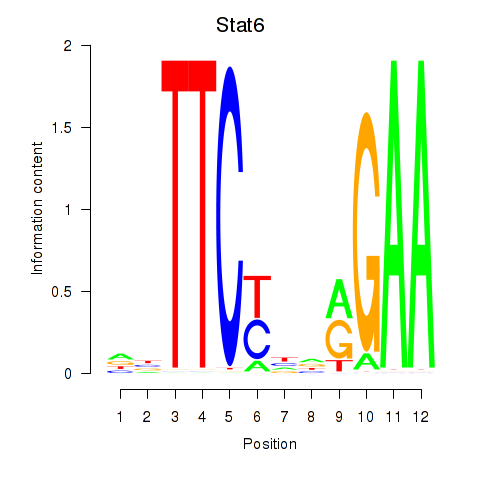
| Stat6 | 0.76 |
|

|
|
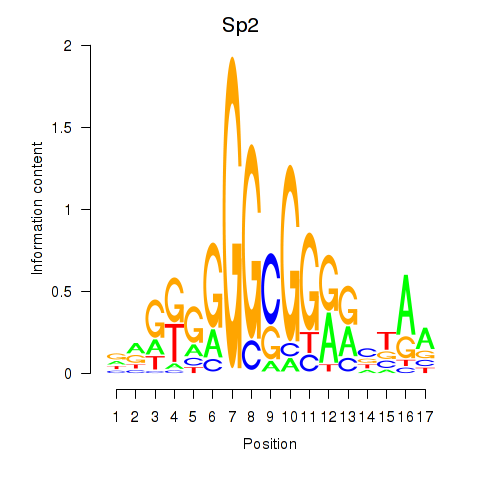
| Sp2 | 0.76 |
|
|
|
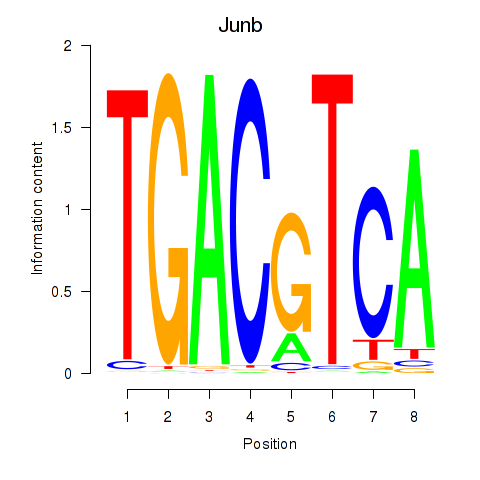
| Junb_Jund | 0.76 |
|
|
|
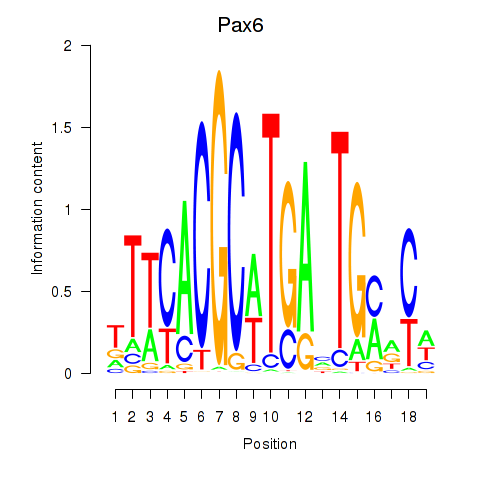
| Pax6 | 0.75 |
|
|
|
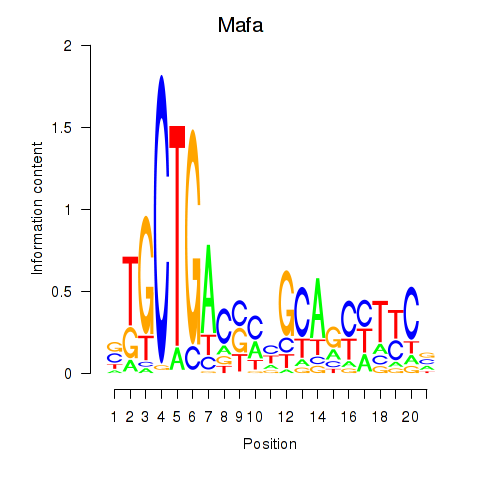
| Mafa | 0.75 |
|
|
|
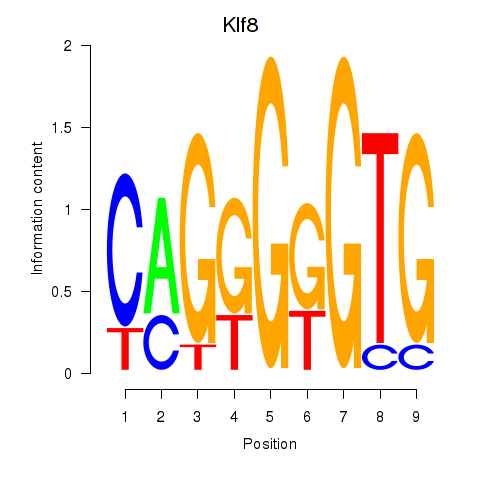
| Klf8 | 0.75 |
|
|
|
| CAGUGCA | 0.75 |
|

|

|
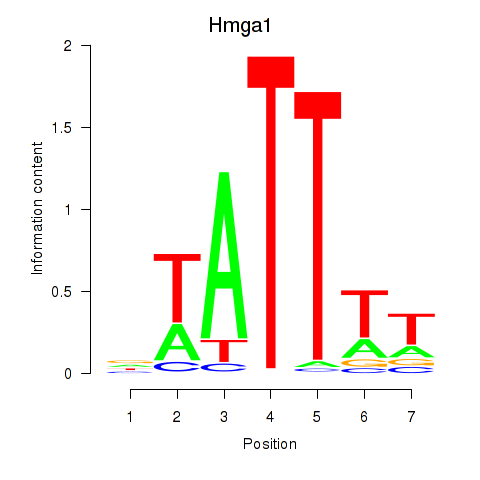
| Hmga1 | 0.74 |
|
|
|
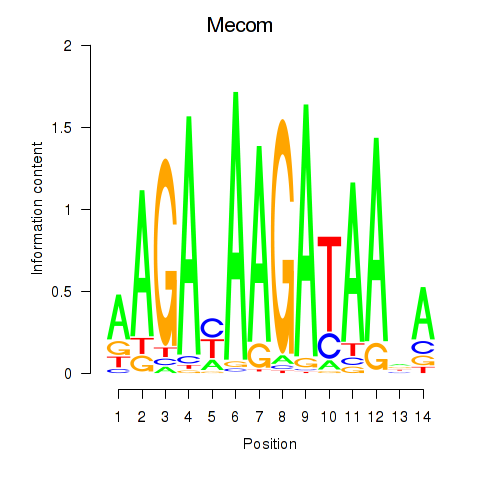
| Mecom | 0.74 |
|
|
|
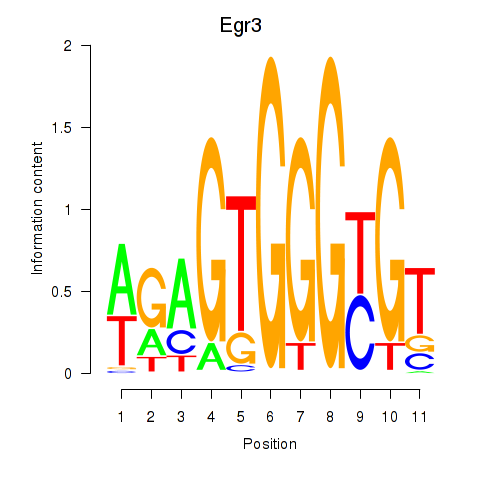
| Egr3 | 0.74 |
|
|
|
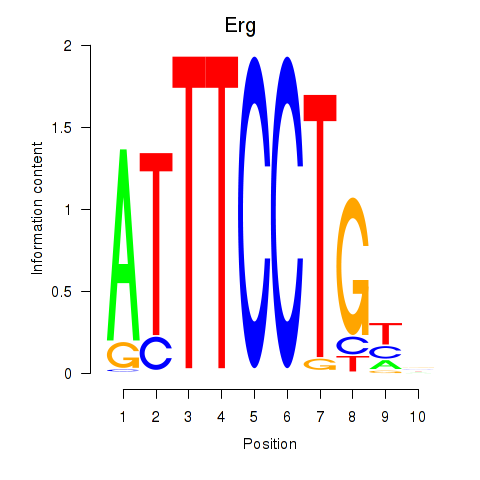
| Erg | 0.74 |
|
|
|
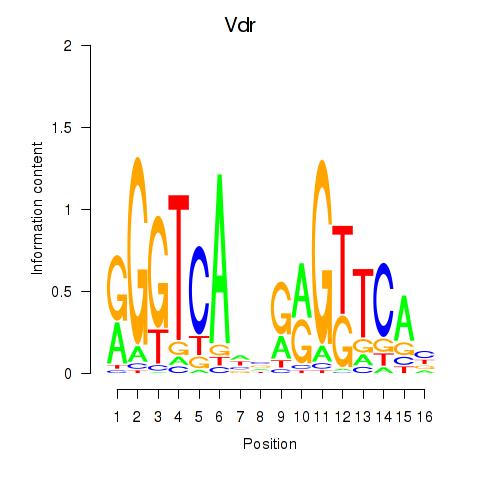
| Vdr | 0.74 |
|
|
|
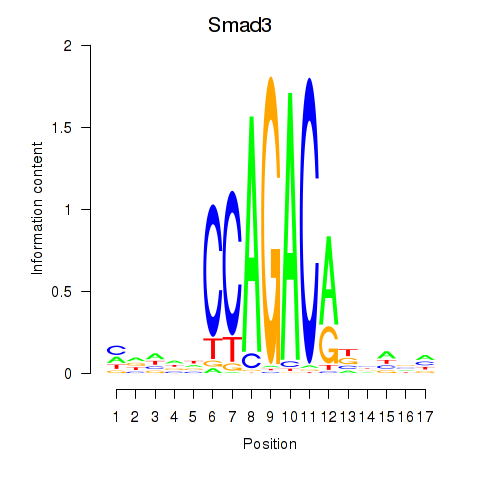
| Smad3 | 0.74 |
|
|
|
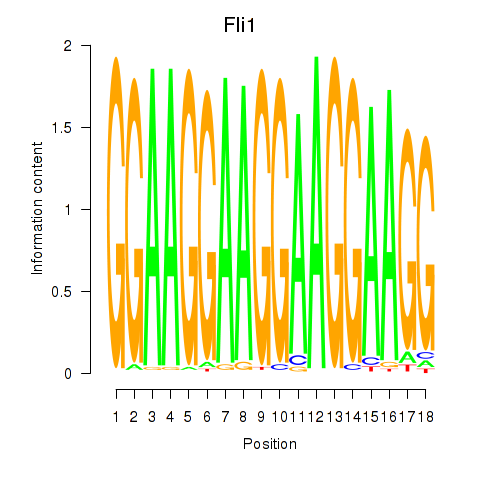
| Fli1 | 0.73 |
|
|
|
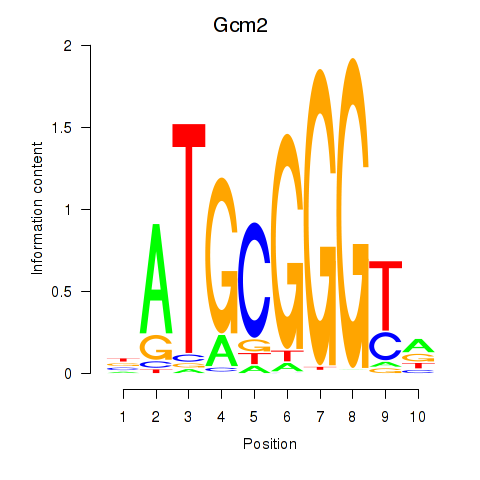
| Gcm2 | 0.73 |
|

|
|
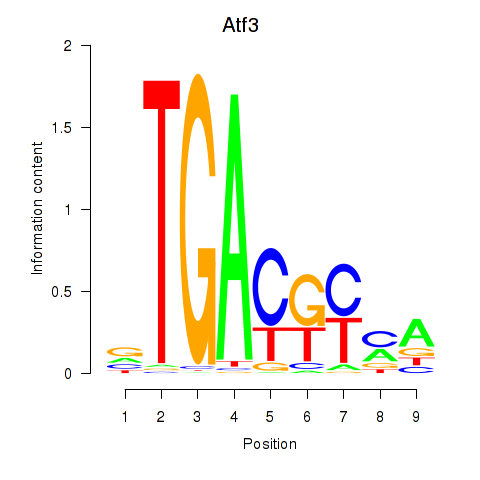
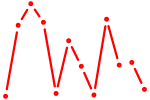
| Atf3 | 0.73 |
|

|
|
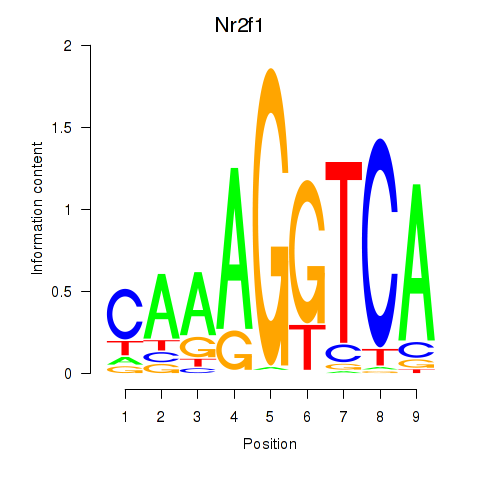
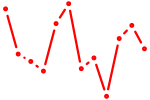
| Nr2f1_Nr4a1 | 0.73 |
|
|
|
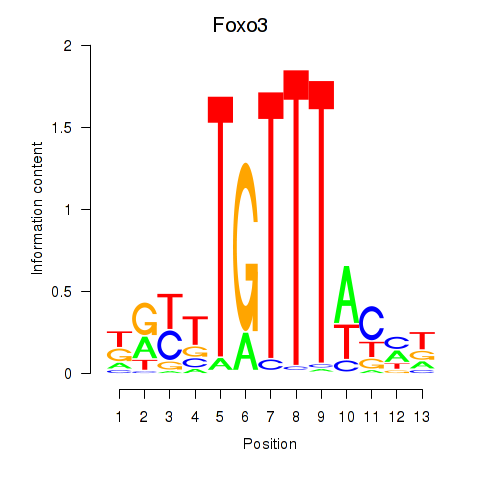
| Foxo3 | 0.73 |
|
|
|
| UUGGCAA | 0.72 |
|

|

|
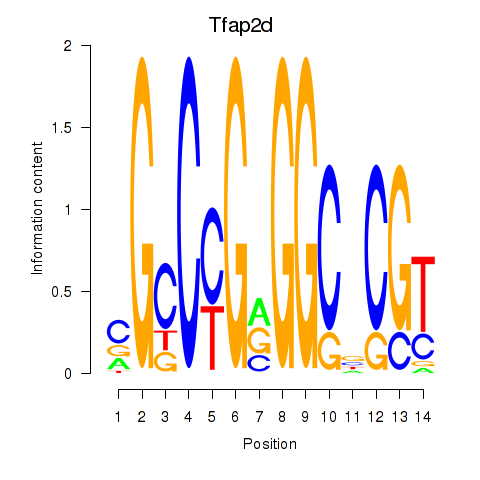
| Tfap2d | 0.72 |
|
|
|
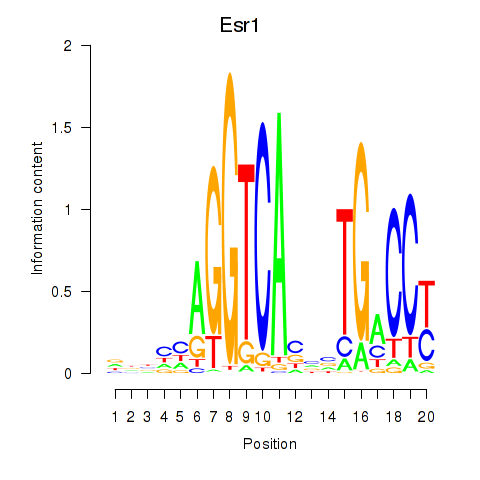
| Esr1 | 0.72 |
|
|
|
| UGCAUAG | 0.72 |
|
|

|
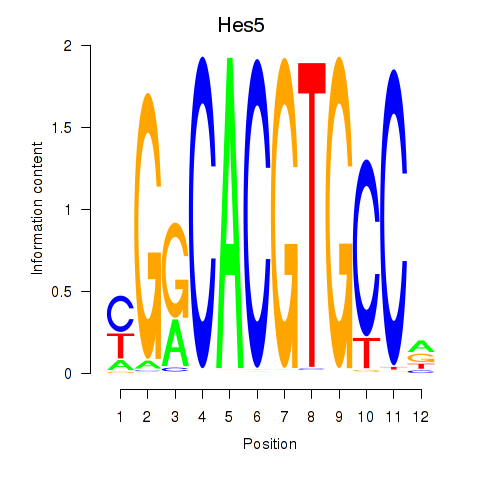
| Hes5_Hes7 | 0.71 |
|
|
|
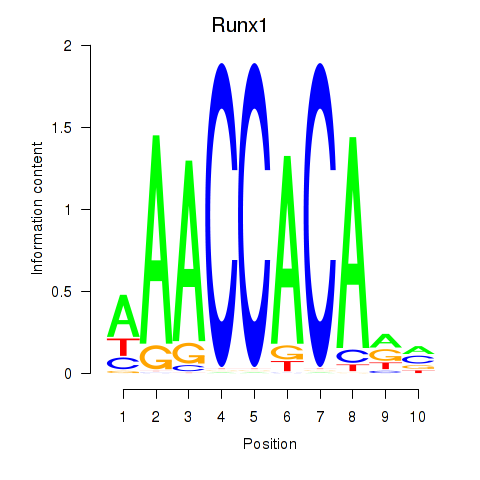
| Runx1 | 0.71 |
|
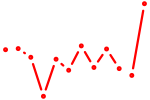
|
|
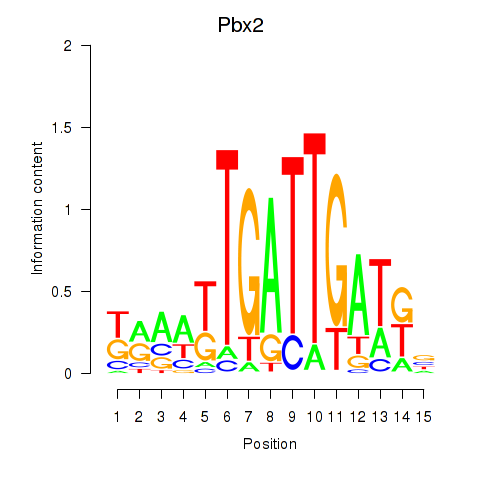
| Pbx2 | 0.71 |
|
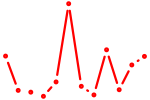
|
|
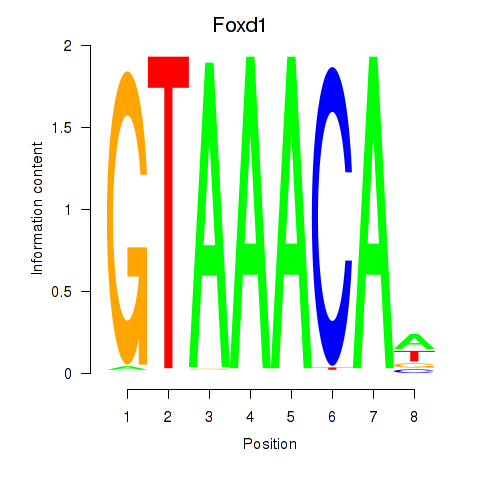
| Foxd1 | 0.71 |
|
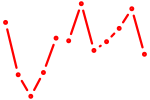
|
|
| GGAAUGU | 0.70 |
|
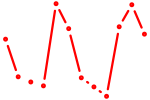
|

|
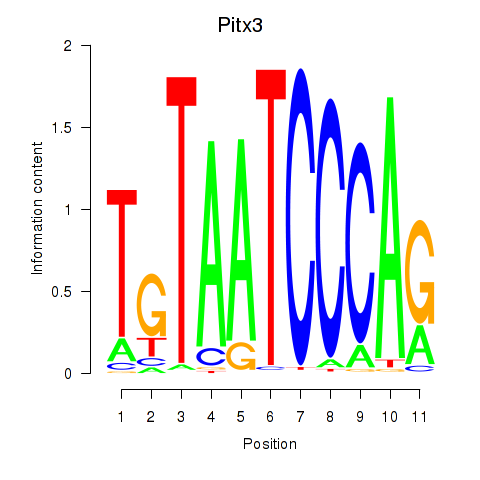
| Pitx3 | 0.70 |
|
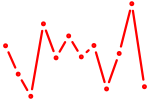
|
|
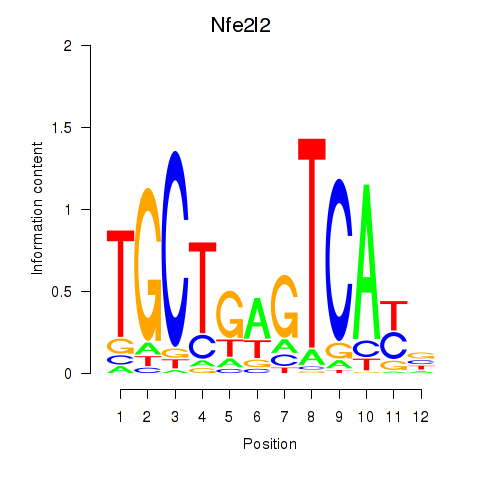
| Nfe2l2 | 0.70 |
|
|
|
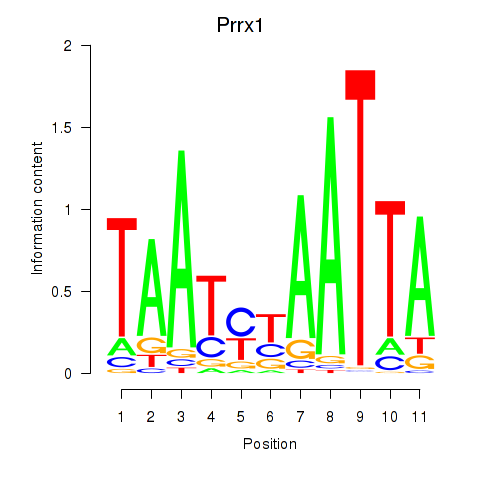
| Prrx1_Isx_Prrxl1 | 0.70 |
|
|
|
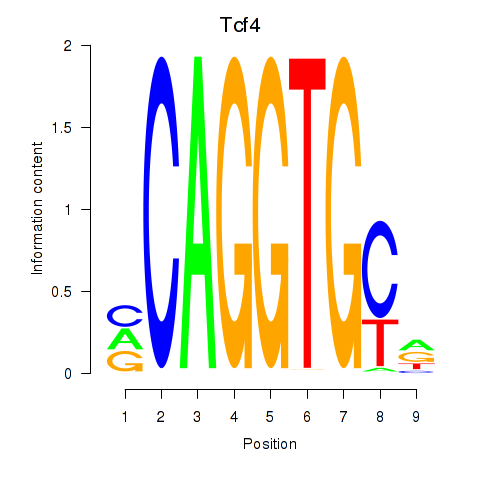
| Tcf4_Mesp1 | 0.70 |
|
|
|
| AUUGCAC | 0.69 |
|

|

|
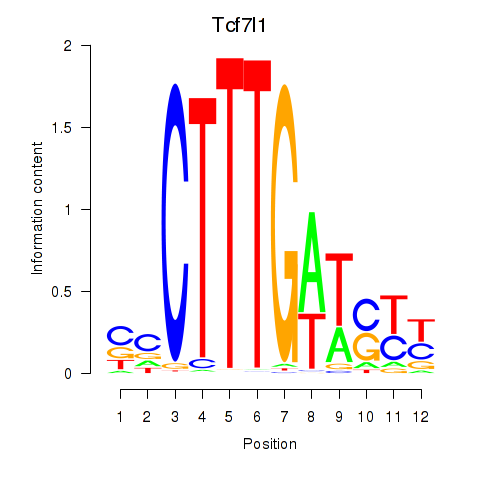
| Tcf7l1 | 0.69 |
|
|
|
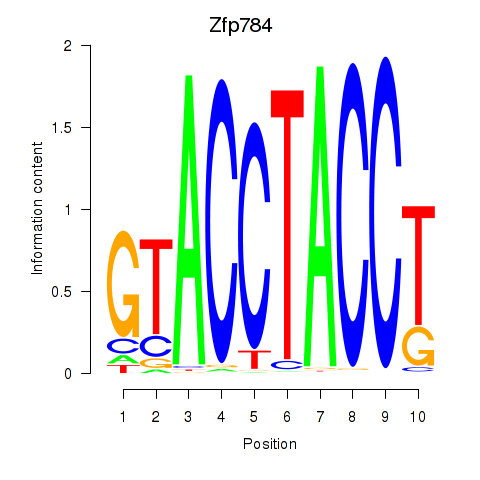
| Zfp784 | 0.69 |
|

|
|
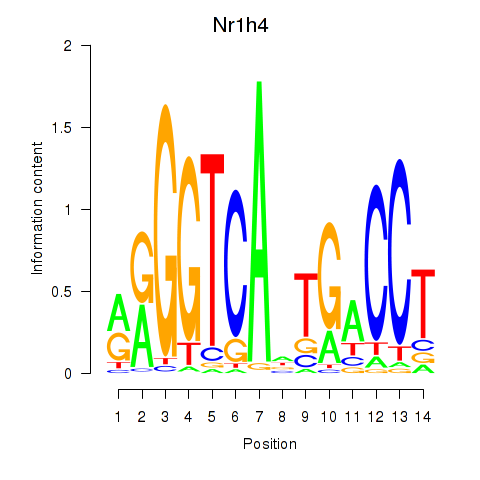
| Nr1h4 | 0.69 |
|
|
|
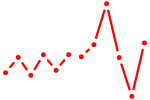
| Tbx21 | 0.68 |
|
|
|
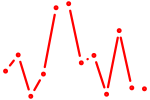
| Nr1i2 | 0.68 |
|
|
|
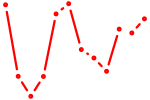
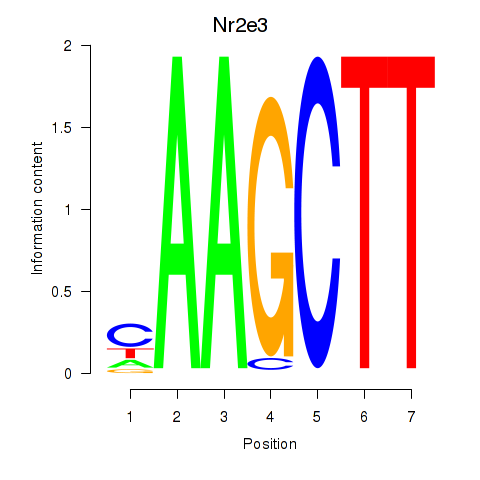
| Nr2e3 | 0.68 |
|
|
|
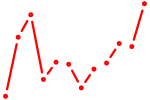
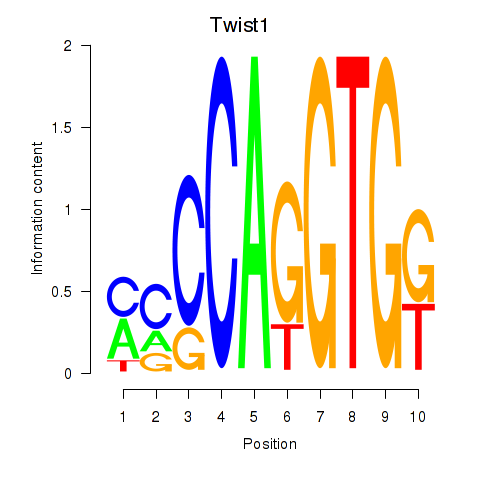
| Twist1 | 0.67 |
|
|
|
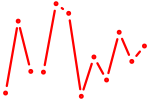
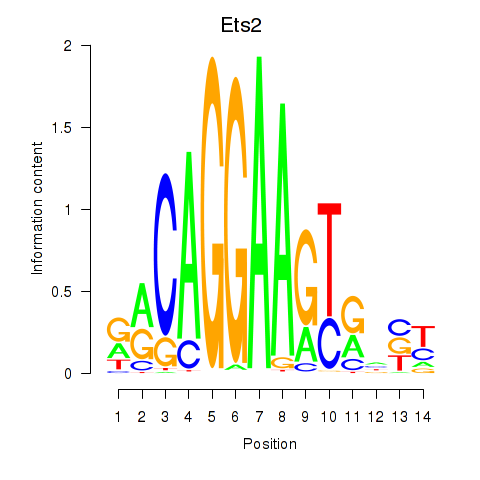
| Ets2 | 0.67 |
|
|
|
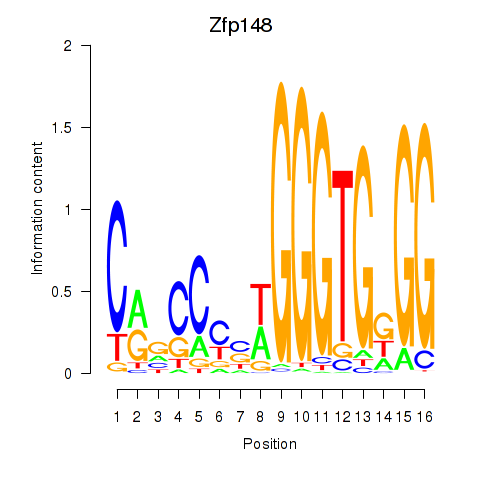
| Zfp148 | 0.67 |
|

|
|
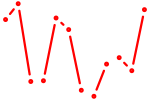
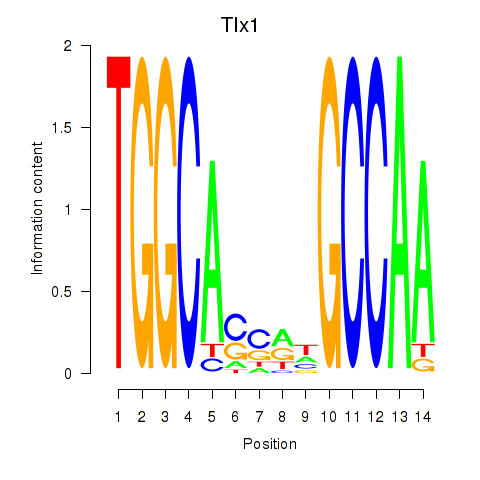
| Tlx1 | 0.67 |
|
|
|
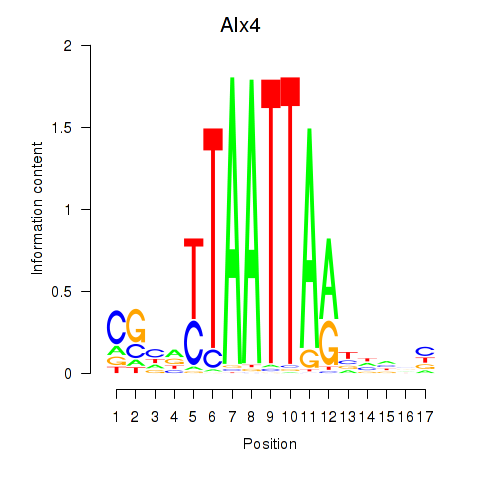
| Alx4 | 0.67 |
|
|
|
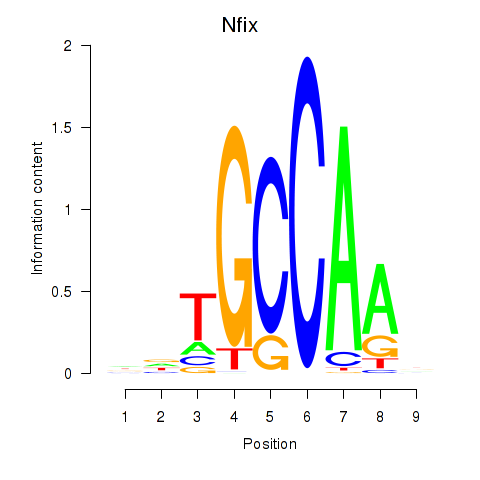
| Nfix | 0.66 |
|
|
|
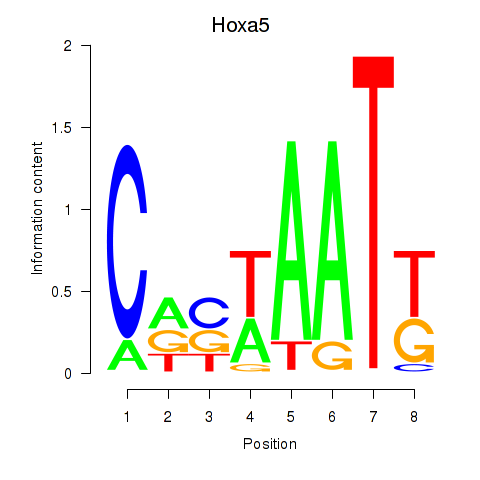
| Hoxa5 | 0.66 |
|
|
|
| AUGGCUU | 0.66 |
|
|

|
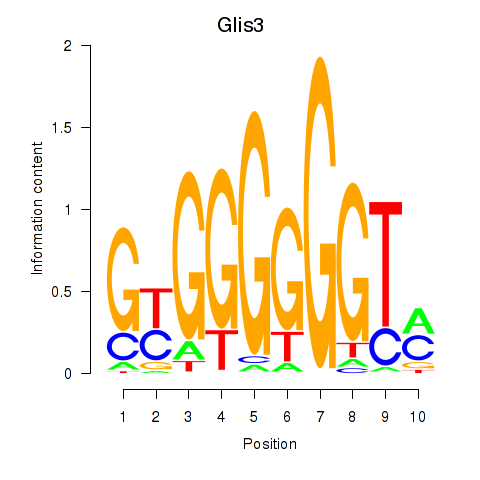
| Glis3 | 0.66 |
|
|
|
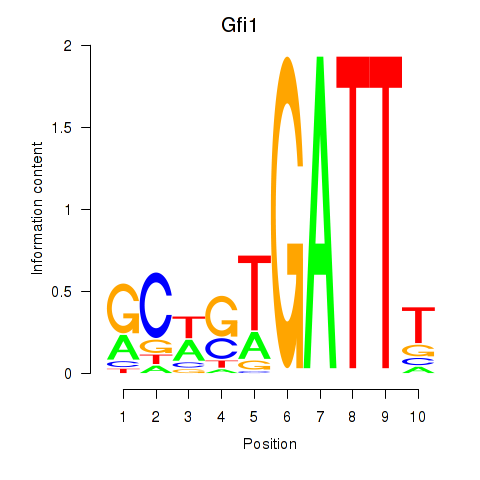
| Gfi1_Gfi1b | 0.66 |
|
|
|
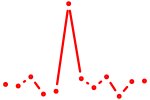
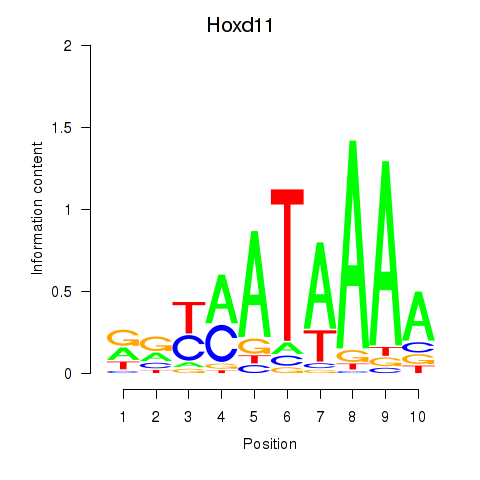
| Hoxd11_Cdx1_Hoxc11 | 0.66 |
|
|
|
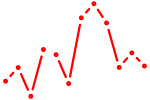
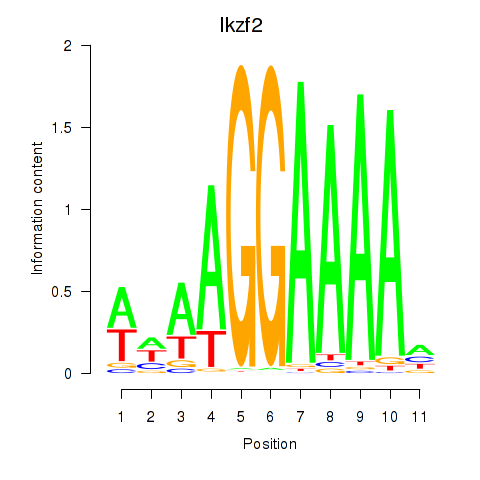
| Ikzf2 | 0.66 |
|
|
|
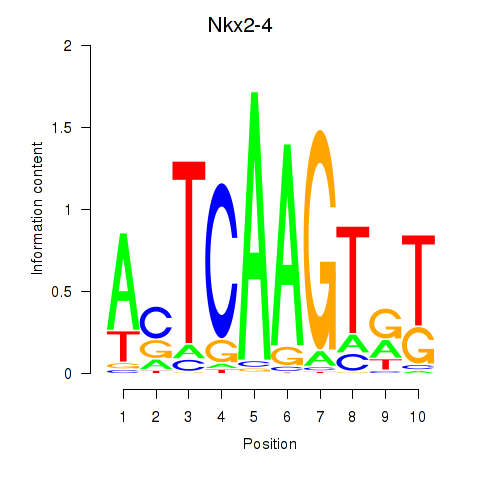
| Nkx2-4 | 0.65 |
|
|
|
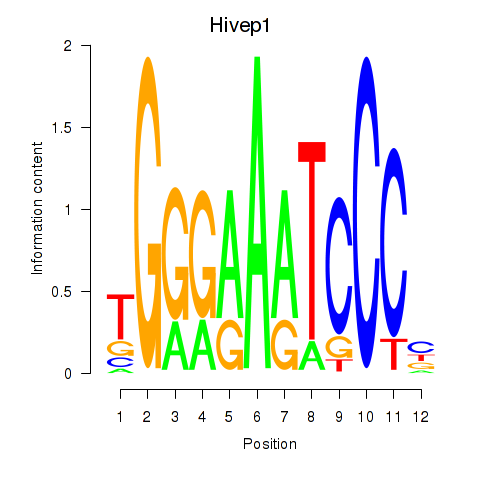
| Hivep1 | 0.65 |
|
|
|
| UCACAGU | 0.65 |
|
|

|
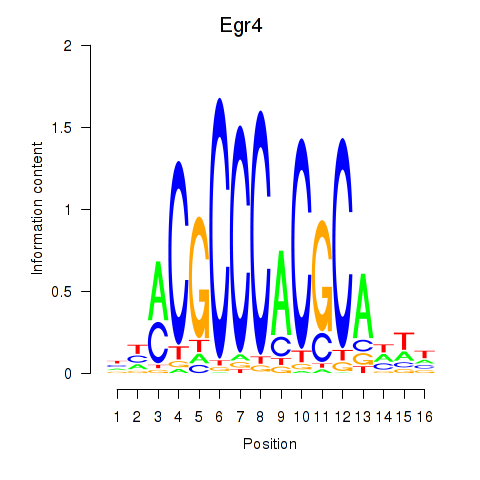
| Egr4 | 0.65 |
|
|
|
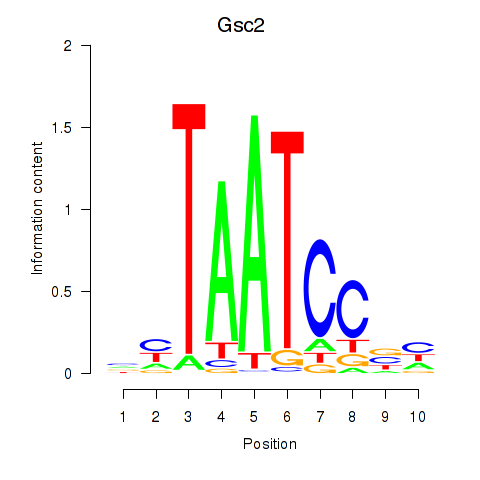
| Gsc2_Dmbx1 | 0.65 |
|
|
|
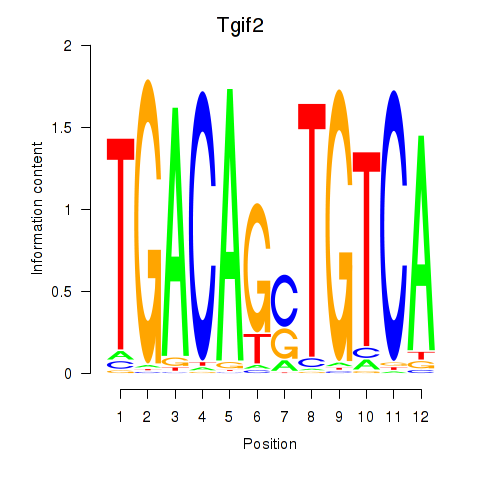
| Tgif2_Tgif2lx1_Tgif2lx2 | 0.65 |
|
|
|
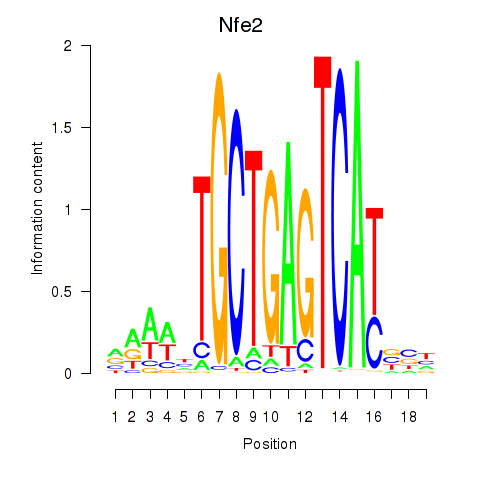
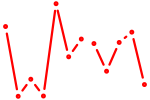
| Nfe2_Bach1_Mafk | 0.65 |
|
|
|
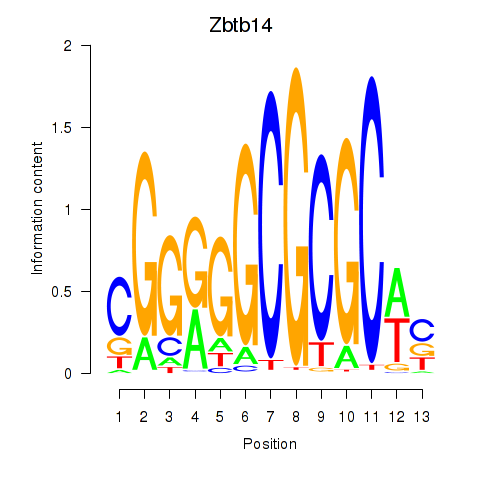
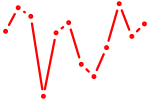
| Zbtb14 | 0.64 |
|
|
|
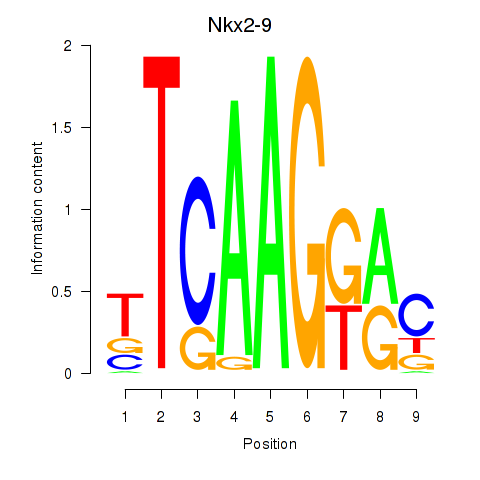
| Nkx2-9 | 0.64 |
|
|
|
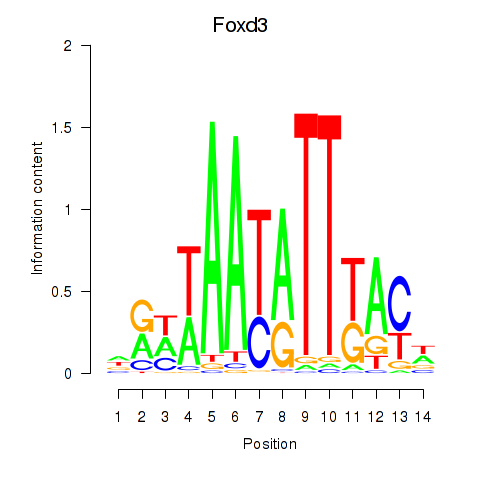
| Foxd3 | 0.64 |
|

|
|
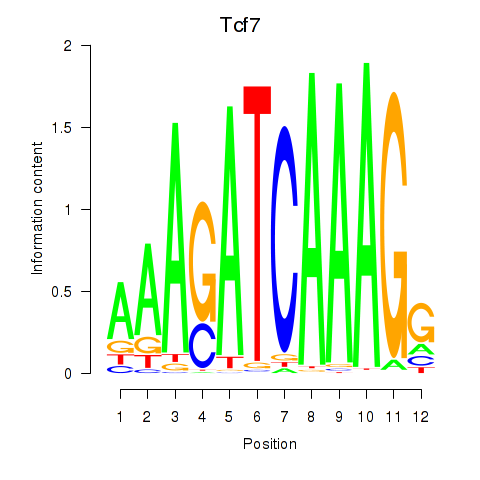
| Tcf7_Tcf7l2 | 0.64 |
|

|
|
| AGCAGCG | 0.63 |
|

|

|
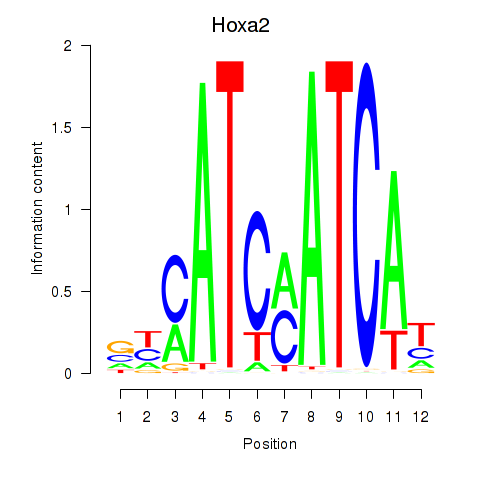
| Hoxa2 | 0.63 |
|

|
|
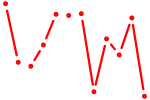
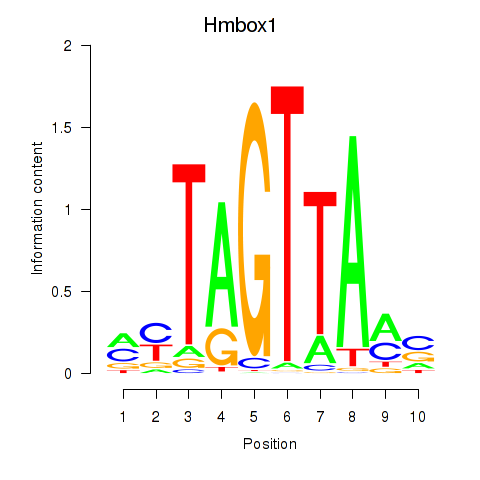
| Hmbox1 | 0.63 |
|
|
|
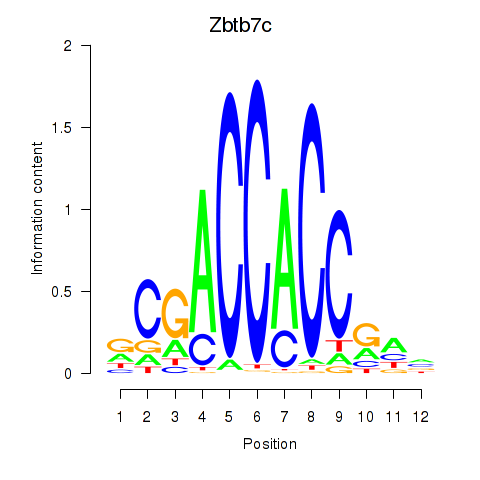
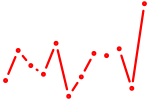
| Zbtb7c | 0.63 |
|

|
|
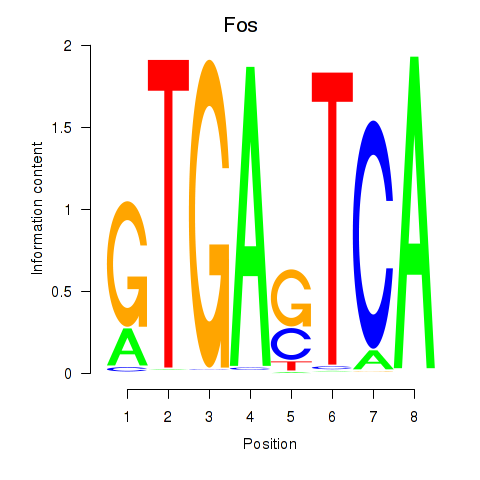
| Fos | 0.63 |
|
|
|
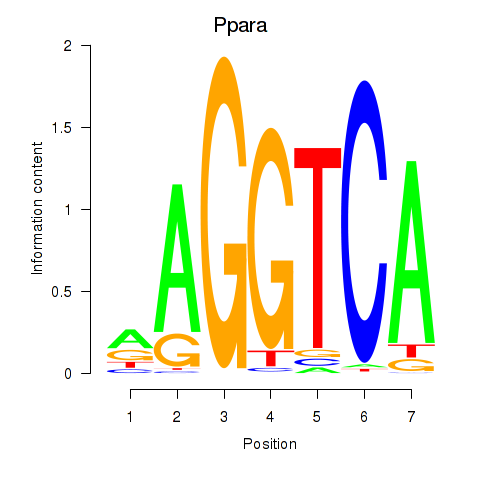
| Ppara | 0.63 |
|

|
|
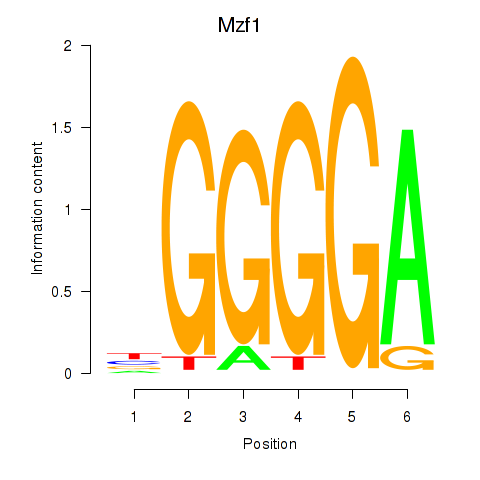
| Mzf1 | 0.63 |
|
|
|
| Arntl_Tfe3_Mlx_Mitf_Mlxipl_Tfec | 0.63 |
|

|
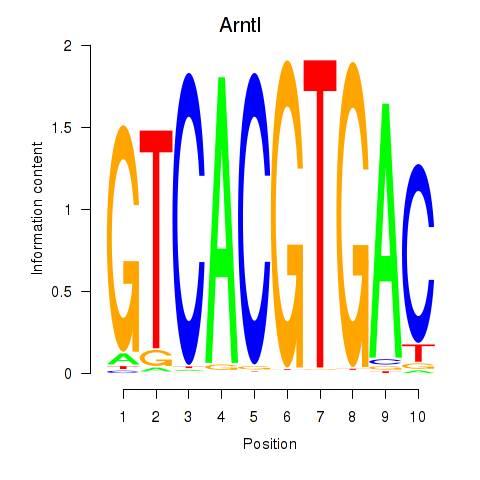
|
| Cenpb | 0.63 |
|

|
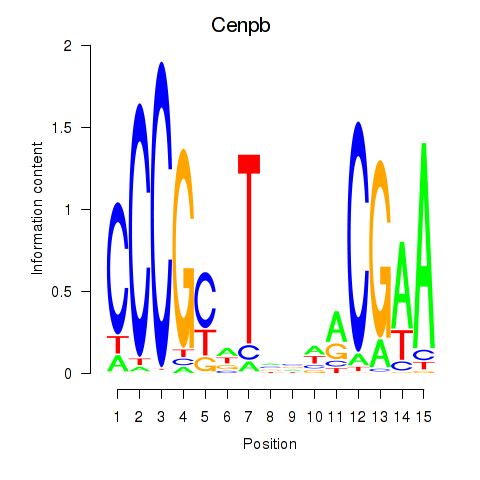
|
| Pou3f2 | 0.62 |
|
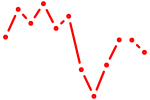
|
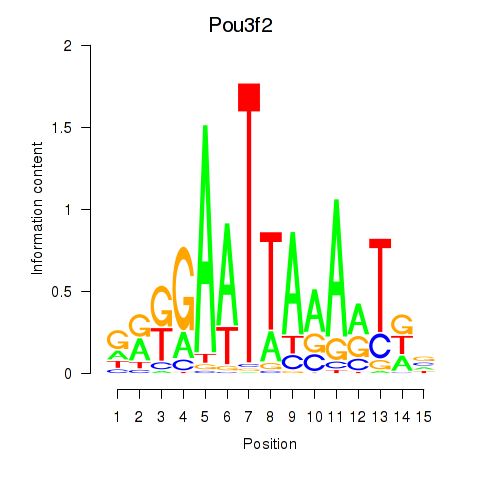
|
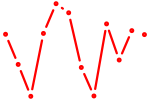
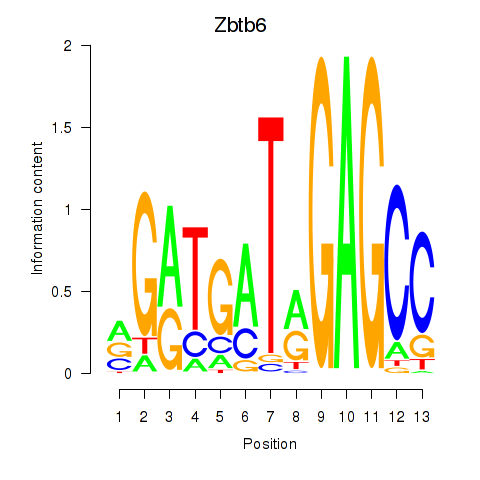
| Zbtb6 | 0.62 |
|
|
|
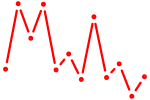
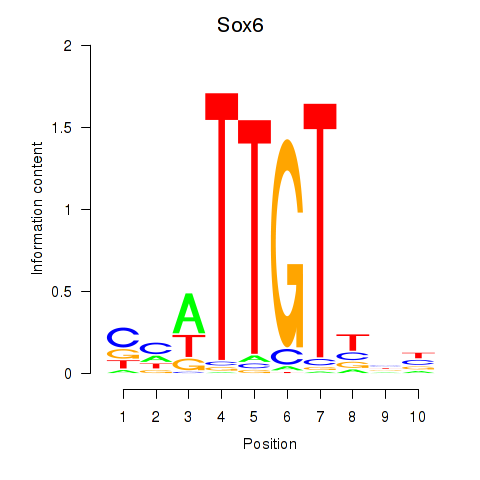
| Sox6_Sox9 | 0.62 |
|
|
|
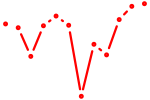
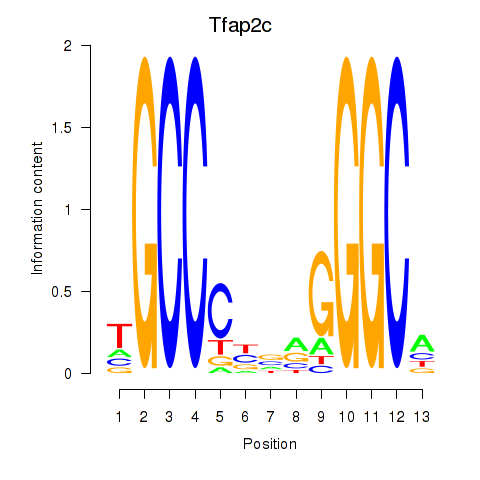
| Tfap2c | 0.62 |
|
|
|
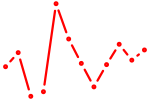
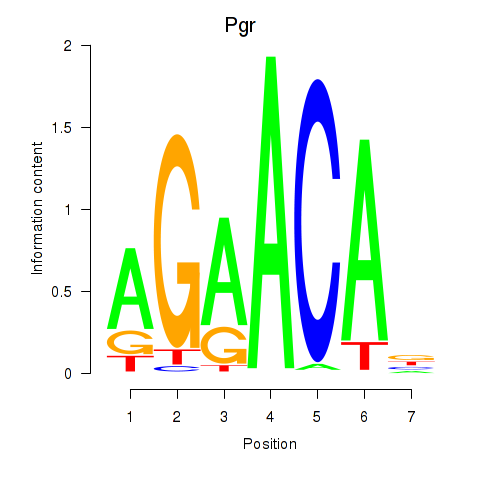
| Pgr_Nr3c1 | 0.62 |
|
|
|
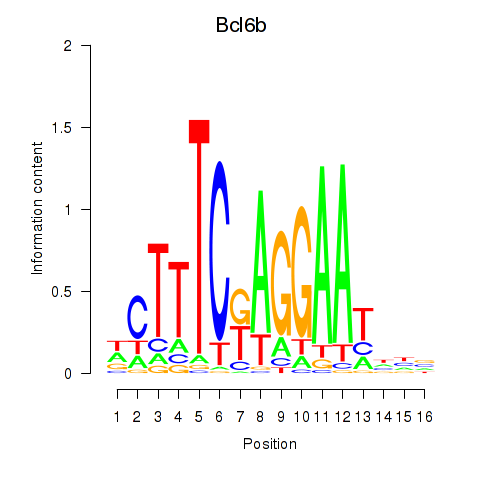
| Bcl6b | 0.62 |
|

|
|
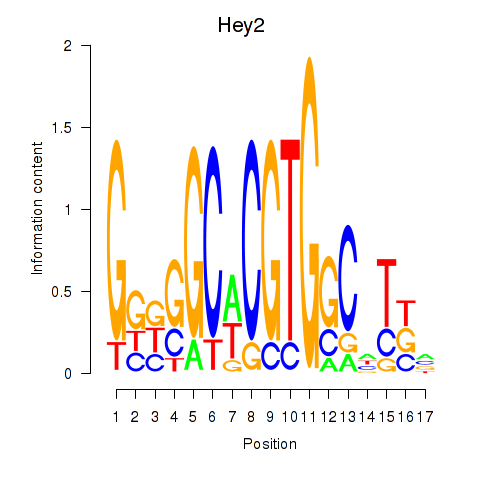
| Hey2 | 0.62 |
|
|
|
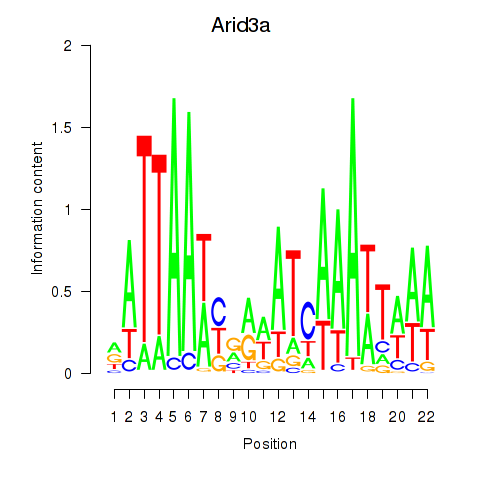
| Arid3a | 0.62 |
|
|
|
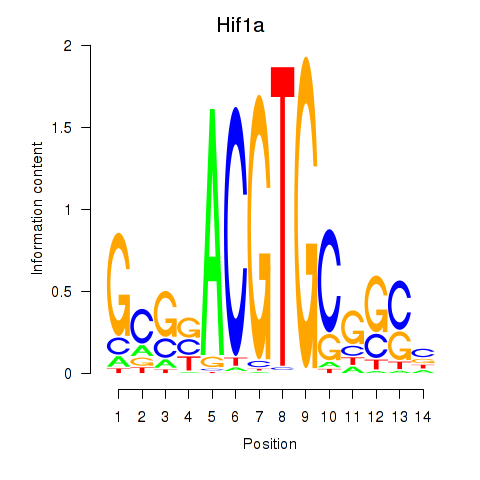
| Hif1a | 0.62 |
|
|
|
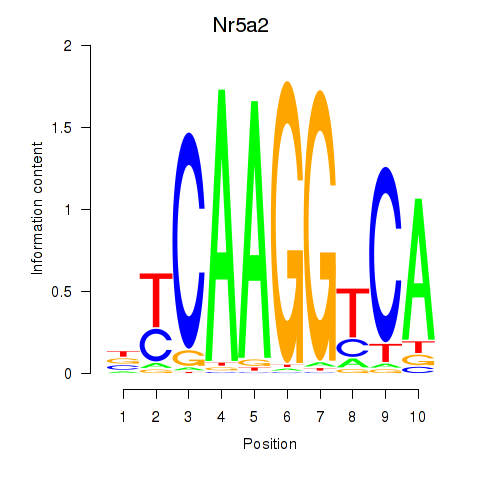
| Nr5a2 | 0.62 |
|

|
|
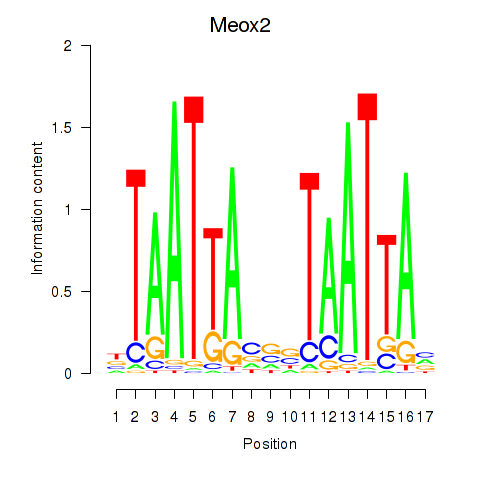
| Meox2 | 0.61 |
|
|
|
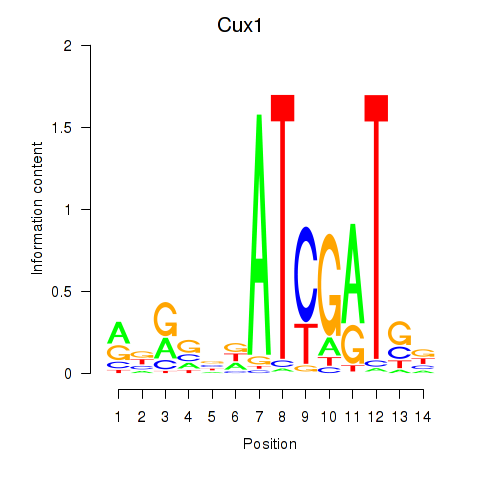
| Cux1 | 0.61 |
|
|
|
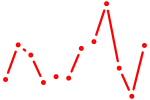
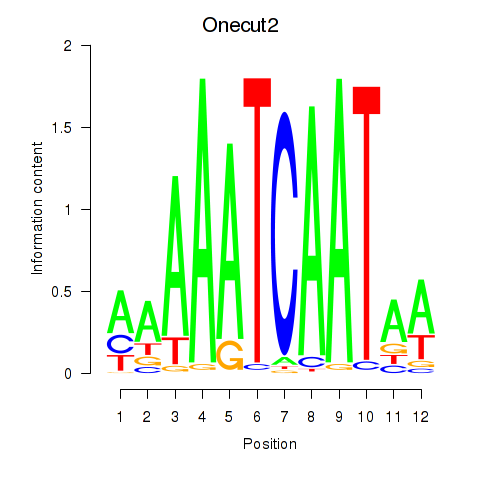
| Onecut2_Onecut3 | 0.61 |
|
|
|
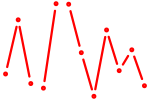
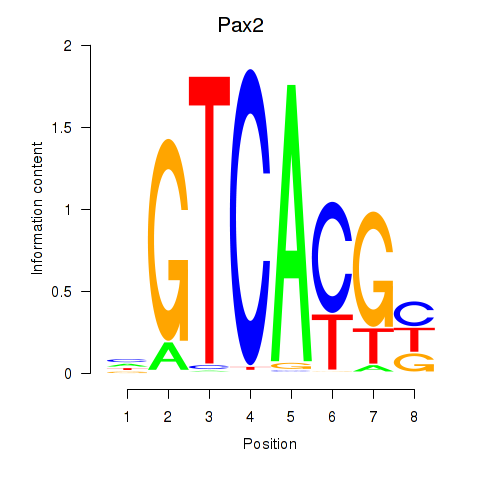
| Pax2 | 0.61 |
|
|
|
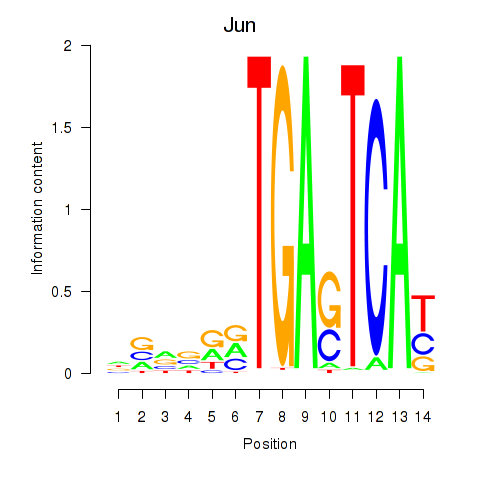
| Jun | 0.61 |
|
|
|
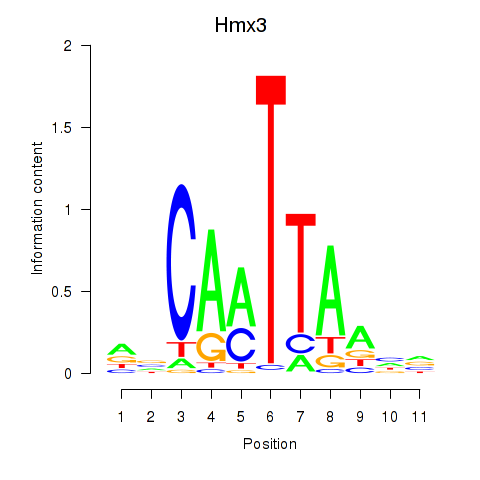
| Hmx3 | 0.61 |
|
|
|
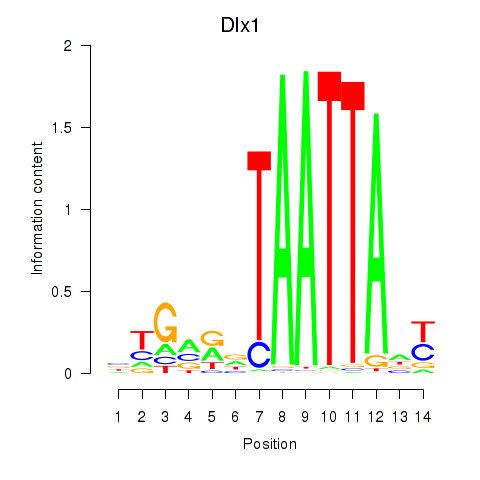
| Dlx1 | 0.61 |
|

|
|
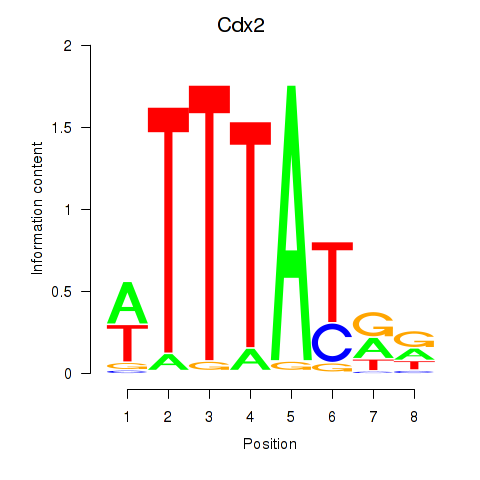
| Cdx2 | 0.61 |
|
|
|
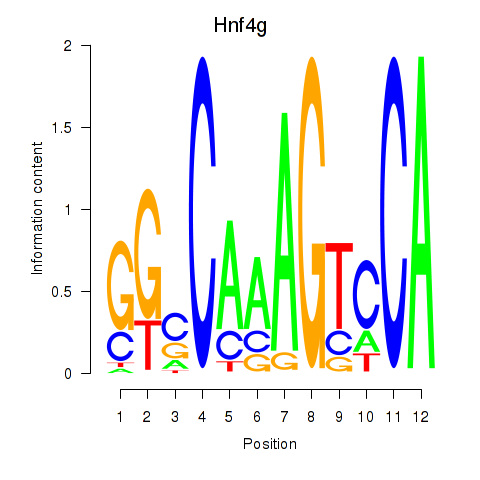
| Hnf4g | 0.60 |
|
|
|
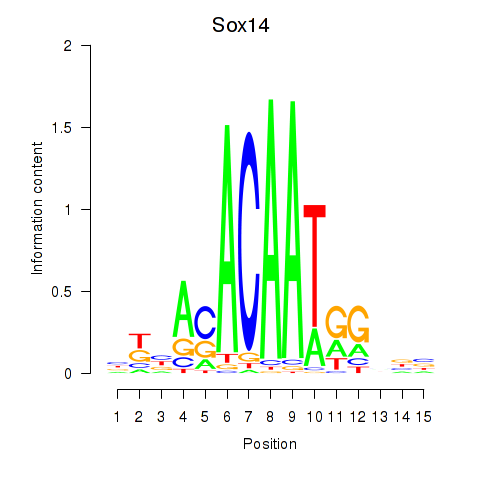
| Sox14 | 0.60 |
|
|
|
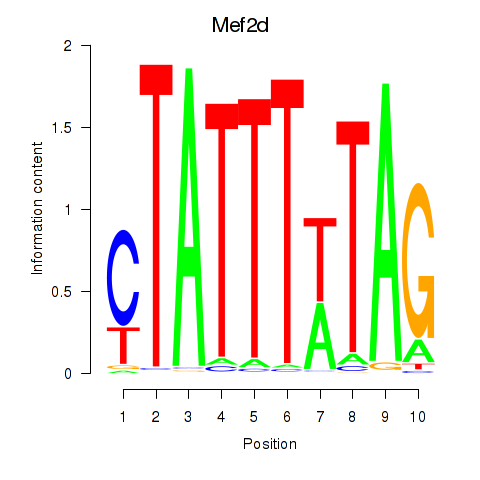
| Mef2d_Mef2a | 0.60 |
|
|
|
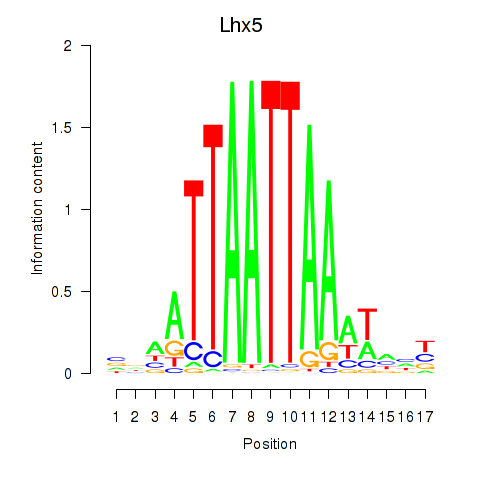
| Lhx5_Lmx1b_Lhx1 | 0.60 |
|
|
|
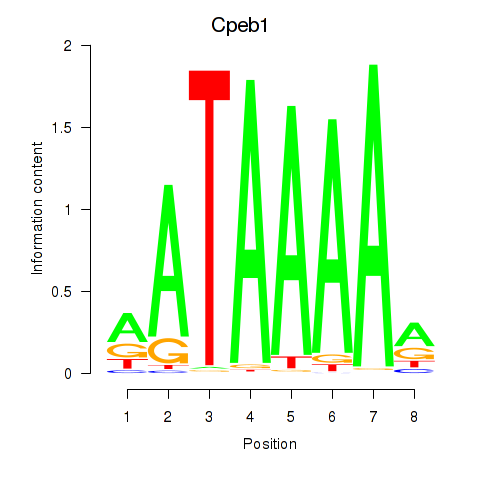
| Cpeb1 | 0.60 |
|

|
|
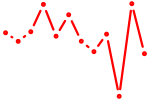
| Aire | 0.59 |
|
|
|
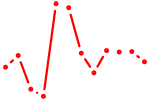
| ACAGUAC | 0.59 |
|
|

|
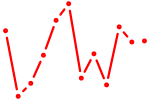
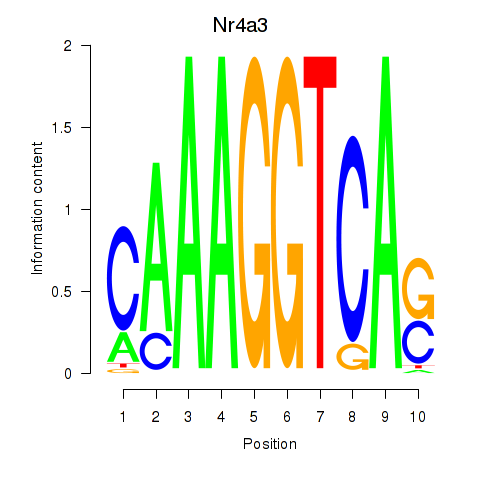
| Nr4a3 | 0.59 |
|
|
|
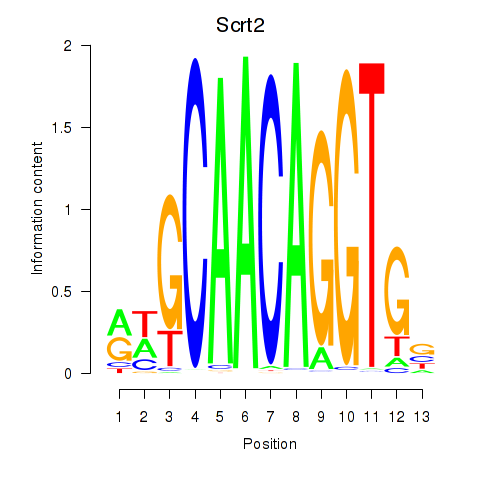
| Scrt2 | 0.59 |
|
|
|
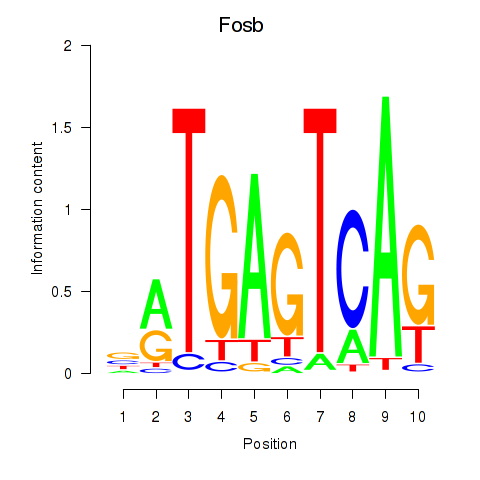
| Fosb | 0.59 |
|

|
|
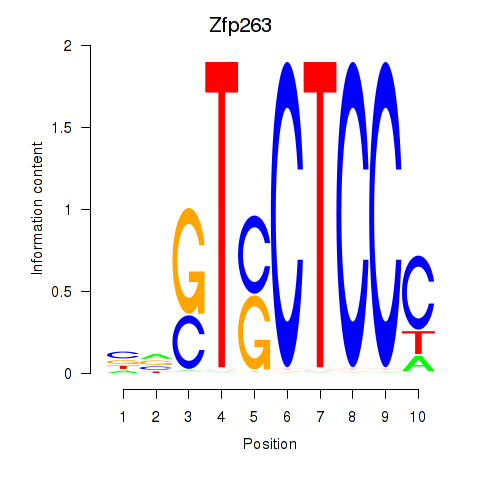
| Zfp263 | 0.59 |
|
|
|
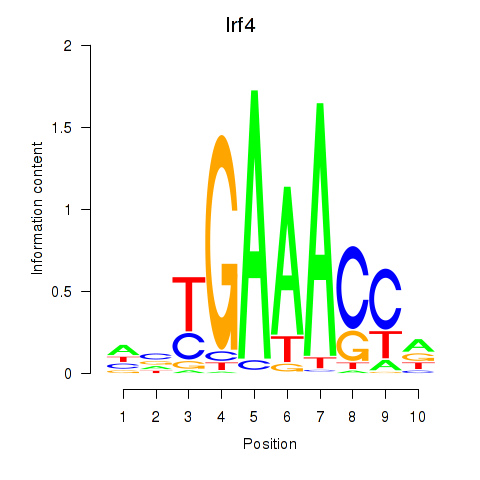
| Irf4 | 0.59 |
|
|
|
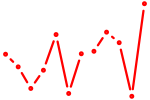
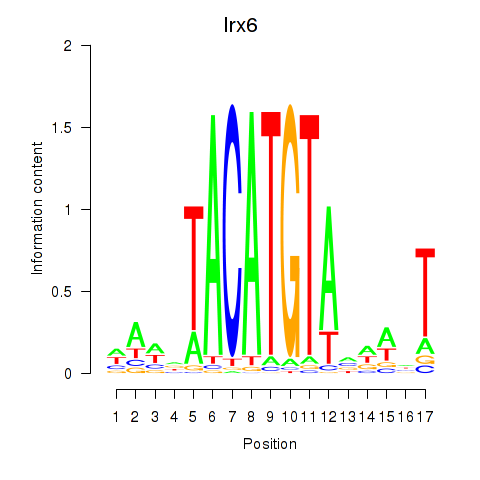
| Irx6_Irx2_Irx3 | 0.59 |
|
|
|
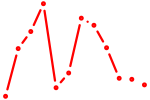
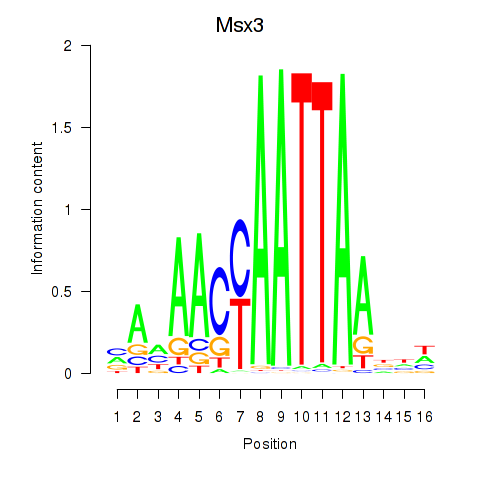
| Msx3 | 0.59 |
|
|
|
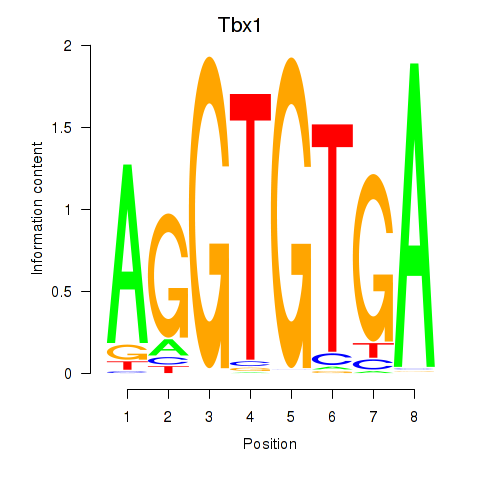
| Tbx1_Eomes | 0.58 |
|
|
|
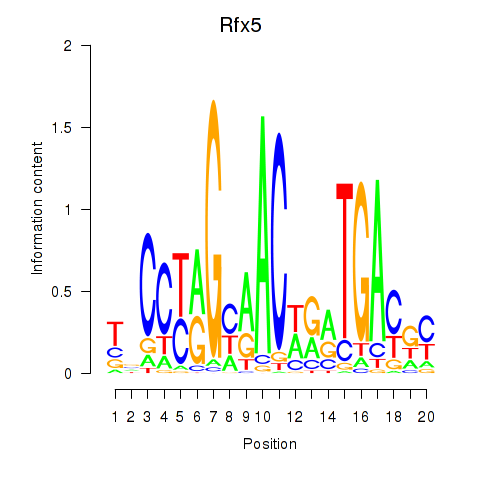
| Rfx5 | 0.58 |
|
|
|
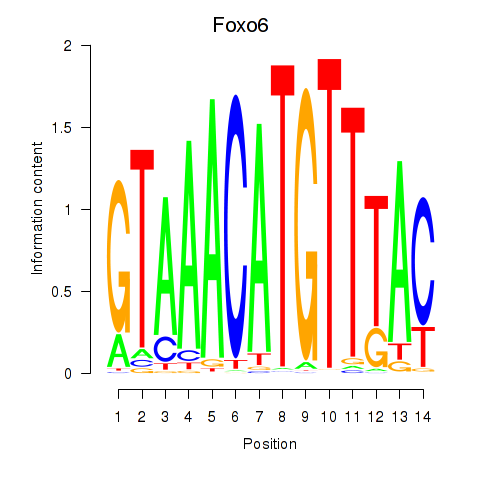
| Foxo6 | 0.58 |
|

|
|
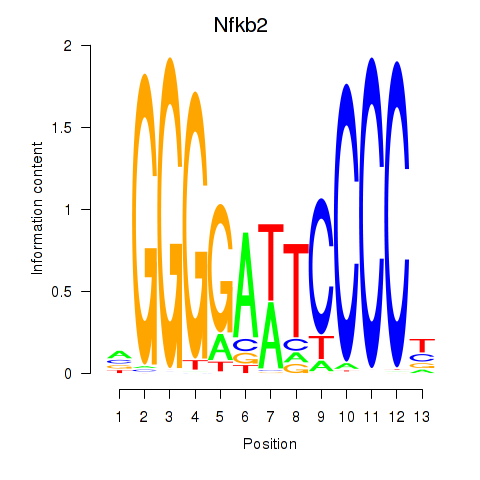
| Nfkb2 | 0.58 |
|
|
|
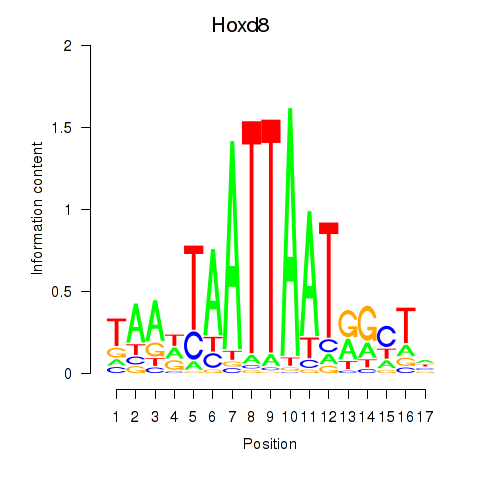
| Hoxd8 | 0.57 |
|
|
|
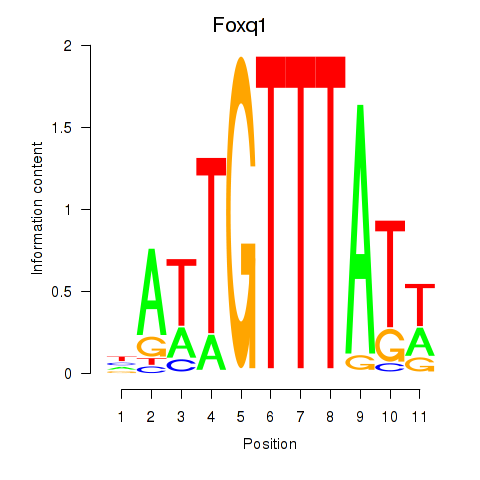
| Foxq1 | 0.57 |
|

|
|
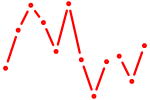
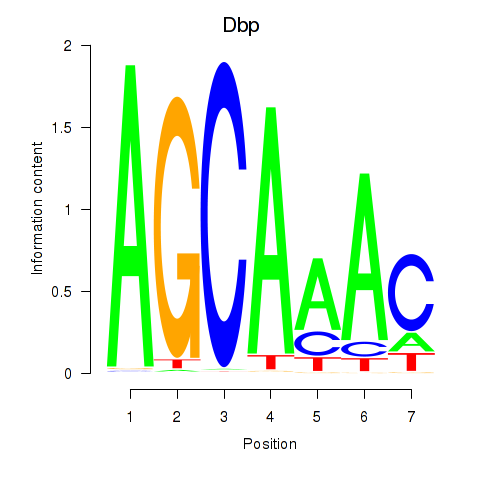
| Dbp | 0.57 |
|
|
|
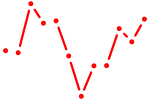
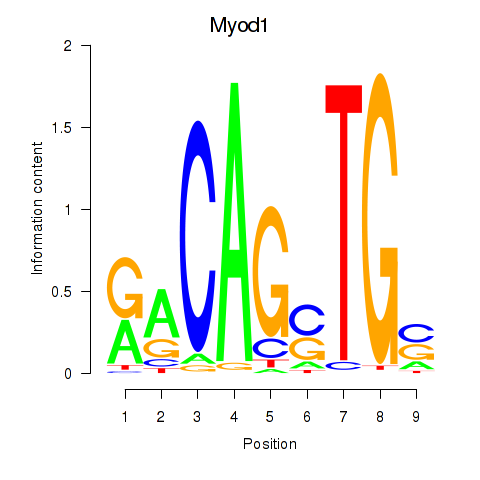
| Myod1 | 0.57 |
|
|
|
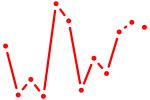
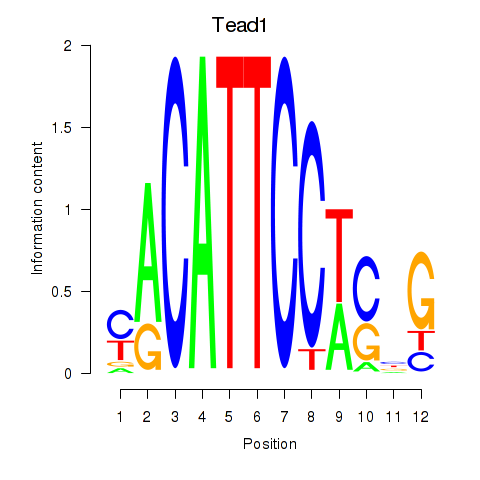
| Tead1 | 0.56 |
|
|
|
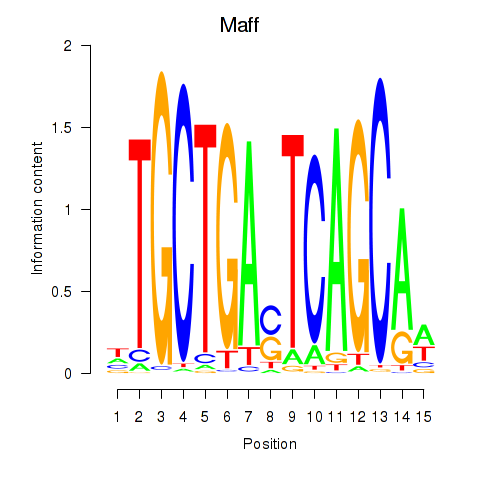
| Maff | 0.56 |
|
|
|
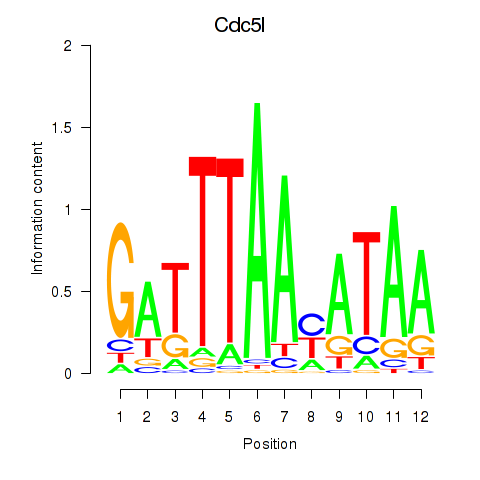
| Cdc5l | 0.56 |
|
|
|
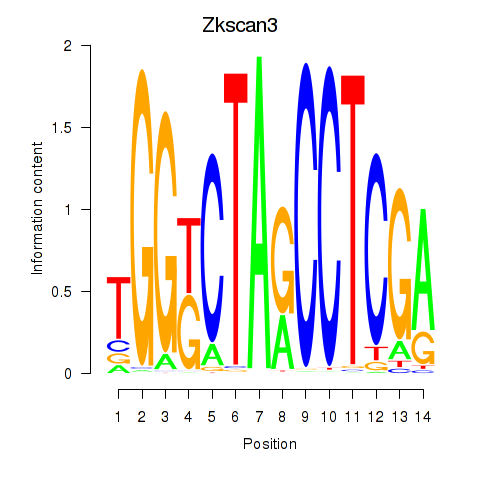
| Zkscan3 | 0.56 |
|
|
|
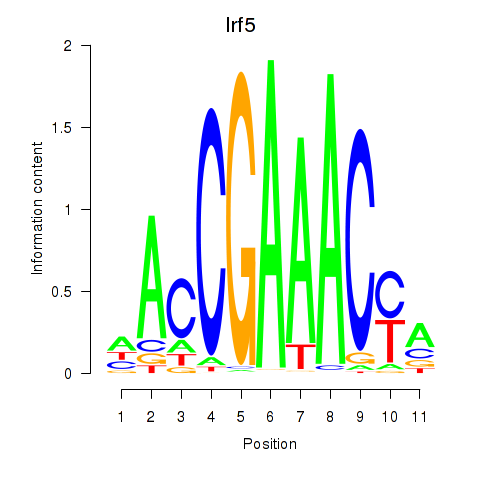
| Irf5_Irf6 | 0.56 |
|
|
|
| GUCAGUU | 0.55 |
|

|

|
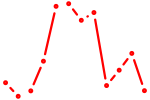
| AGCAGCA | 0.55 |
|
|

|
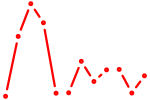
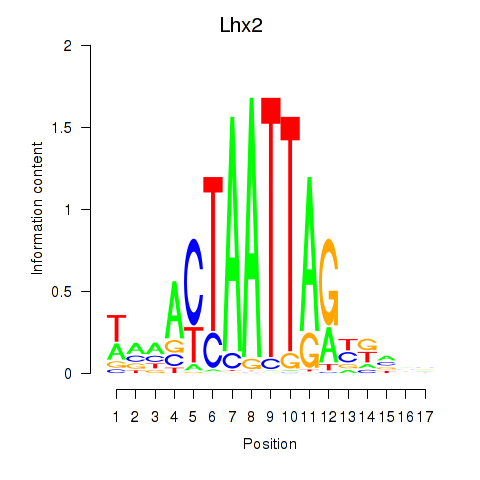
| Lhx2_Hoxc5 | 0.55 |
|
|
|
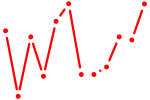
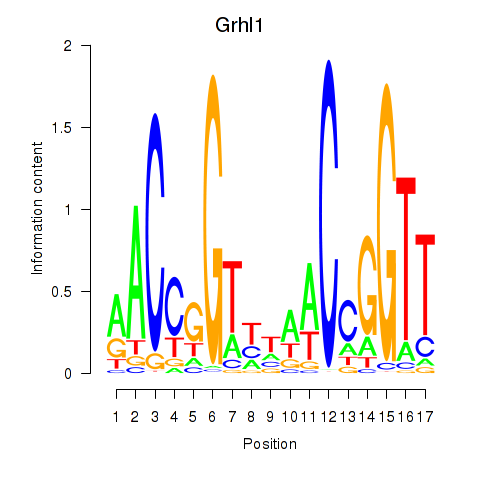
| Grhl1 | 0.55 |
|
|
|
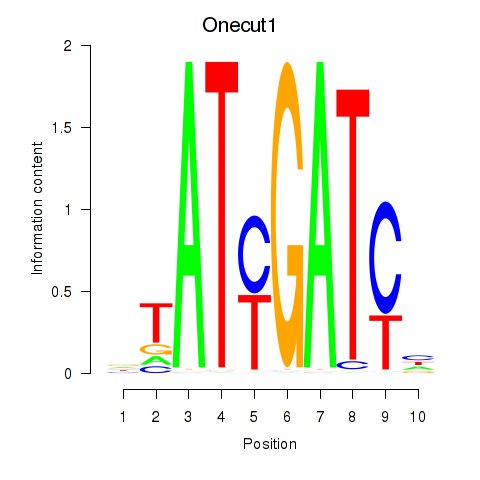
| Onecut1_Cux2 | 0.54 |
|
|
|
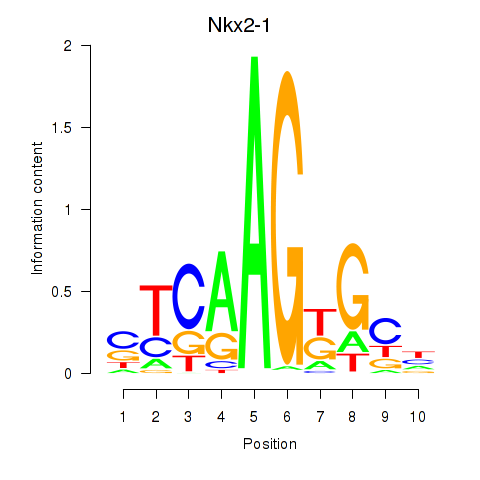
| Nkx2-1 | 0.54 |
|
|
|
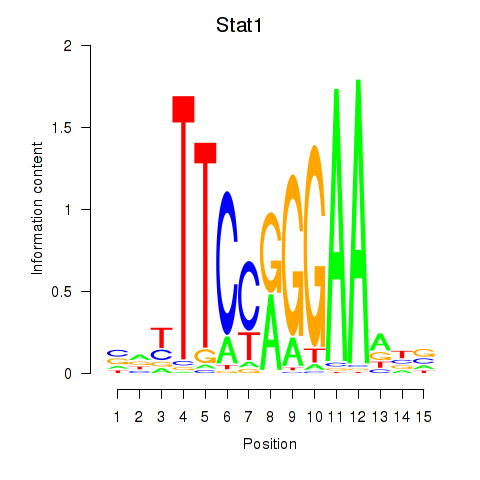
| Stat1 | 0.54 |
|

|
|
| AAGUGCU | 0.54 |
|
|

|
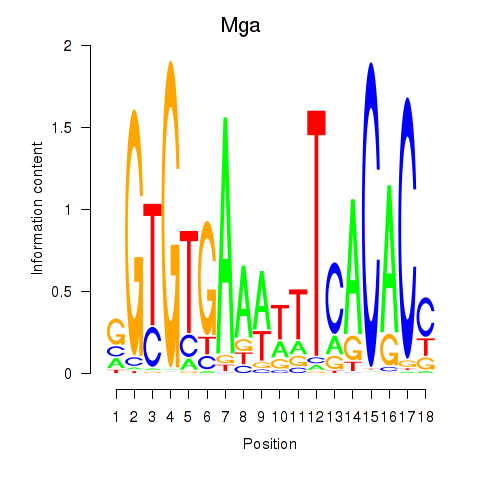
| Mga | 0.54 |
|
|
|
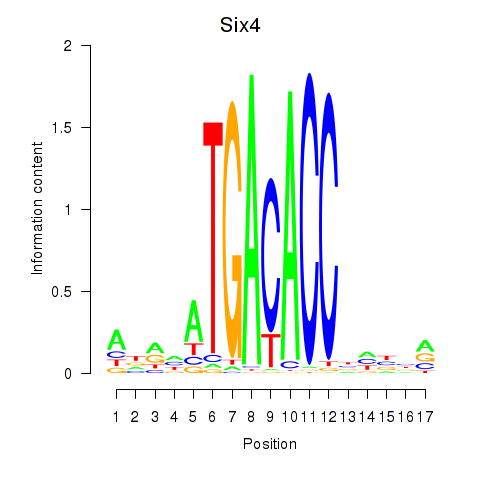
| Six4 | 0.54 |
|
|
|
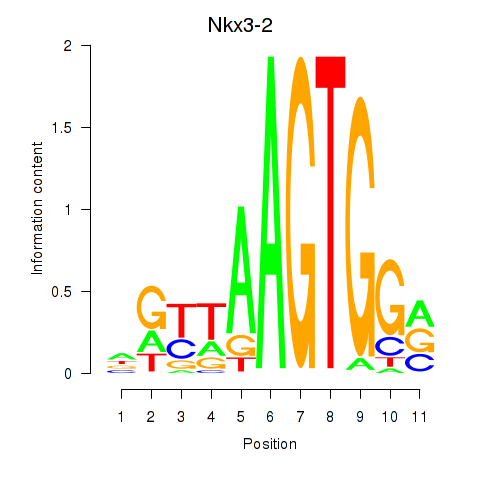
| Nkx3-2 | 0.54 |
|
|
|
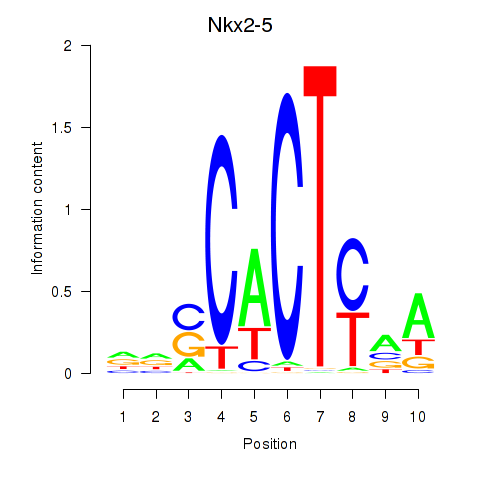
| Nkx2-5 | 0.54 |
|
|
|
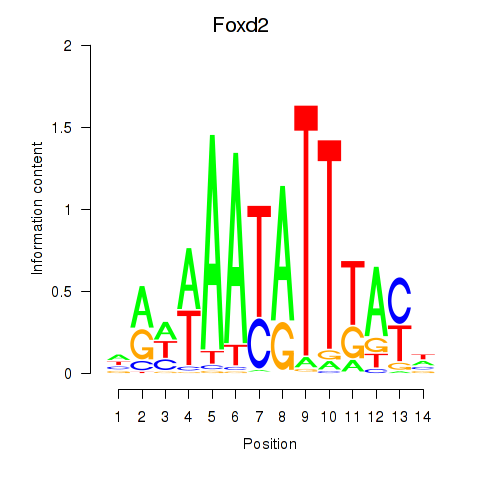
| Foxd2 | 0.53 |
|
|
|
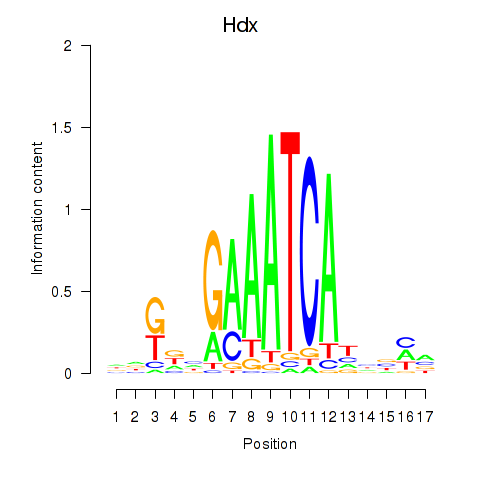
| Hdx | 0.53 |
|

|
|
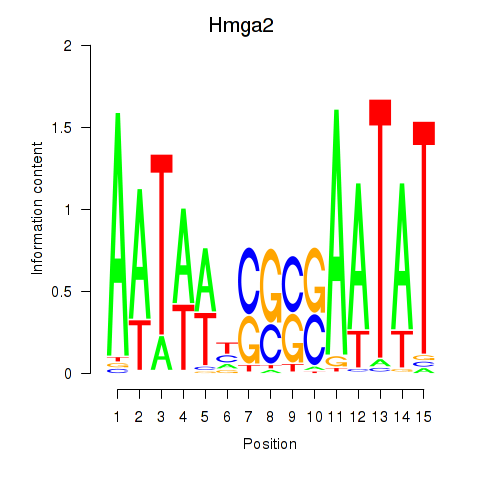
| Hmga2 | 0.53 |
|
|
|
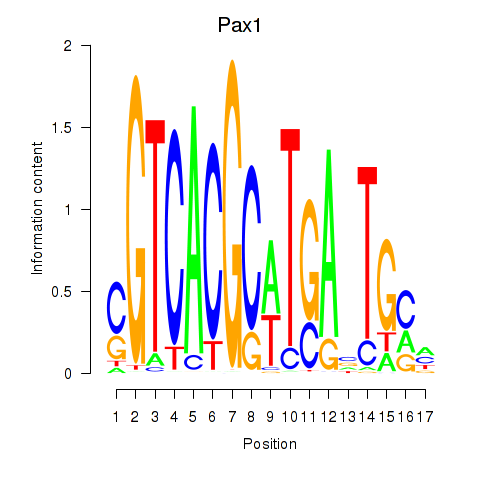
| Pax1_Pax9 | 0.53 |
|
|
|
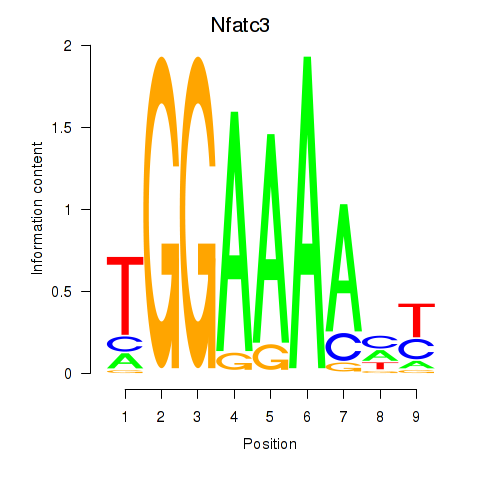
| Nfatc3 | 0.53 |
|
|
|
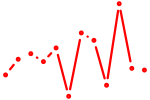
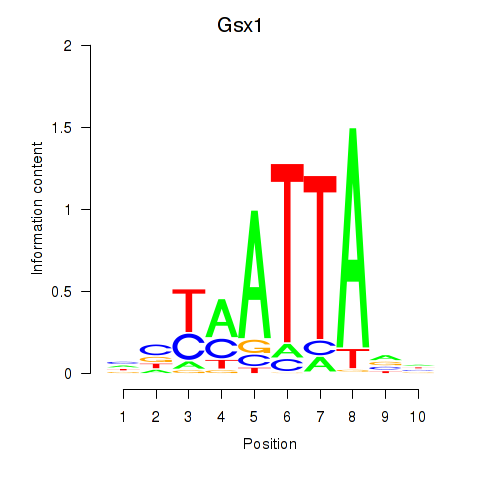
| Gsx1_Alx1_Mixl1_Lbx2 | 0.53 |
|
|
|
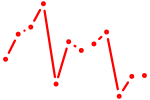
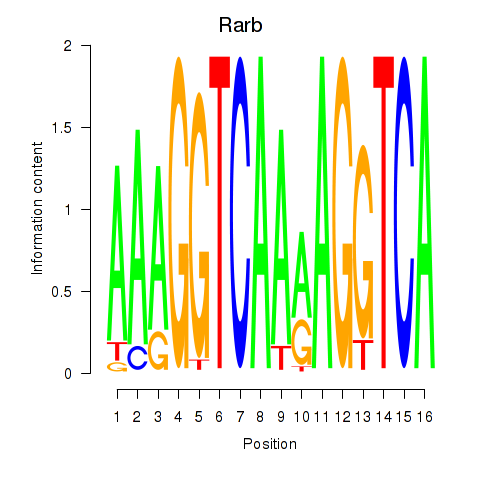
| Rarb | 0.53 |
|
|
|
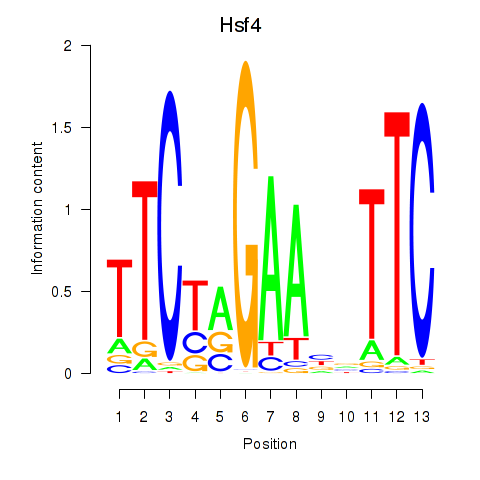
| Hsf4 | 0.53 |
|
|
|
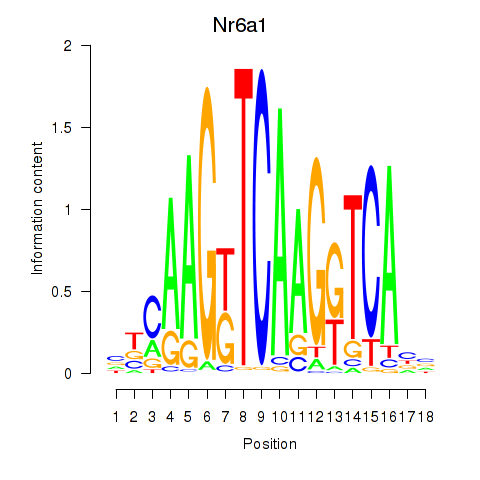
| Nr6a1 | 0.53 |
|
|
|
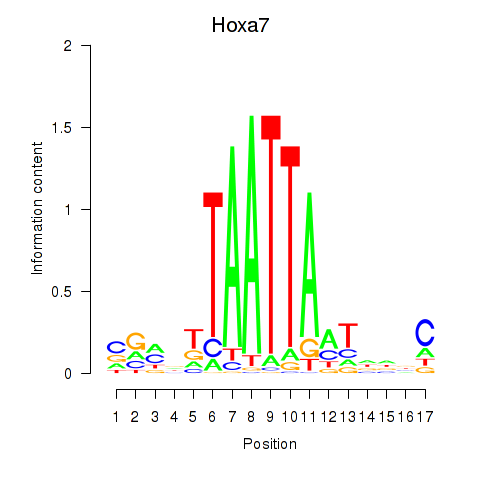
| Hoxa7_Hoxc8 | 0.53 |
|
|
|
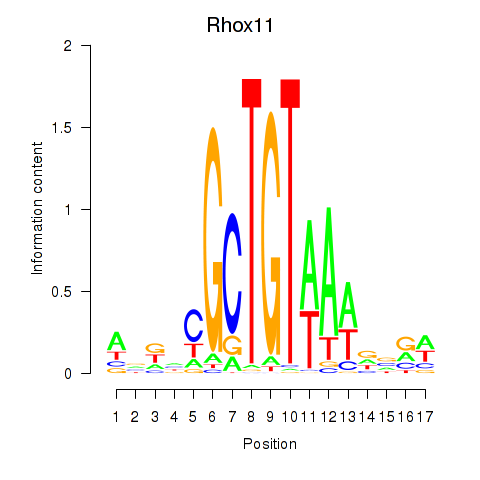
| Rhox11 | 0.53 |
|
|
|
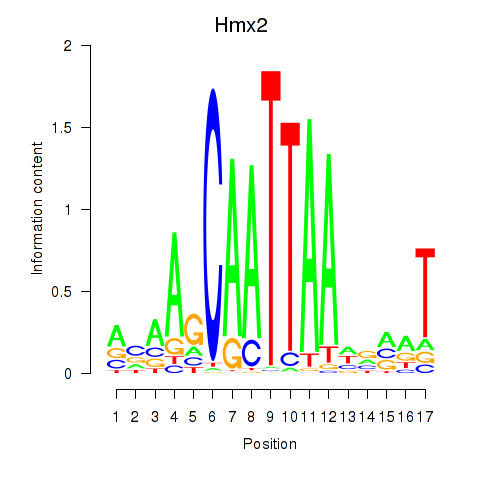
| Hmx2 | 0.52 |
|

|
|
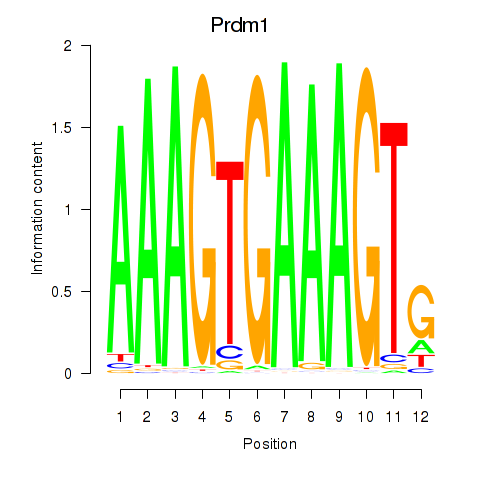
| Prdm1 | 0.52 |
|
|
|
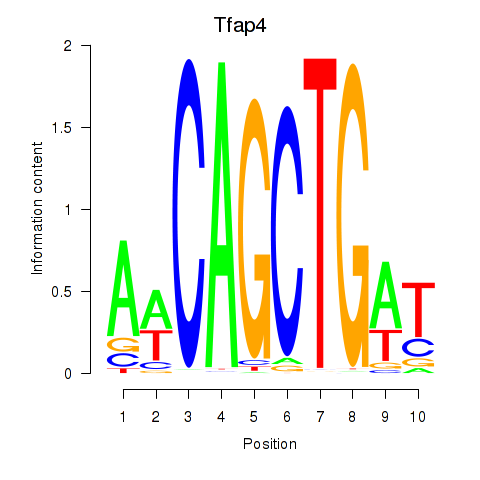
| Tfap4 | 0.52 |
|
|
|
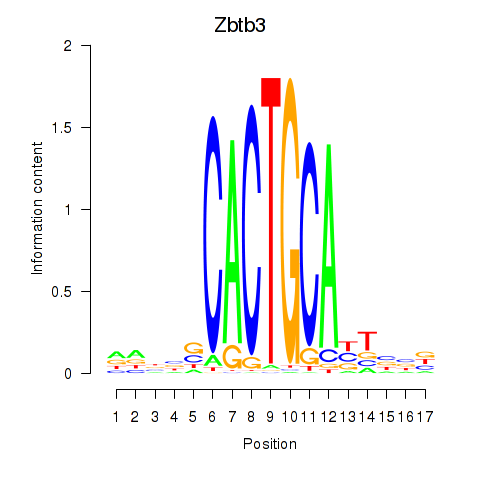
| Zbtb3 | 0.52 |
|

|
|
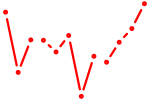
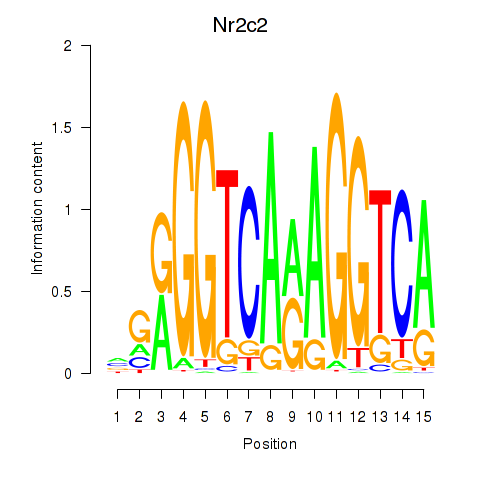
| Nr2c2 | 0.52 |
|
|
|
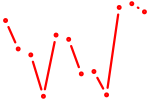
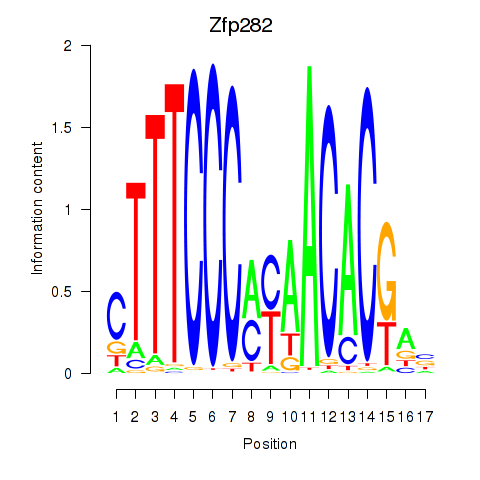
| Zfp282 | 0.52 |
|
|
|
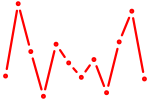
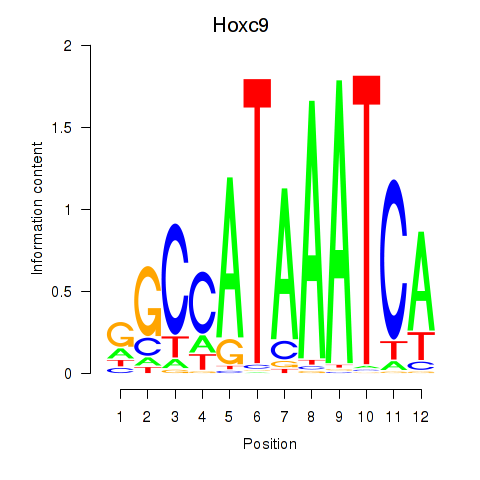
| Hoxc9 | 0.52 |
|
|
|
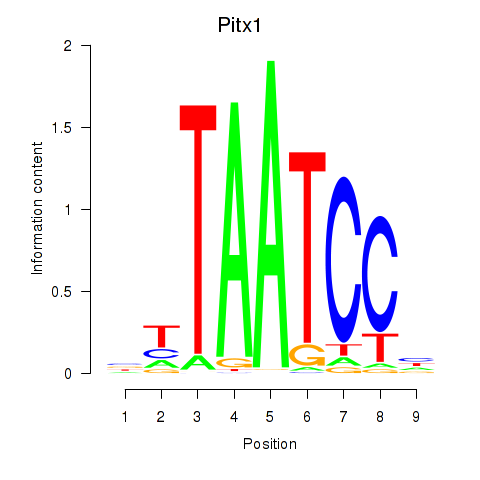
| Pitx1 | 0.52 |
|
|
|
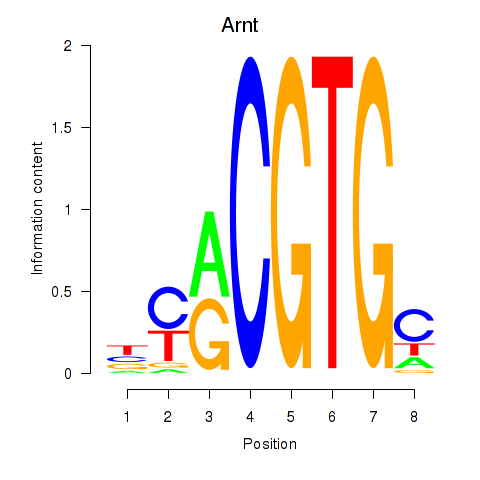
| Arnt | 0.52 |
|
|
|
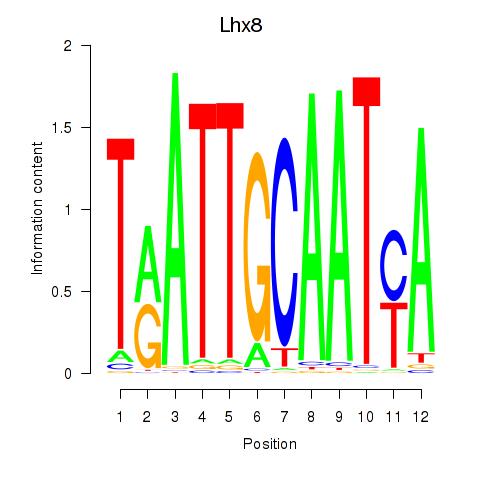
| Lhx8 | 0.51 |
|

|
|
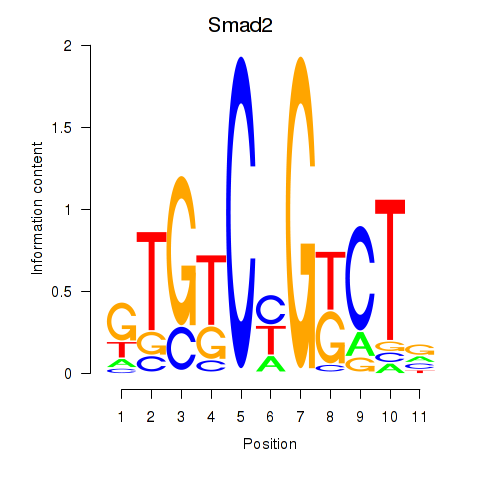
| Smad2 | 0.51 |
|
|
|
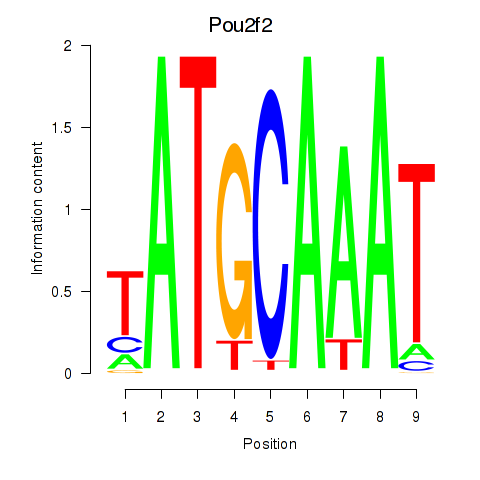
| Pou2f2_Pou3f1 | 0.51 |
|
|
|
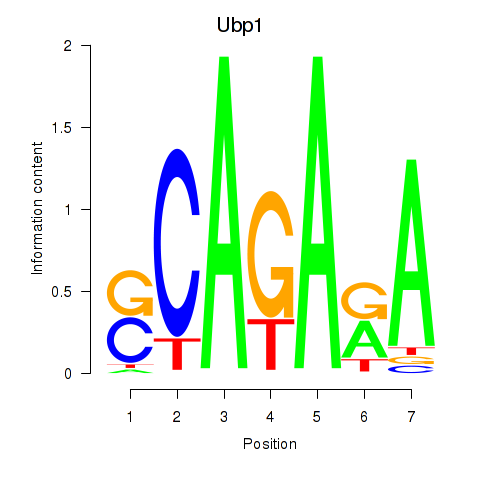
| Ubp1 | 0.51 |
|
|
|
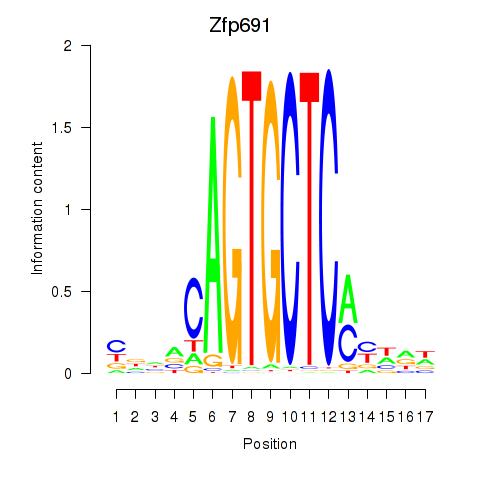
| Zfp691 | 0.50 |
|
|
|
| GCUGGUG | 0.50 |
|
|

|
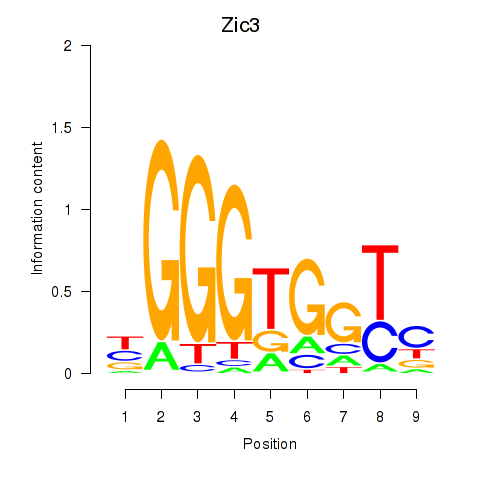
| Zic3 | 0.50 |
|
|
|
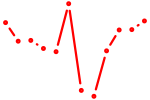
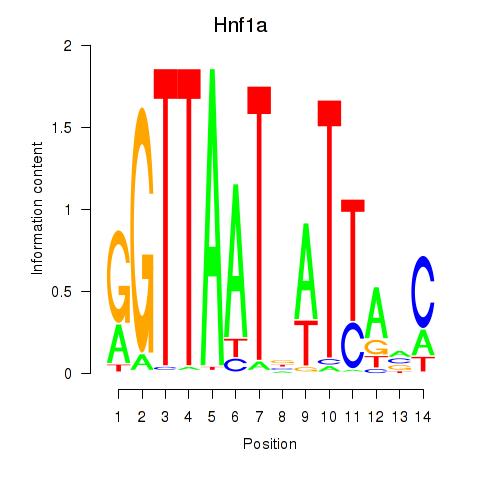
| Hnf1a | 0.50 |
|
|
|
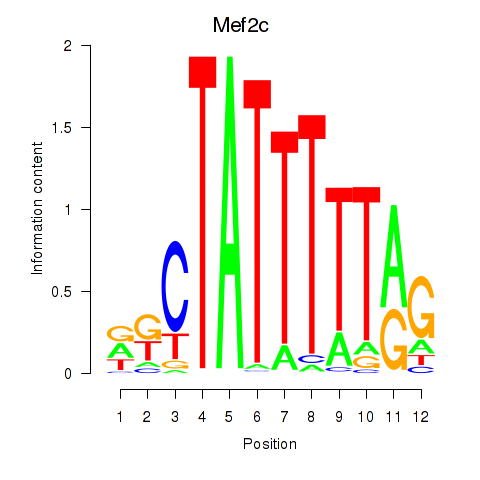
| Mef2c | 0.50 |
|

|
|
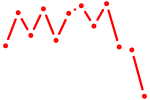
| AACACUG | 0.50 |
|
|

|
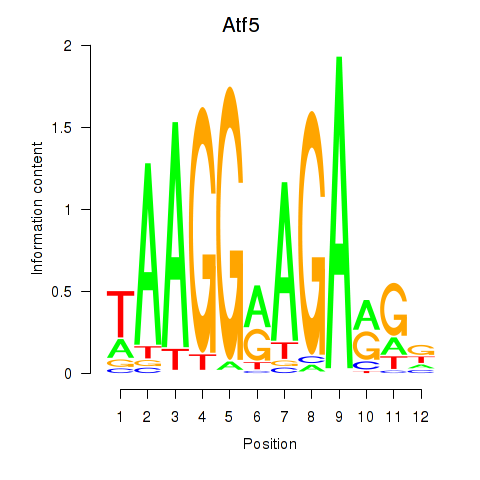
| Atf5 | 0.50 |
|
|
|
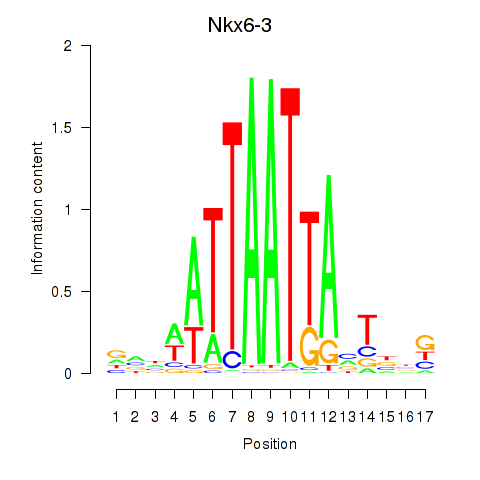
| Nkx6-3_Dbx2_Barx2 | 0.50 |
|
|
|
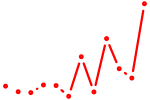
| Tbx19 | 0.49 |
|
|
|
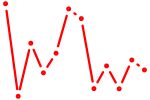
| Rorc_Nr1d1 | 0.49 |
|
|
|
| ACUGCAU | 0.49 |
|
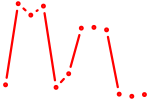
|

|
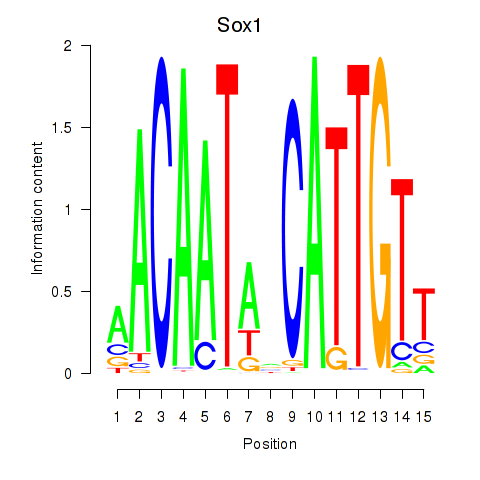
| Sox1 | 0.48 |
|
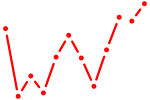
|
|
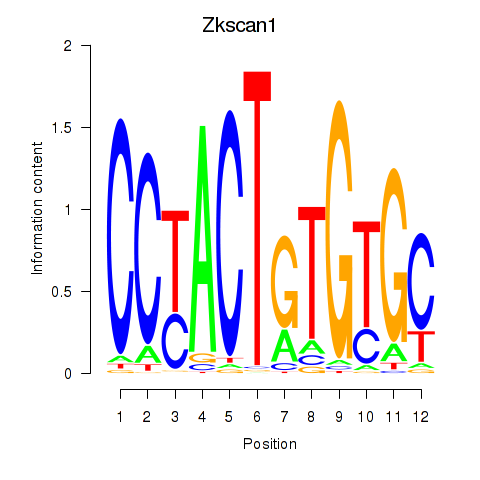
| Zkscan1 | 0.48 |
|
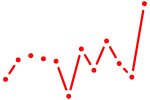
|
|
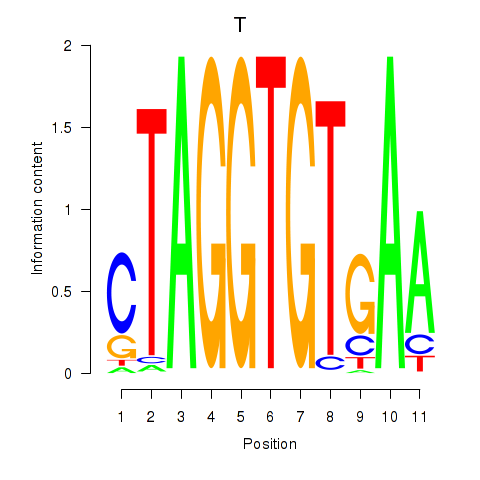
| T | 0.48 |
|
|
|
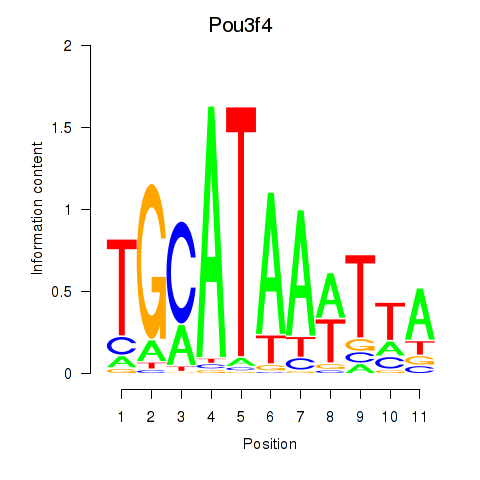
| Pou3f4 | 0.48 |
|
|
|
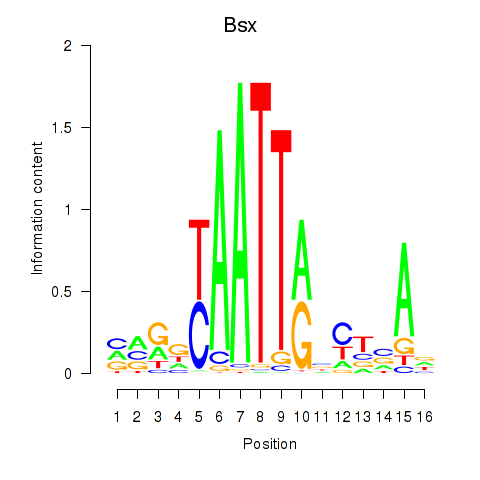
| Bsx | 0.47 |
|
|
|
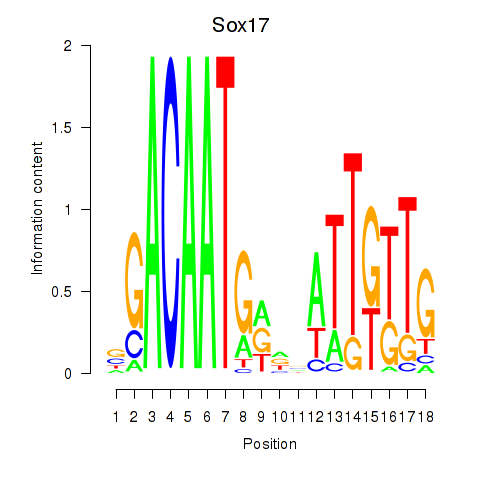
| Sox17 | 0.47 |
|
|
|
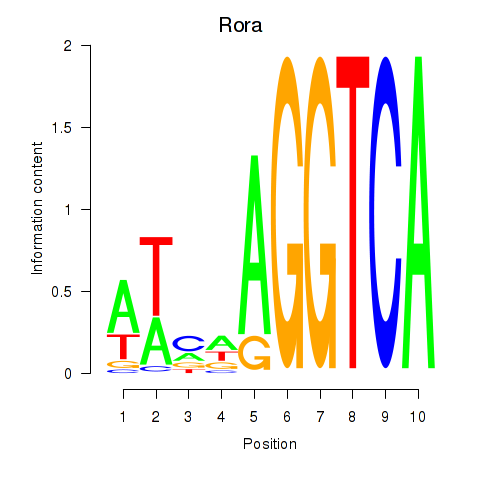
| Rora | 0.47 |
|
|
|
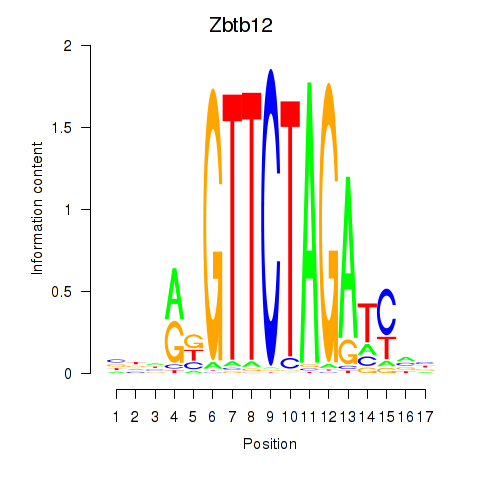
| Zbtb12 | 0.47 |
|
|
|
| UUGGCAC | 0.47 |
|
|

|
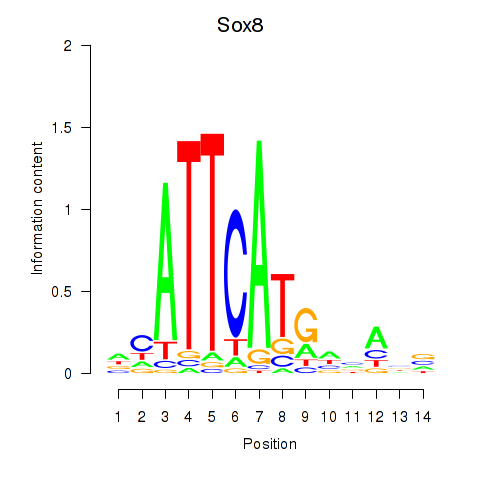
| Sox8 | 0.47 |
|
|
|
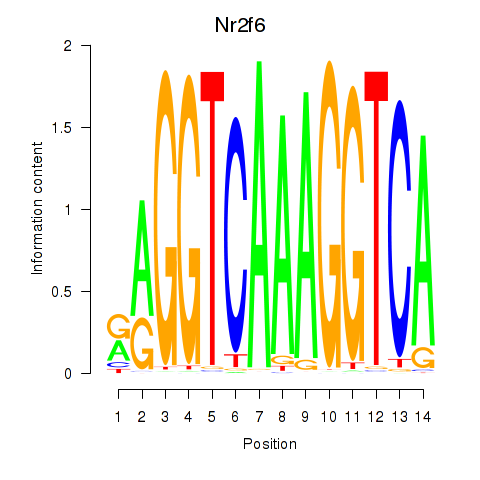
| Nr2f6 | 0.47 |
|
|
|
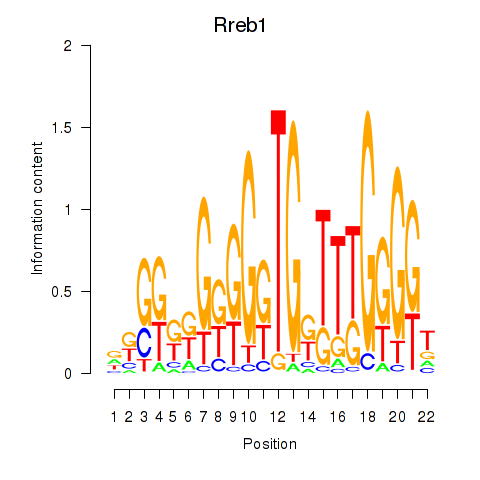
| Rreb1 | 0.47 |
|
|
|
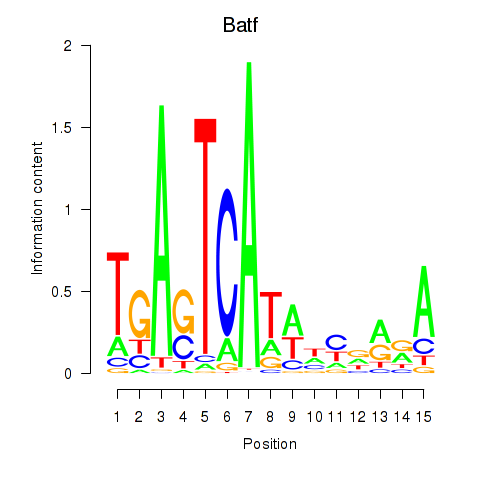
| Batf | 0.46 |
|

|
|
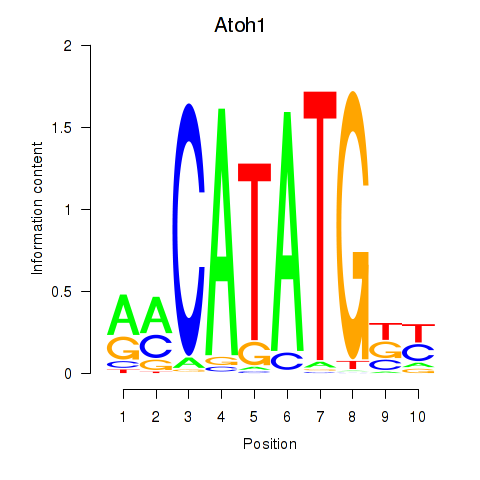
| Atoh1_Bhlhe23 | 0.46 |
|

|
|
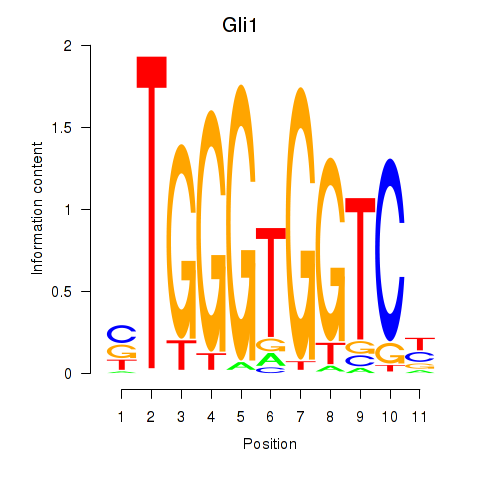
| Gli1 | 0.46 |
|
|
|
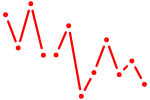
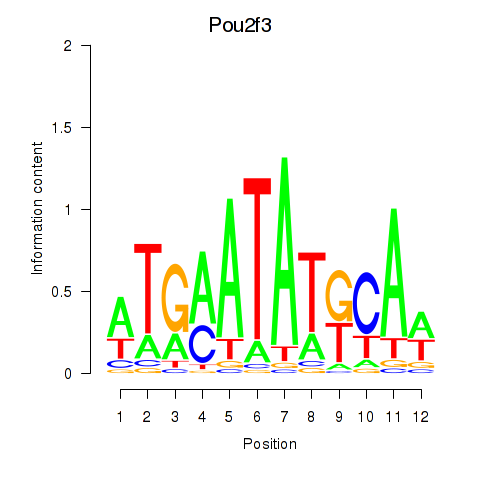
| Pou2f3 | 0.46 |
|
|
|
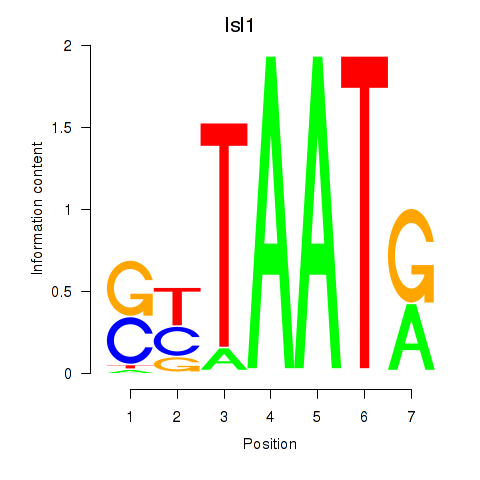
| Isl1 | 0.46 |
|
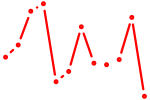
|
|
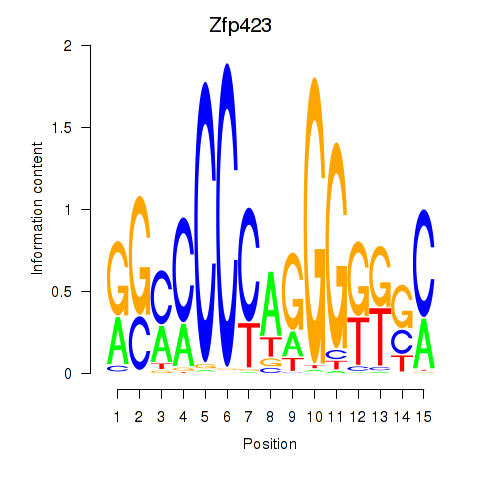
| Zfp423 | 0.46 |
|
|
|
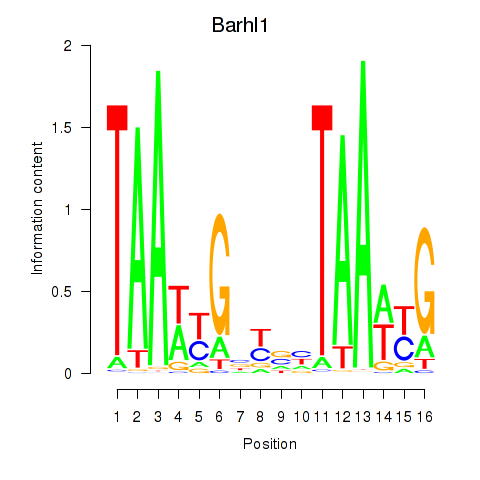
| Barhl1 | 0.46 |
|
|
|
| Barhl2 | 0.45 |
|
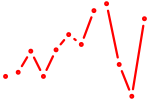
|
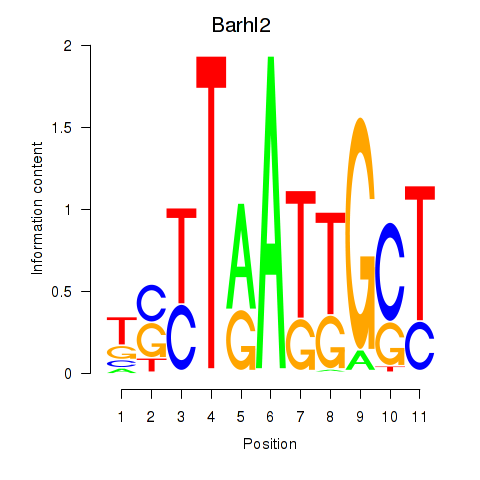
|
| Lhx3 | 0.45 |
|
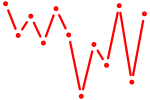
|
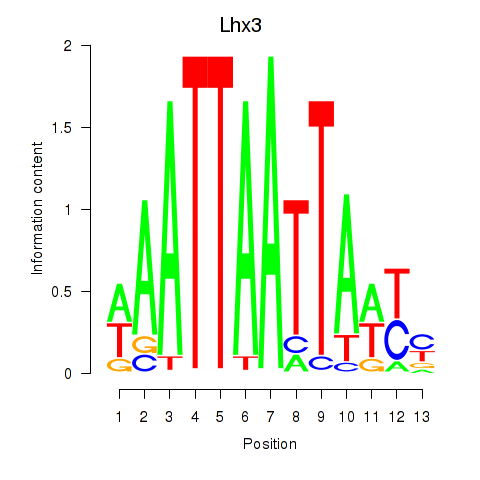
|
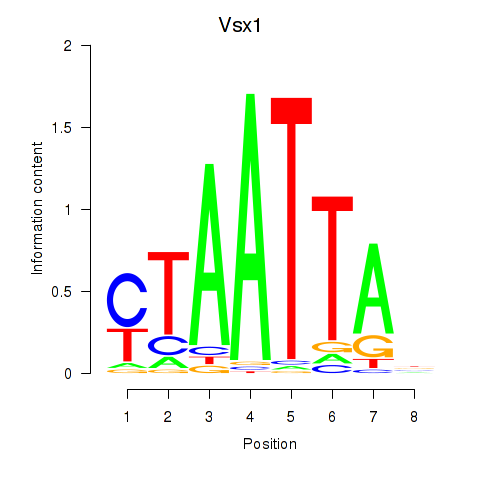
| Vsx1_Uncx_Prrx2_Shox2_Noto | 0.45 |
|
|
|
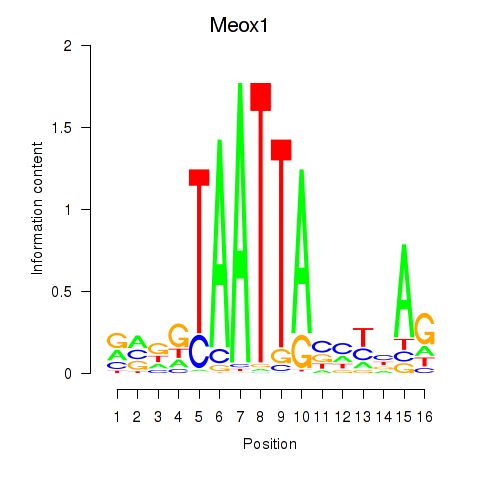
| Meox1 | 0.45 |
|
|
|
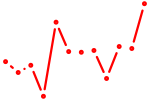
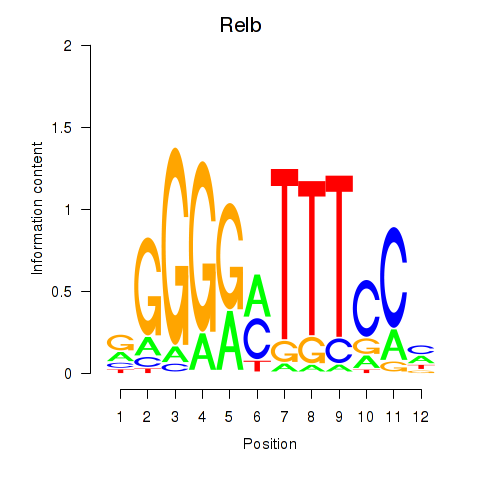
| Relb | 0.45 |
|
|
|
| ACAGUAU | 0.45 |
|

|

|
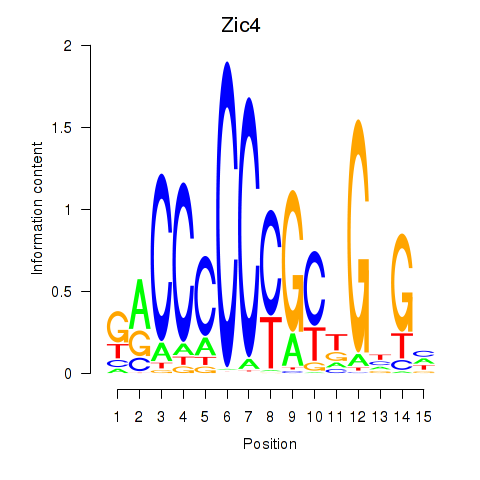
| Zic4 | 0.45 |
|

|
|
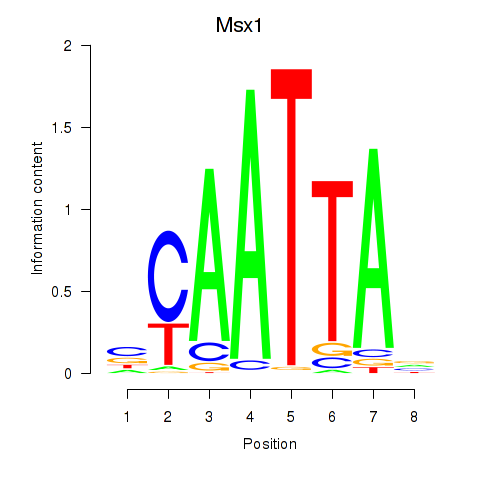
| Msx1_Lhx9_Barx1_Rax_Dlx6 | 0.45 |
|

|
|
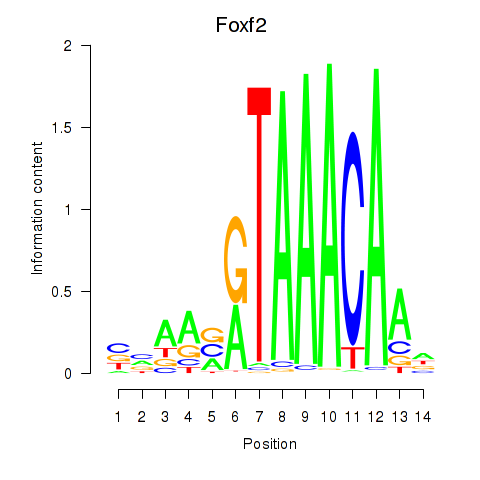
| Foxf2 | 0.45 |
|
|
|
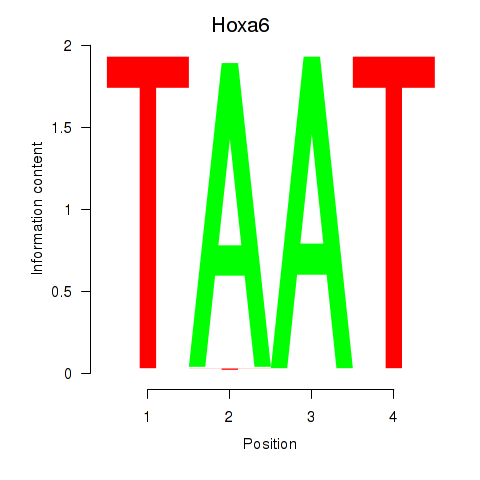
| Hoxa6 | 0.44 |
|
|
|
| Tbx2 | 0.44 |
|
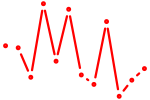
|
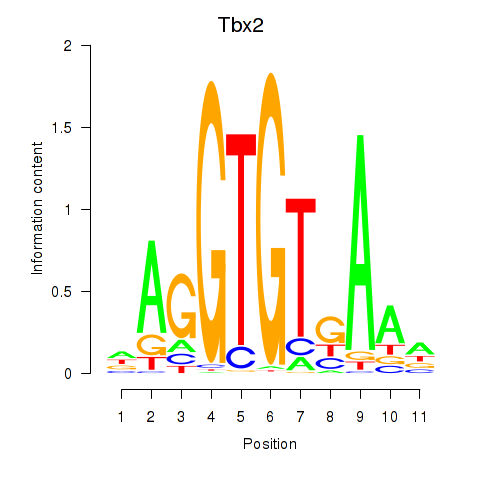
|
| Ppard | 0.44 |
|
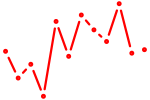
|
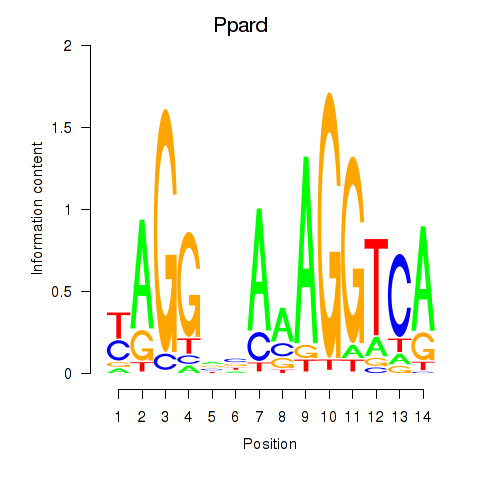
|
| Pou6f2_Pou4f2 | 0.44 |
|
|

|
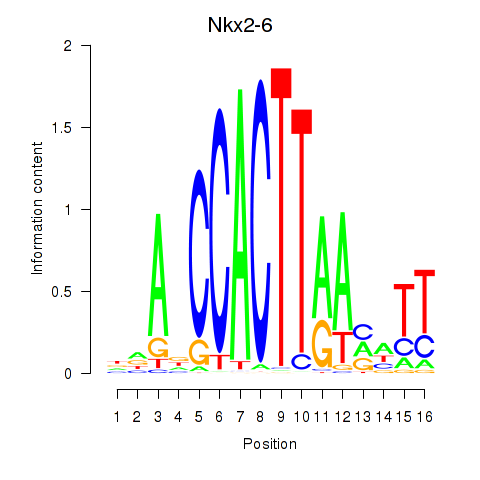
| Nkx2-6 | 0.43 |
|

|
|
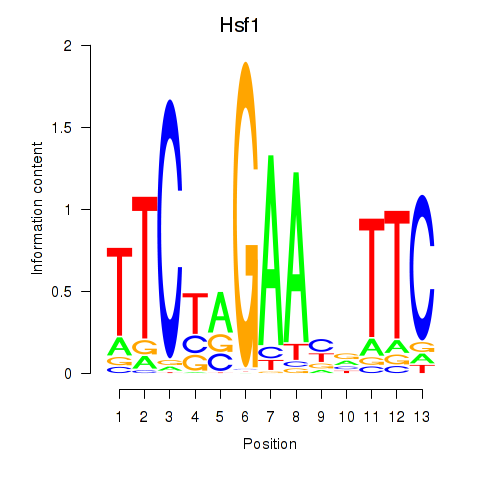
| Hsf1 | 0.43 |
|
|
|
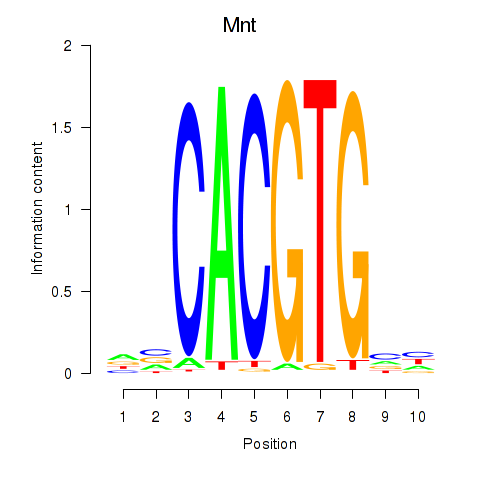
| Mnt | 0.43 |
|

|
|
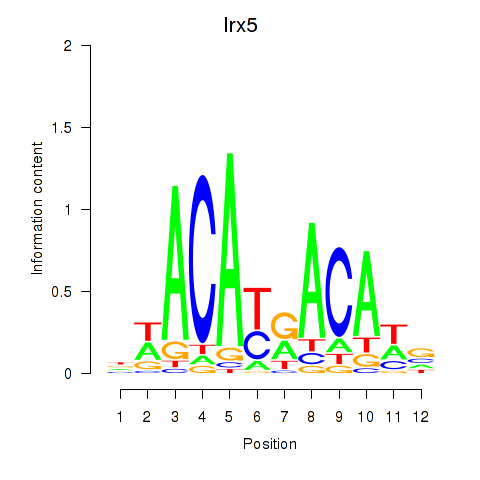
| Irx5 | 0.43 |
|
|
|
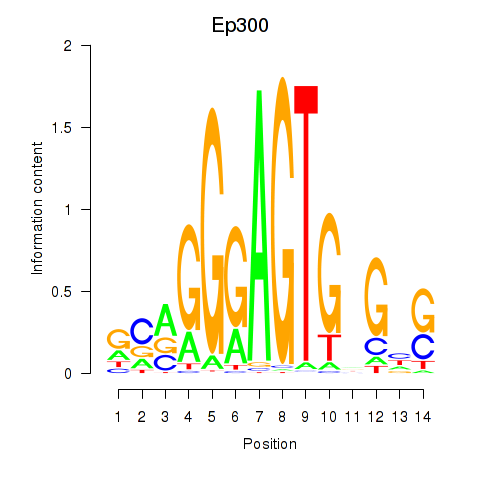
| Ep300 | 0.43 |
|

|
|
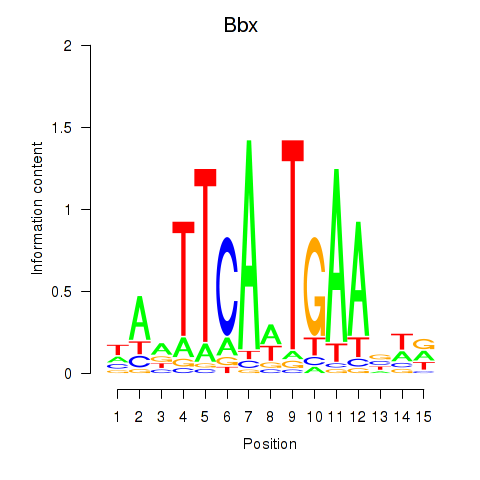
| Bbx | 0.43 |
|
|
|
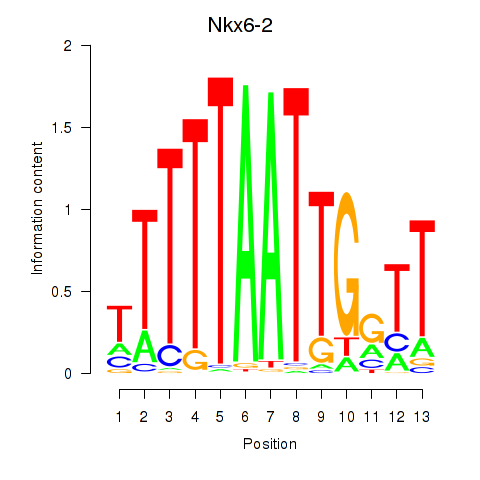
| Nkx6-2 | 0.42 |
|
|
|
| Nr2c1 | 0.42 |
|
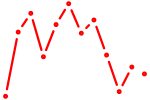
|
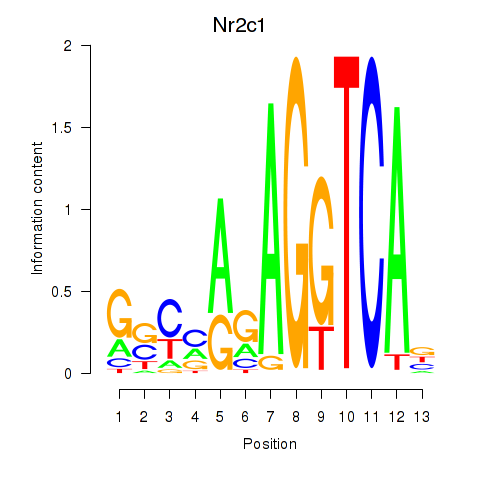
|
| Foxo4 | 0.41 |
|
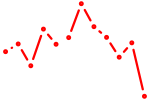
|
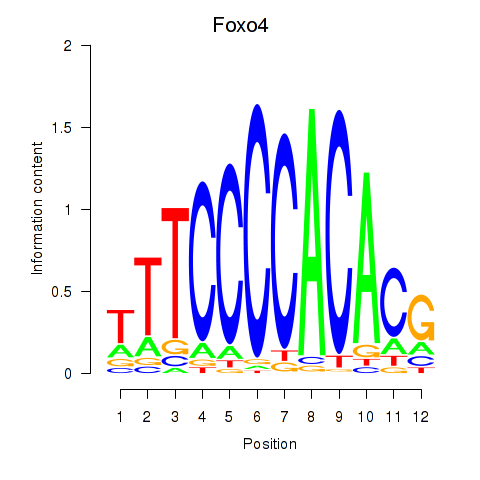
|
| Rbpj | 0.41 |
|
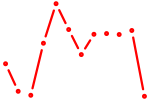
|
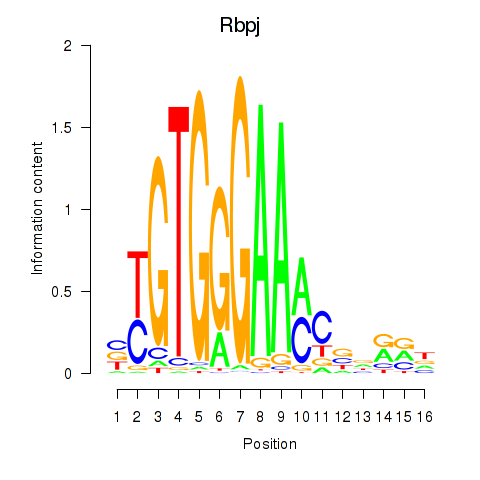
|
| GUAGUGU | 0.41 |
|
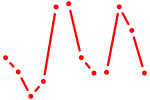
|

|
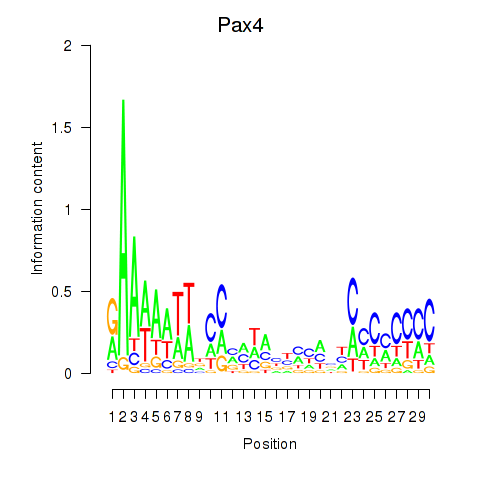
| Pax4 | 0.41 |
|
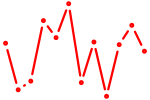
|
|
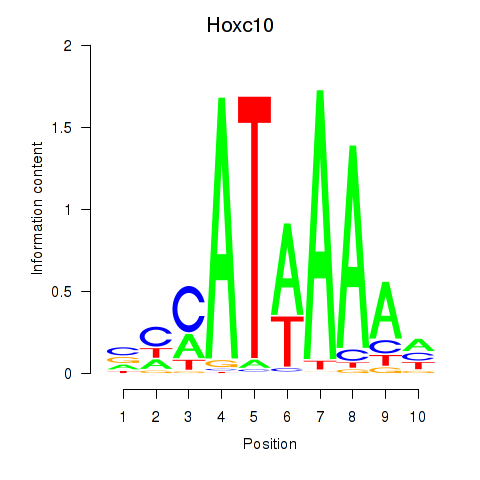
| Hoxc10 | 0.41 |
|
|
|
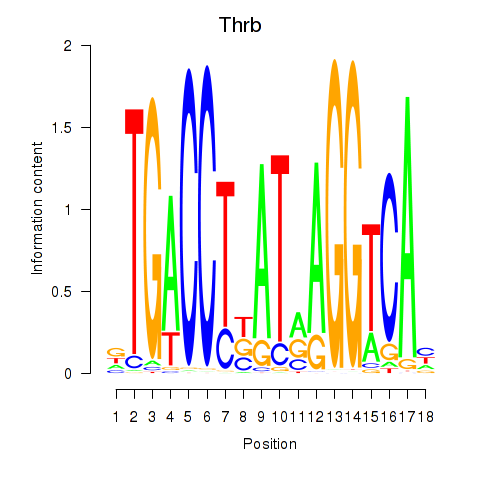
| Thrb | 0.41 |
|
|
|
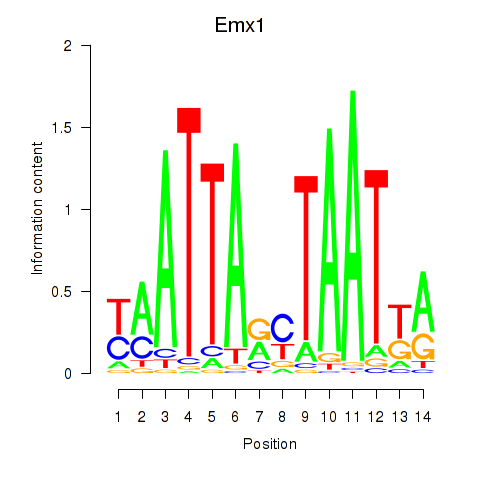
| Emx1_Emx2 | 0.41 |
|
|
|
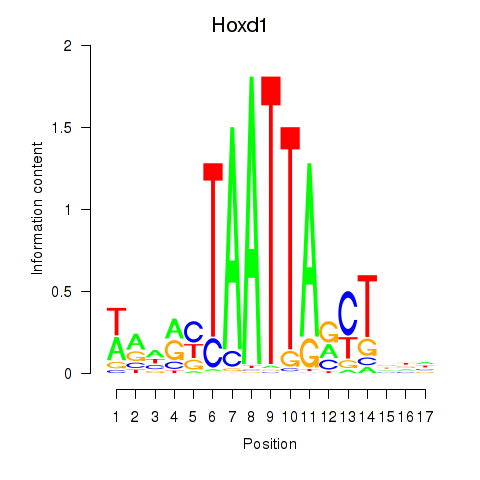
| Hoxd1 | 0.40 |
|
|
|
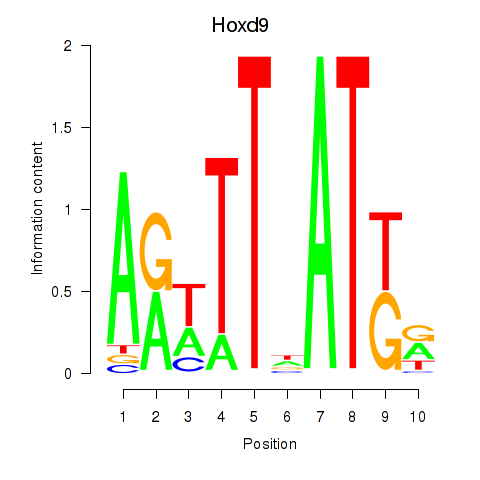
| Hoxd9 | 0.40 |
|
|
|
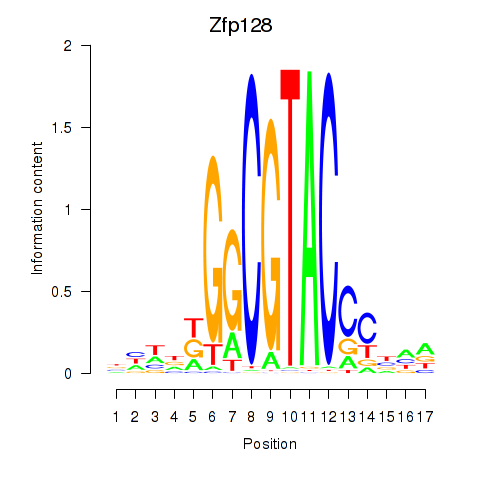
| Zfp128 | 0.40 |
|
|
|
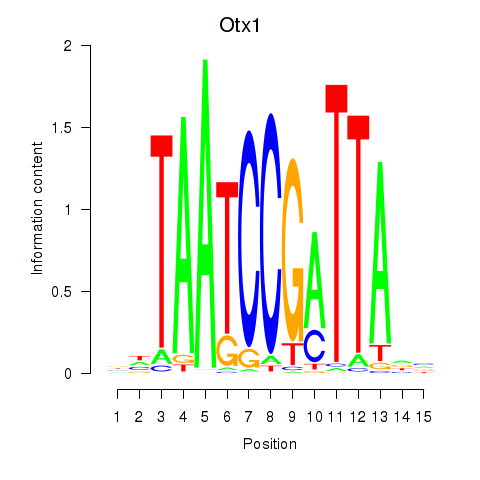
| Otx1 | 0.40 |
|
|
|
| AUGGCAC | 0.40 |
|
|

|
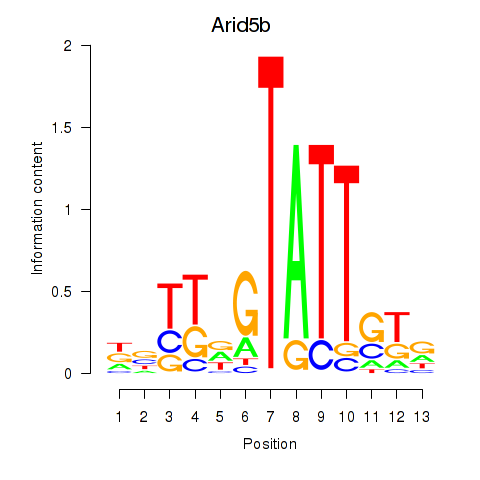
| Arid5b | 0.40 |
|
|
|
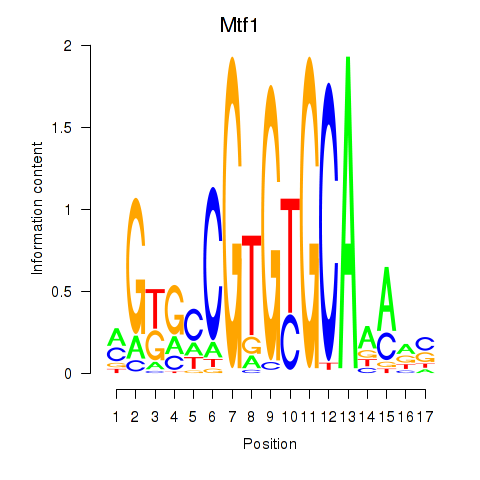
| Mtf1 | 0.40 |
|
|
|
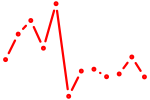
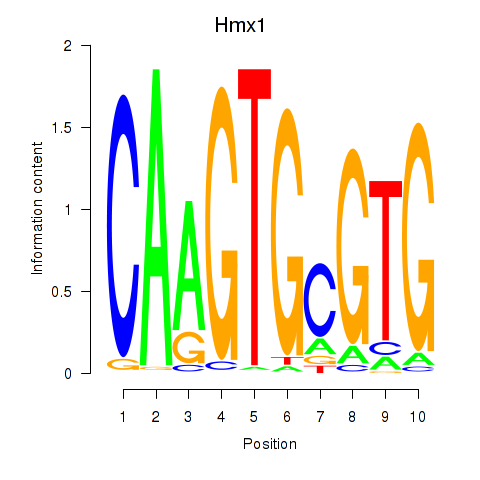
| Hmx1 | 0.40 |
|
|
|
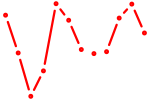
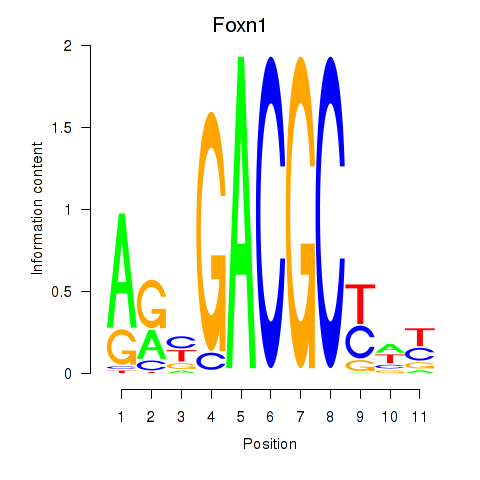
| Foxn1 | 0.39 |
|
|
|
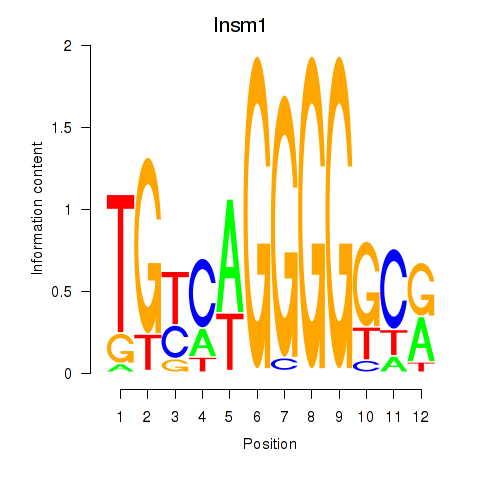
| Insm1 | 0.38 |
|
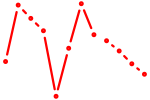
|
|
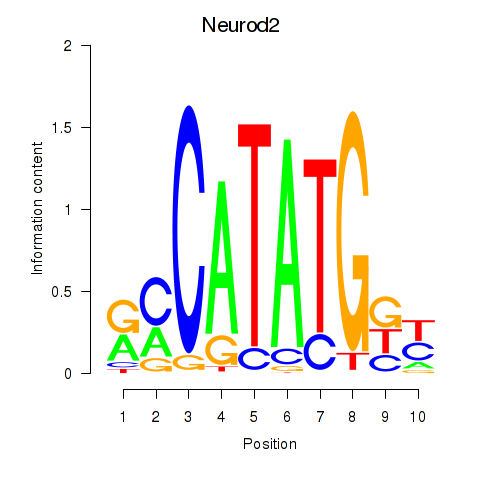
| Neurod2_Bhlha15_Bhlhe22_Olig1 | 0.38 |
|
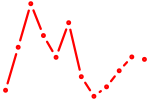
|
|
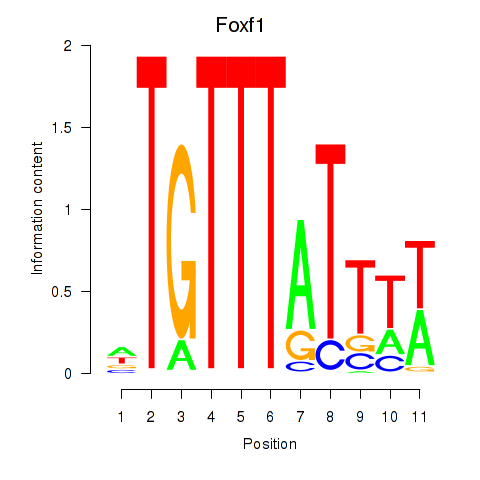
| Foxf1 | 0.38 |
|
|
|
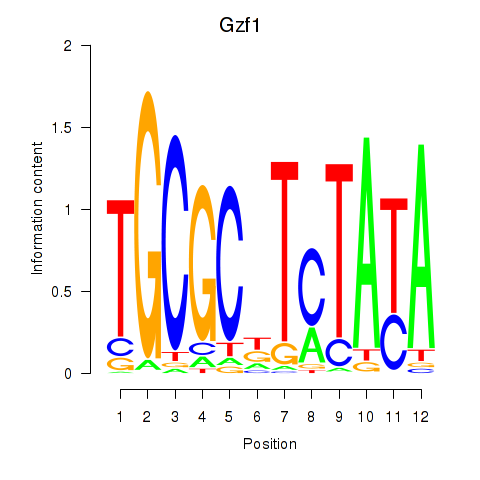
| Gzf1 | 0.38 |
|
|
|
| Clock | 0.38 |
|
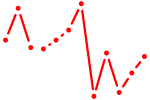
|
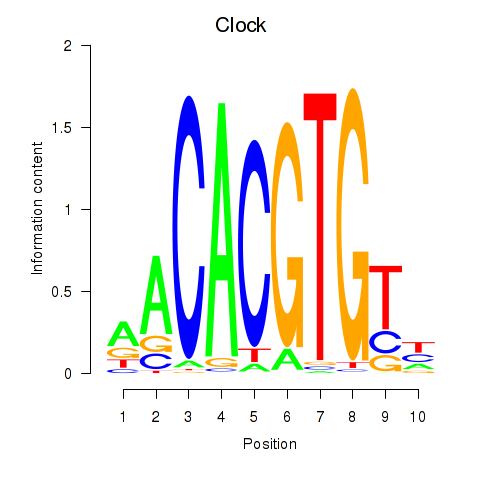
|
| Trp63 | 0.38 |
|
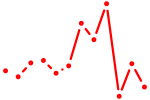
|
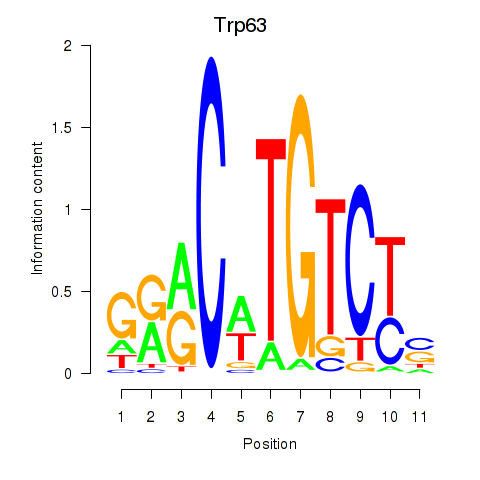
|
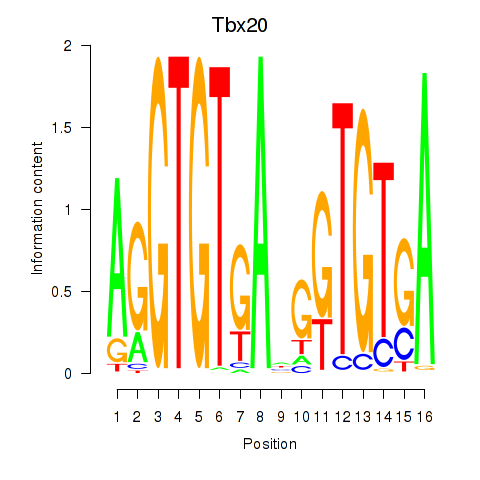
| Tbx20 | 0.37 |
|

|
|
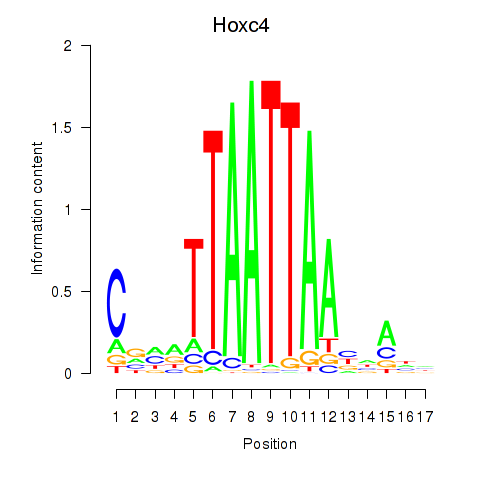
| Hoxc4_Arx_Otp_Esx1_Phox2b | 0.37 |
|

|
|
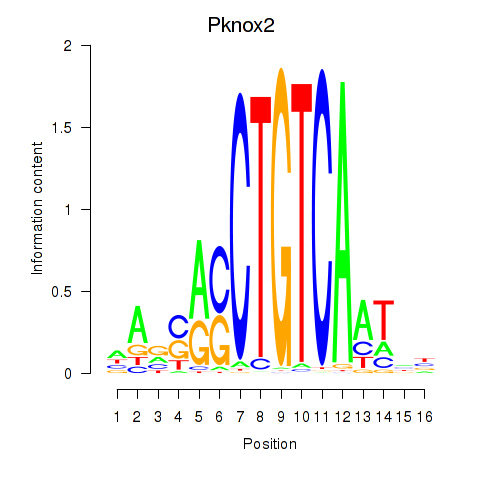
| Pknox2_Pknox1 | 0.37 |
|
|
|
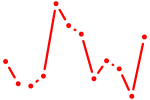
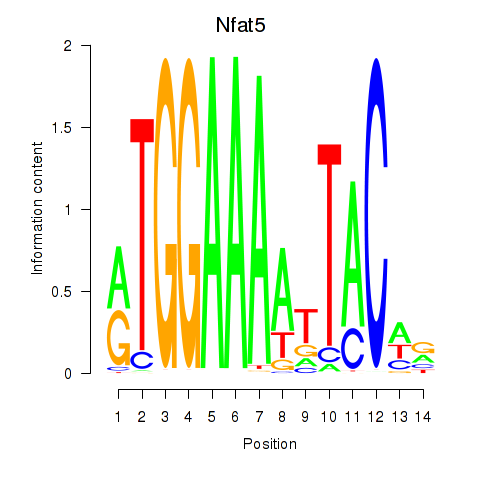
| Nfat5 | 0.37 |
|
|
|
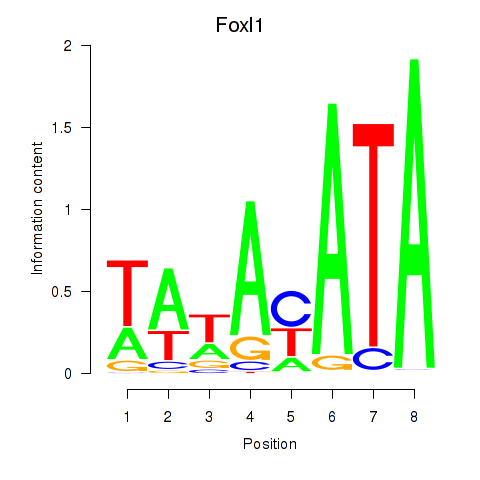
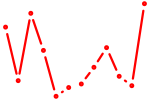
| Foxl1 | 0.37 |
|

|
|
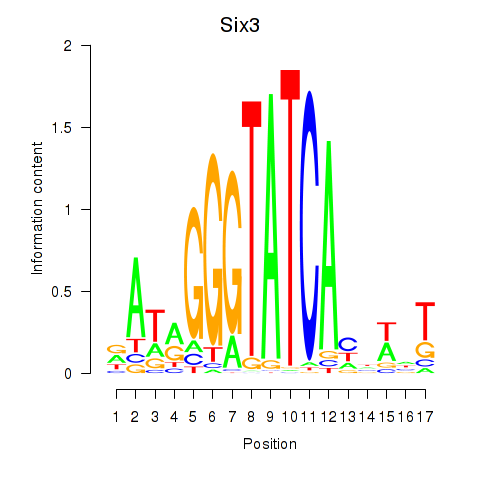
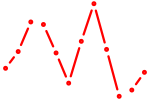
| Six3_Six1_Six2 | 0.37 |
|
|
|
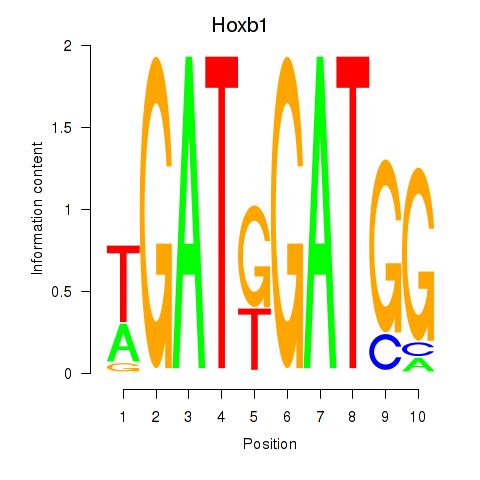
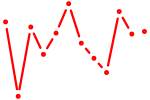
| Hoxb1 | 0.37 |
|
|
|
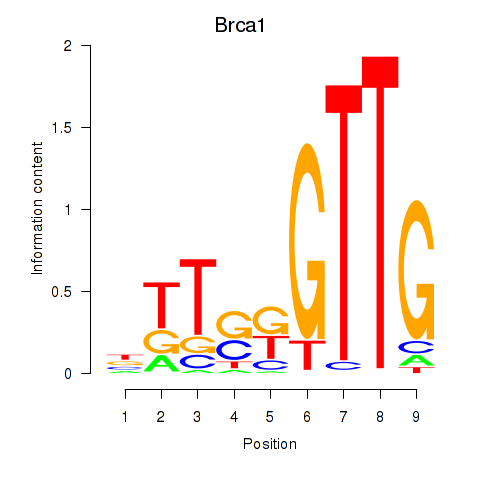
| Brca1 | 0.36 |
|
|
|
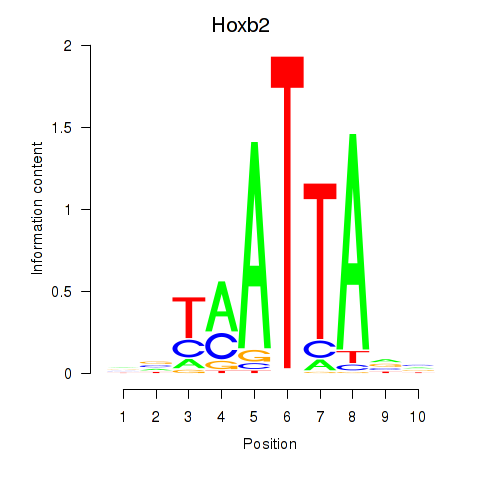
| Hoxb2_Dlx2 | 0.36 |
|
|
|
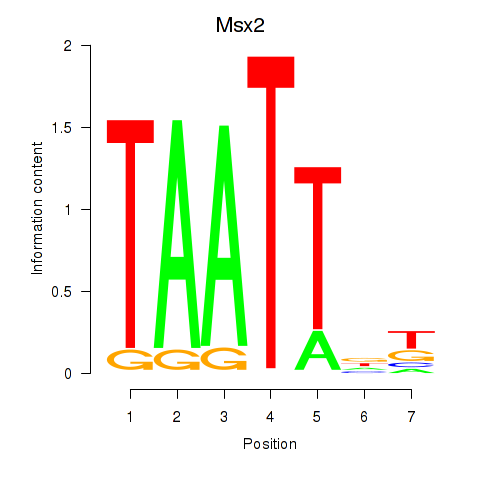
| Msx2_Hoxd4 | 0.36 |
|
|
|
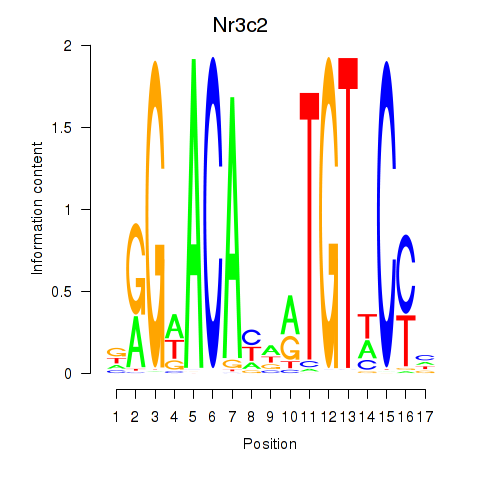
| Nr3c2 | 0.36 |
|
|
|
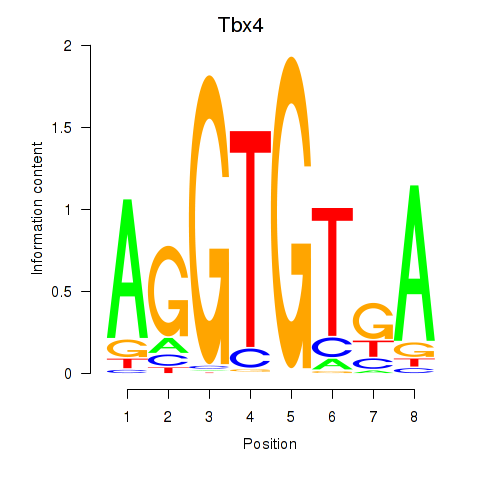
| Tbx4 | 0.36 |
|
|
|
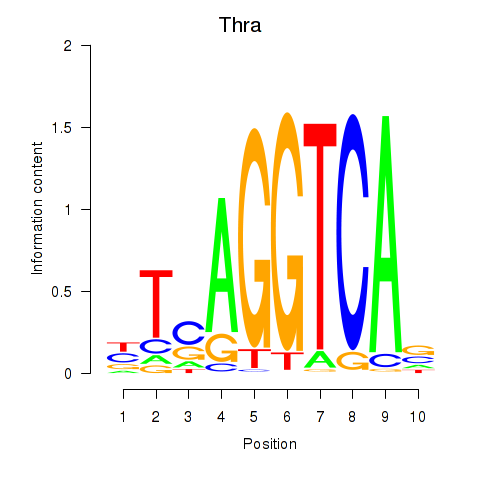
| Thra | 0.36 |
|
|
|
| Zfp524 | 0.36 |
|
|
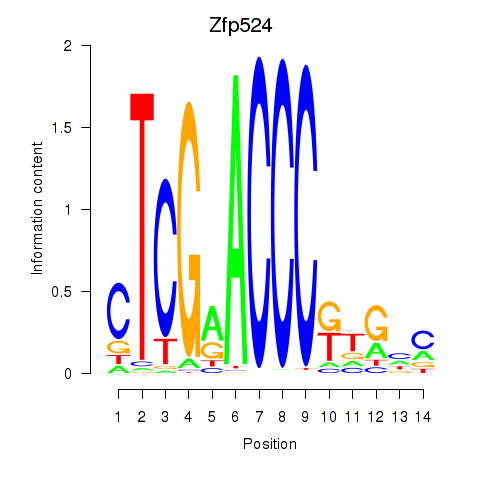
|
| Zfhx3 | 0.36 |
|
|

|
| CCAGUGU | 0.36 |
|
|

|
| UCCAGUU | 0.36 |
|
|

|
| Cebpd | 0.36 |
|
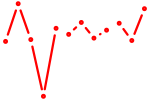
|
|
| GAUUGUC | 0.36 |
|
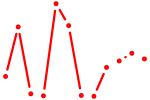
|

|
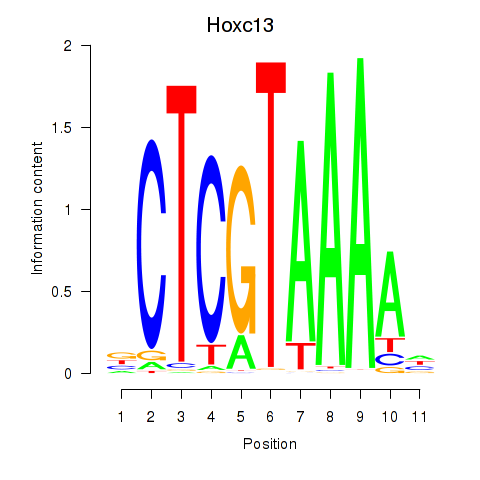
| Hoxc13_Hoxd13 | 0.35 |
|
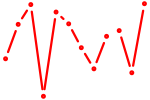
|
|
| ACCCUGU | 0.35 |
|
|
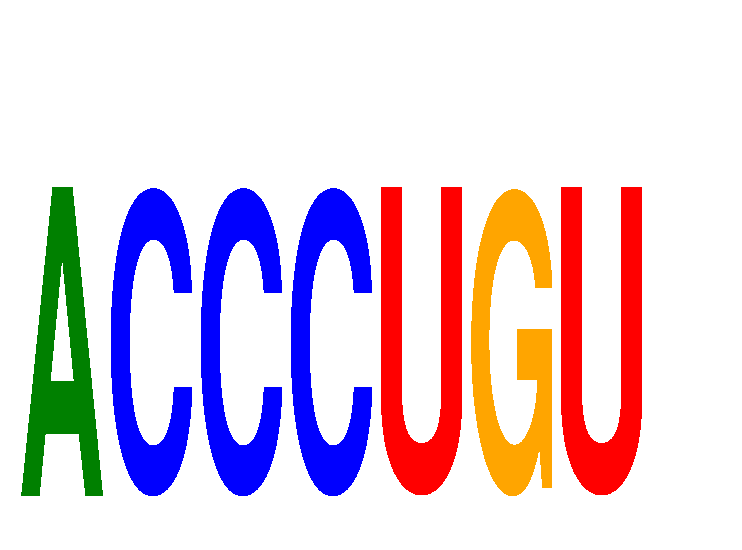
|
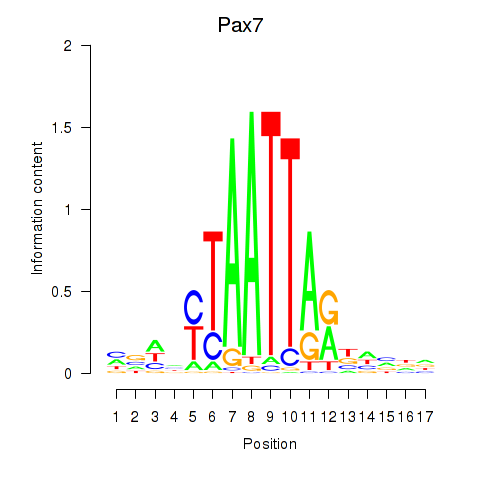
| Pax7 | 0.35 |
|
|
|
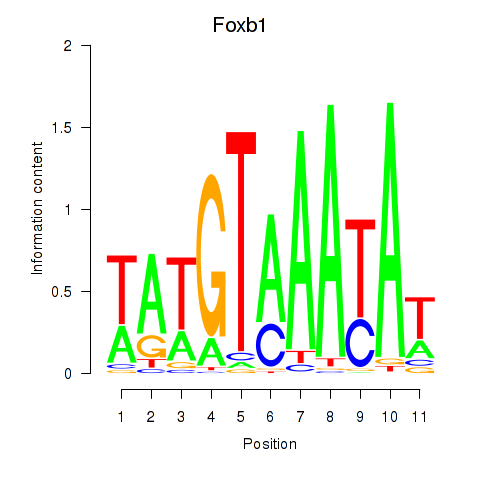
| Foxb1 | 0.34 |
|
|
|
| Nr2e1 | 0.34 |
|
|
|
| Gcm1 | 0.34 |
|
|
|
| Pou4f3 | 0.34 |
|

|
|
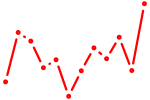
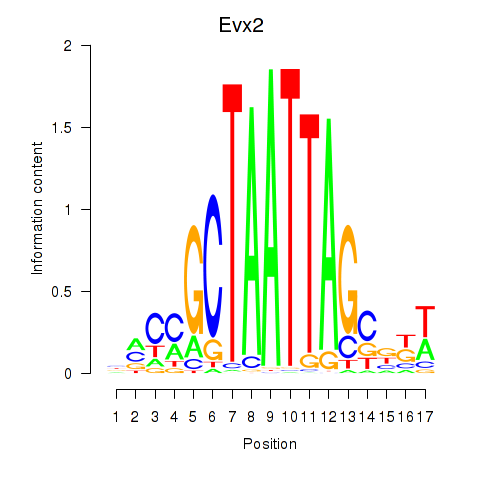
| Evx2 | 0.34 |
|
|
|
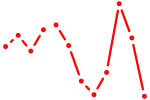
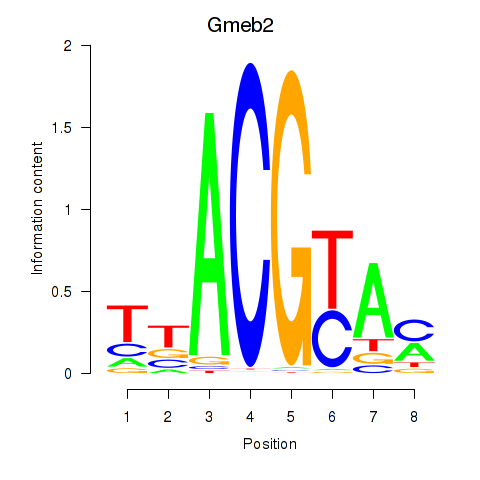
| Gmeb2 | 0.33 |
|
|
|
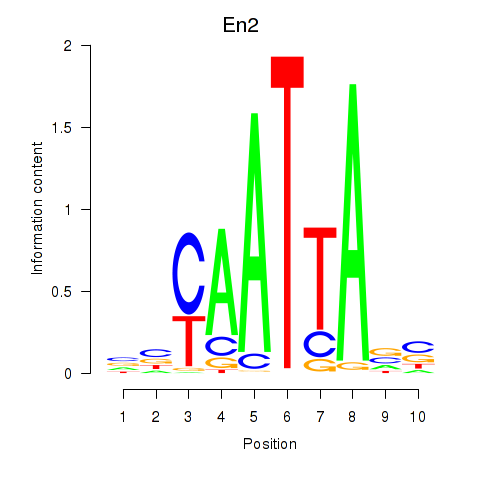
| En2 | 0.33 |
|

|
|
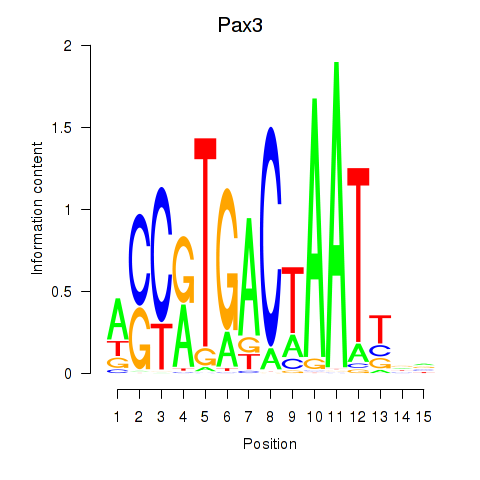
| Pax3 | 0.33 |
|
|
|
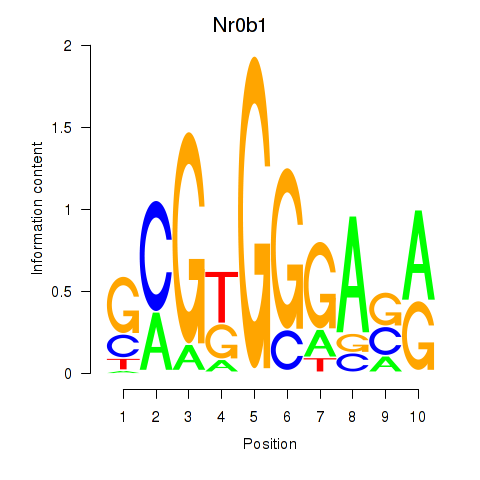
| Nr0b1 | 0.33 |
|
|
|
| GCAGCAU | 0.32 |
|

|

|
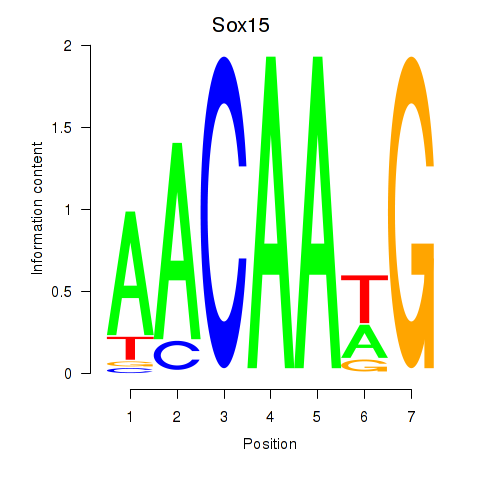
| Sox15 | 0.32 |
|
|
|
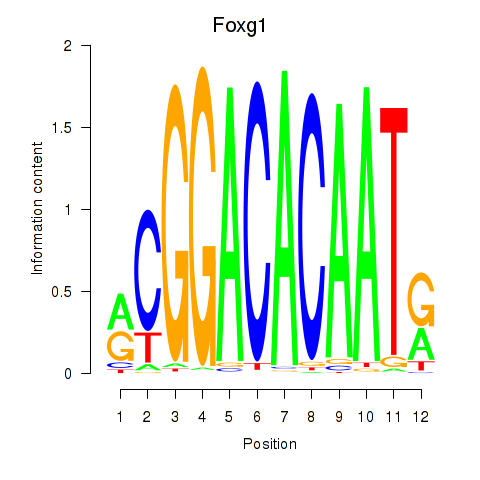
| Foxg1 | 0.32 |
|
|
|
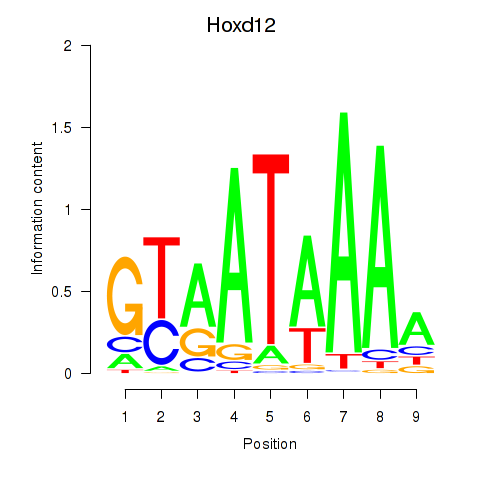
| Hoxd12 | 0.32 |
|
|
|
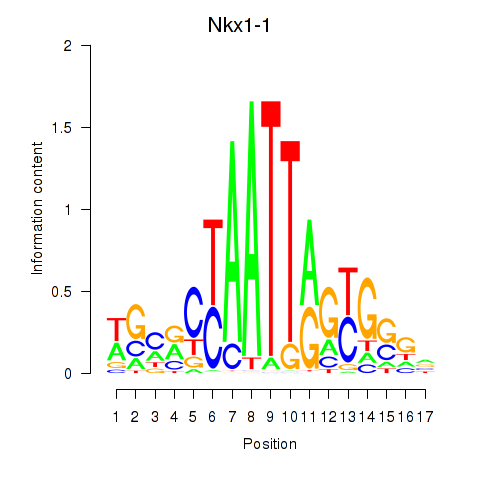
| Nkx1-1_Nkx1-2 | 0.32 |
|
|
|
| CCUUCAU | 0.32 |
|
|

|
| UUGUUCG | 0.31 |
|
|

|
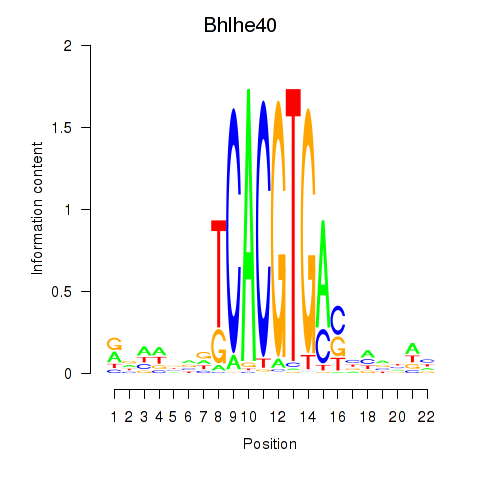
| Bhlhe40 | 0.31 |
|

|
|
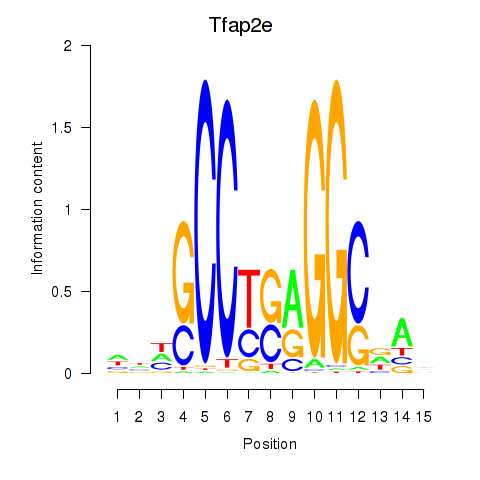
| Tfap2e | 0.31 |
|
|
|
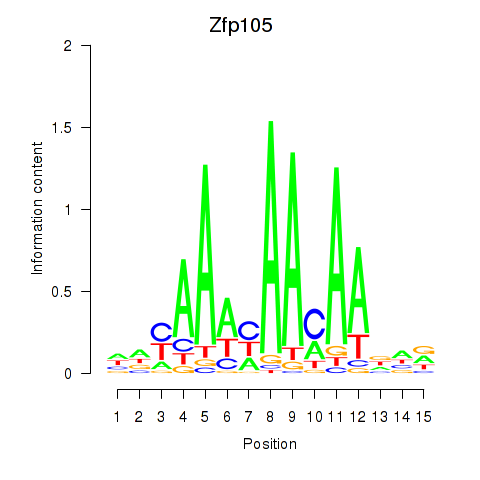
| Zfp105 | 0.31 |
|
|
|
| UCACAUU | 0.31 |
|
|

|
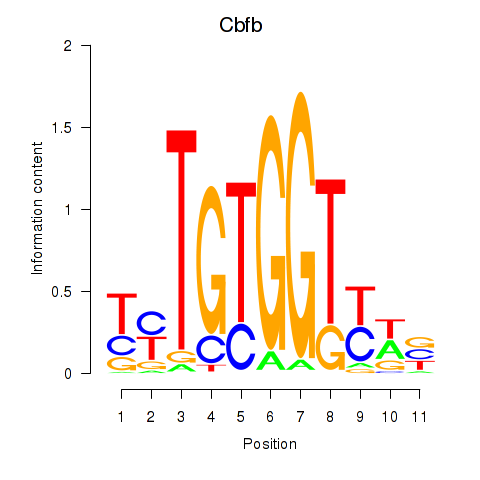
| Cbfb | 0.30 |
|
|
|
| GGAAGAC | 0.30 |
|
|

|
| UGAAAUG | 0.30 |
|
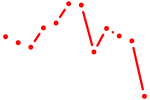
|

|
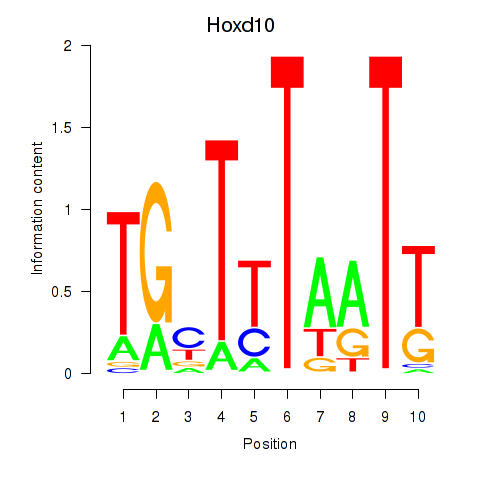
| Hoxd10 | 0.30 |
|
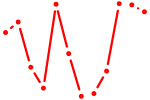
|
|
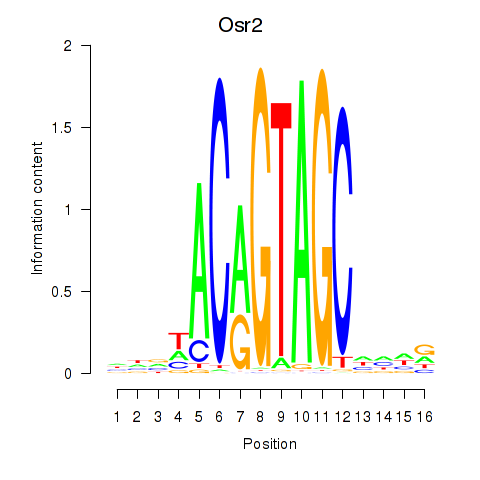
| Osr2_Osr1 | 0.30 |
|
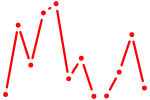
|
|
| CACAGUG | 0.30 |
|
|

|
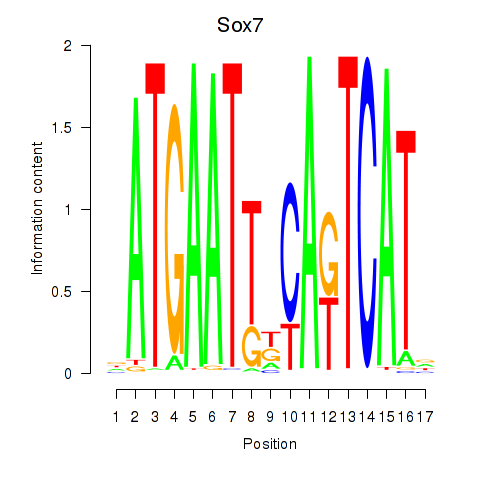
| Sox7 | 0.30 |
|
|
|
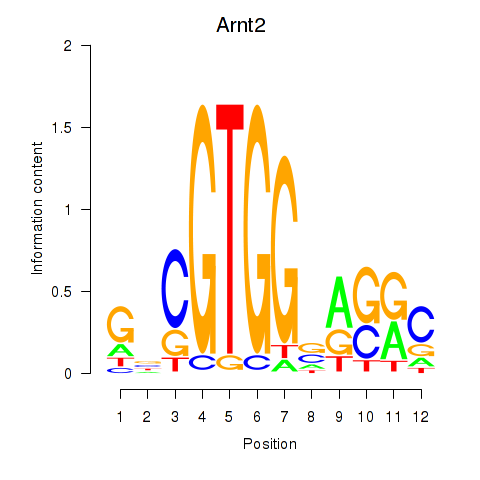
| Arnt2 | 0.30 |
|
|
|
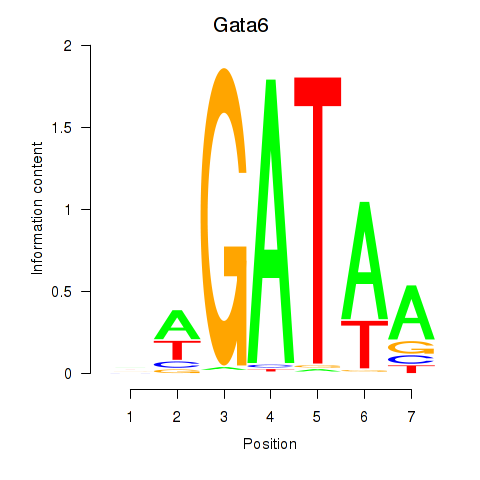
| Gata6 | 0.30 |
|
|
|
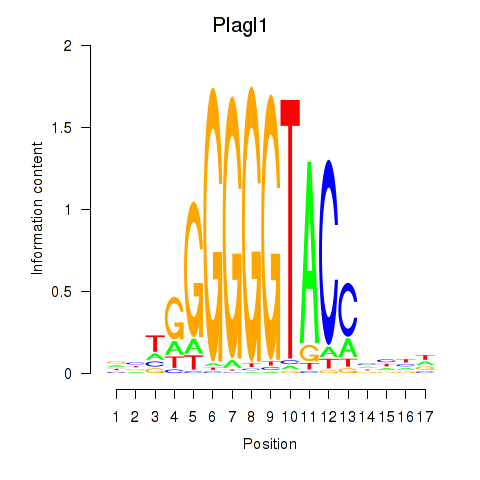
| Plagl1 | 0.29 |
|
|
|
| AACAGUC | 0.29 |
|
|

|
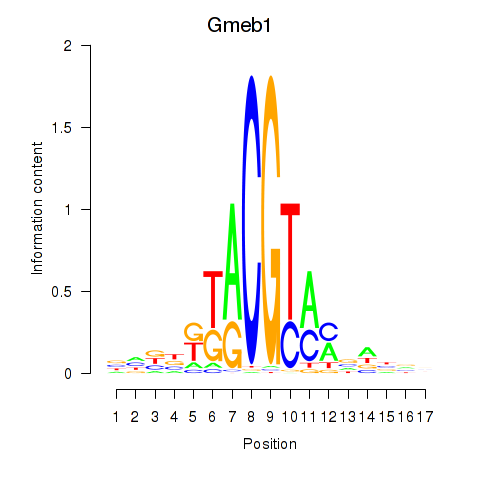
| Gmeb1 | 0.29 |
|
|
|
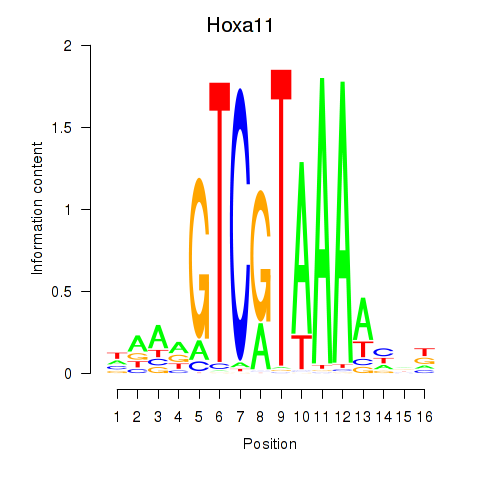
| Hoxa11_Hoxc12 | 0.29 |
|
|
|
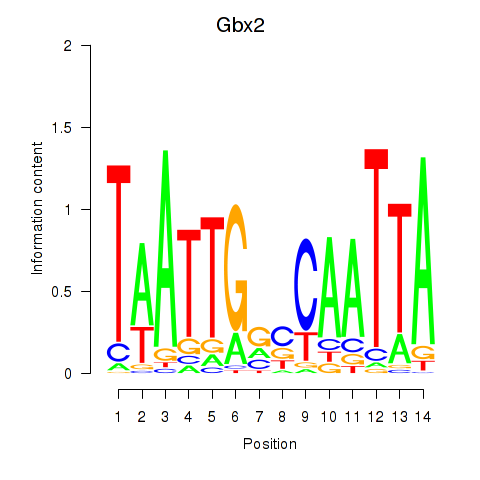
| Gbx2 | 0.29 |
|
|
|
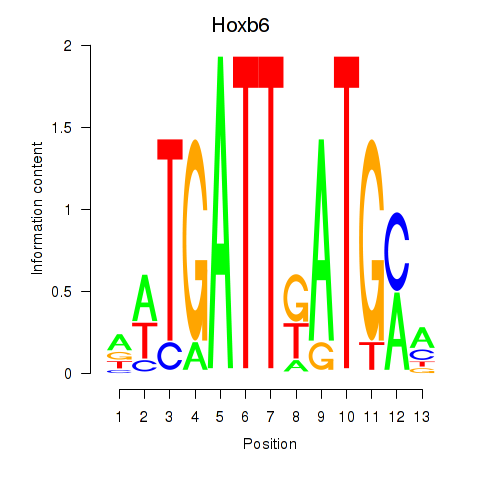
| Hoxb6 | 0.28 |
|

|
|
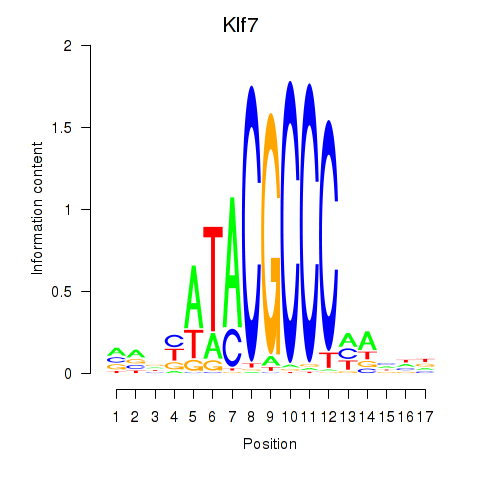
| Klf7 | 0.27 |
|

|
|
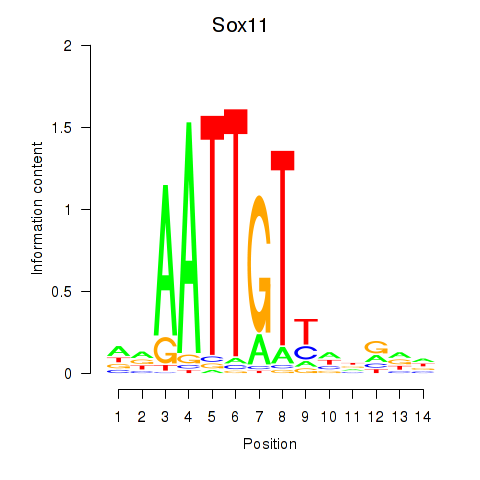
| Sox11 | 0.27 |
|
|
|
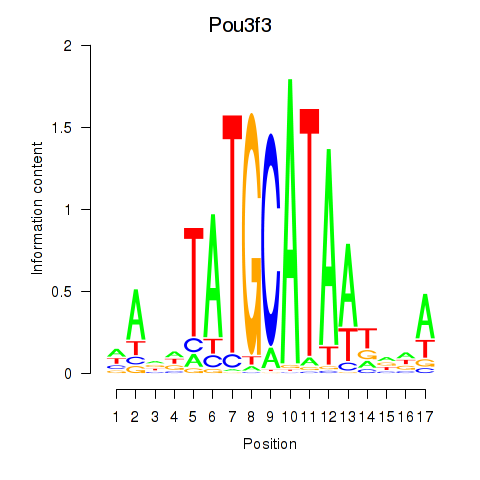
| Pou3f3 | 0.27 |
|
|
|
| UAAGACG | 0.26 |
|
|

|
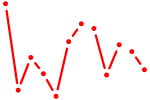
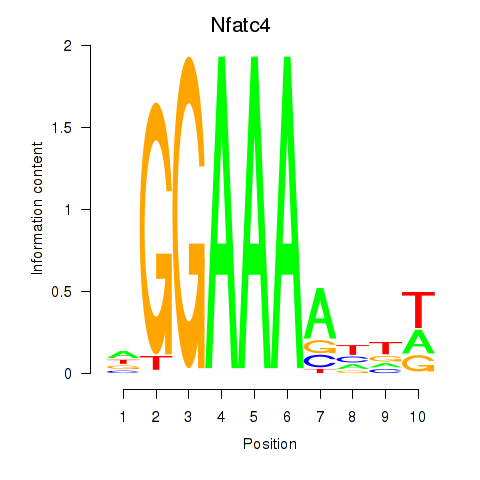
| Nfatc4 | 0.26 |
|
|
|
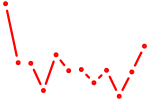
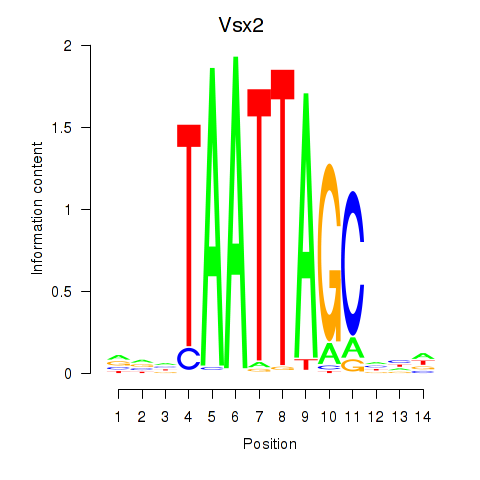
| Vsx2_Dlx3 | 0.26 |
|
|
|
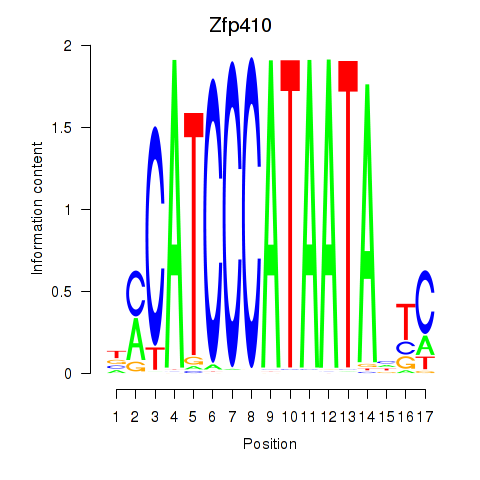
| Zfp410 | 0.26 |
|
|
|
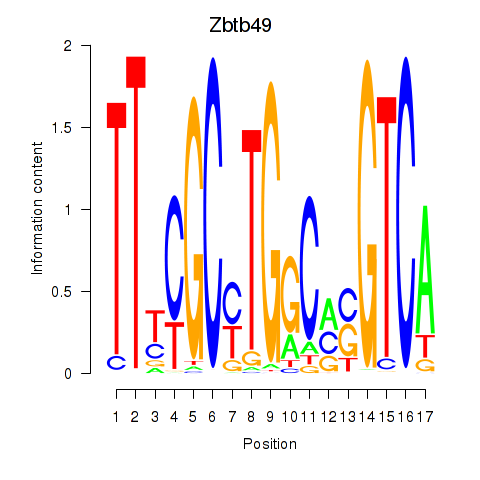
| Zbtb49 | 0.25 |
|
|
|
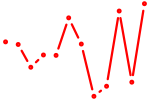
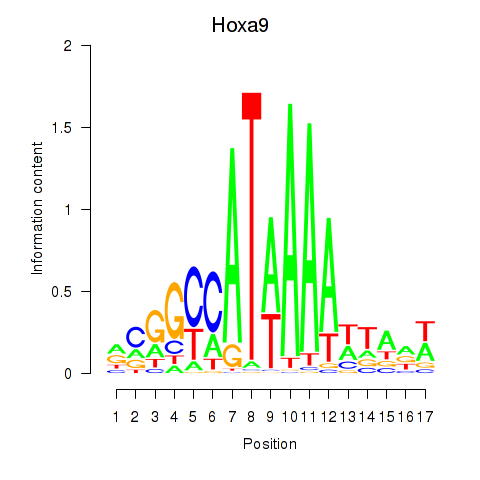
| Hoxa9_Hoxb9 | 0.25 |
|
|
|
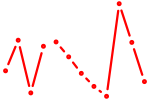
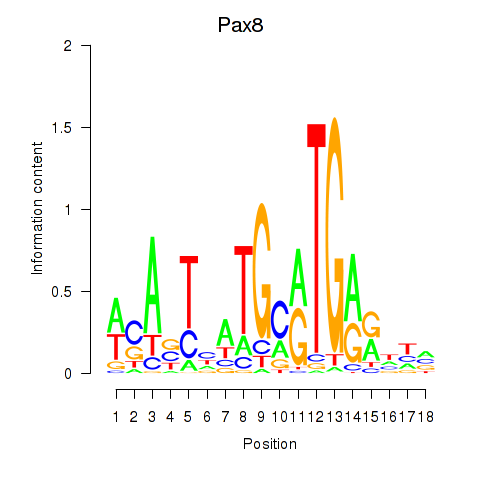
| Pax8 | 0.25 |
|
|
|
| Klf3 | 0.25 |
|
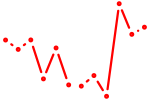
|
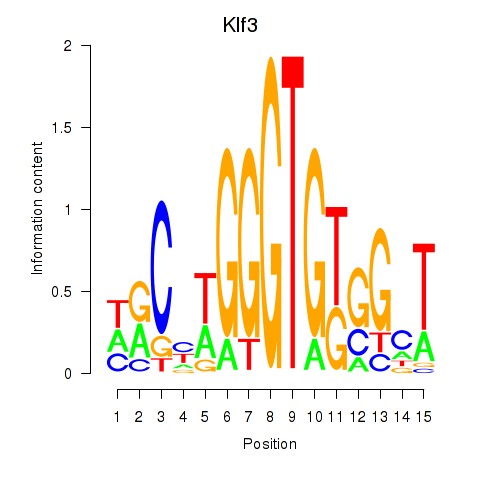
|
| Prdm4 | 0.24 |
|
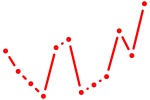
|
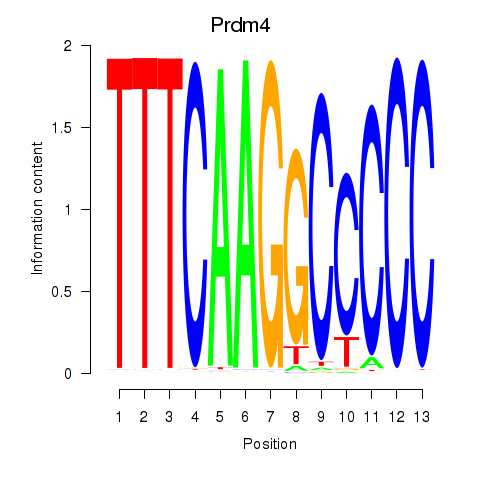
|
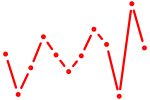
| Rara | 0.24 |
|
|
|
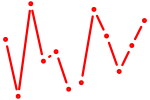
| Prox1 | 0.24 |
|
|
|
| GCUACAU | 0.24 |
|

|

|
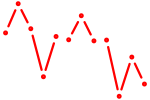
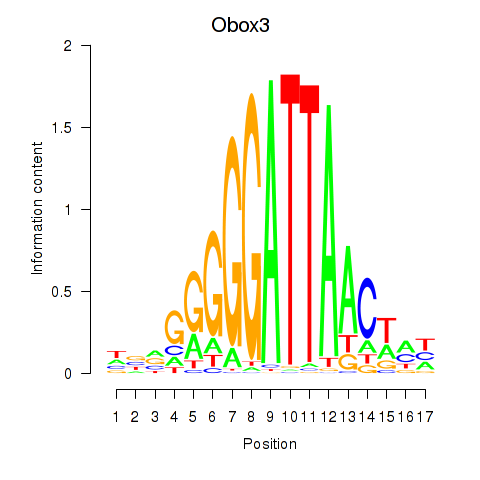
| Obox3 | 0.24 |
|
|
|
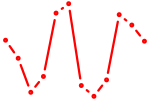
| ACUGGCC | 0.23 |
|
|

|
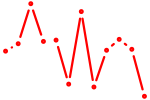
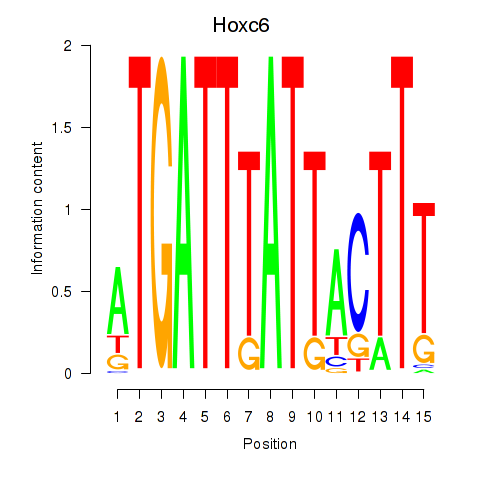
| Hoxc6 | 0.23 |
|
|
|
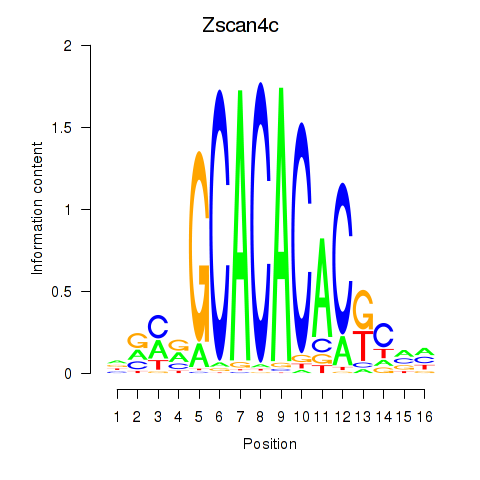
| Zscan4c | 0.22 |
|
|
|
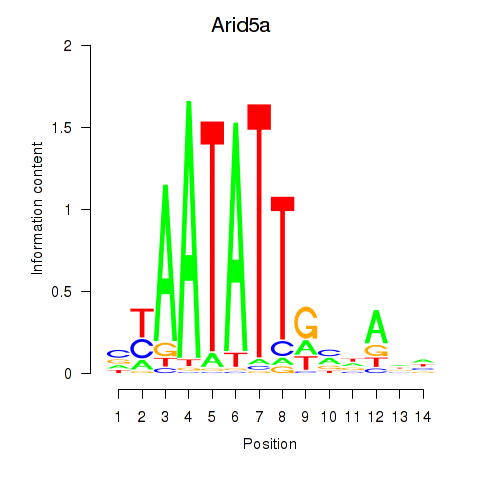
| Arid5a | 0.22 |
|
|
|
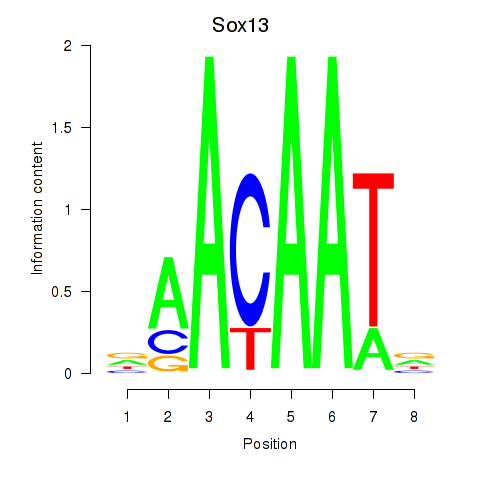
| Sox13 | 0.22 |
|
|
|
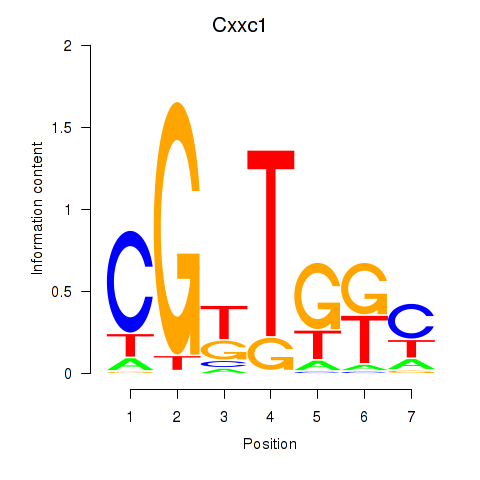
| Cxxc1 | 0.21 |
|
|
|
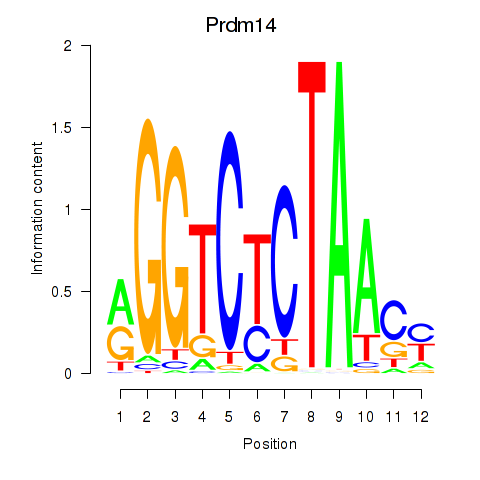
| Prdm14 | 0.21 |
|
|
|
| AAUGCCC | 0.21 |
|

|
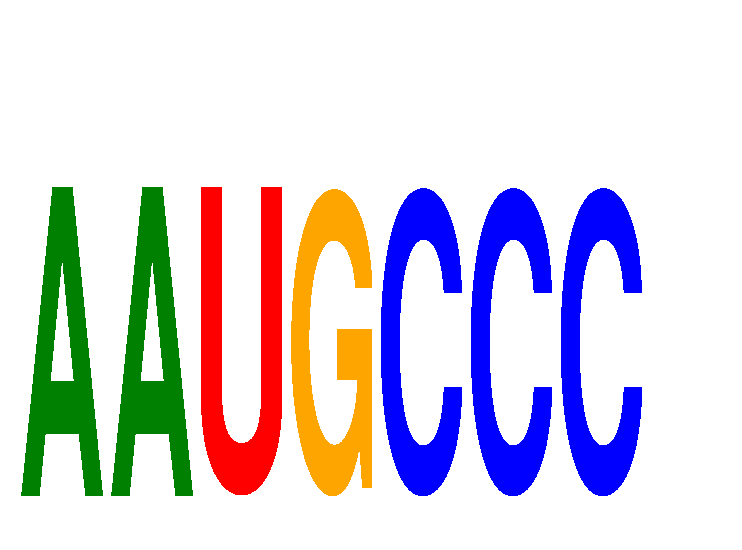
|
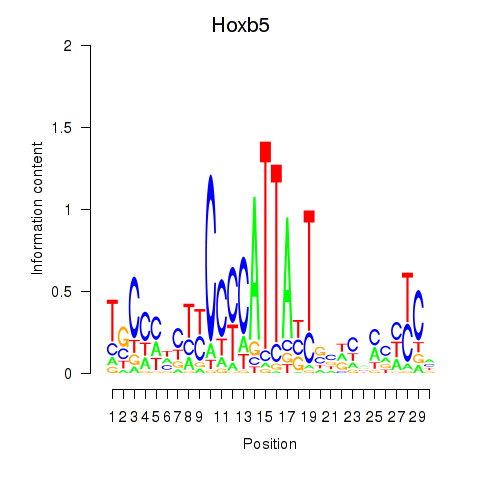
| Hoxb5 | 0.21 |
|
|
|
| GAUCAGA | 0.20 |
|
|

|
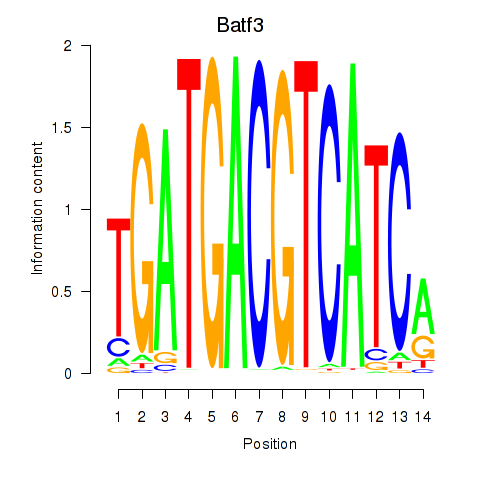
| Batf3 | 0.19 |
|
|
|
| AGCUUAU | 0.19 |
|
|

|
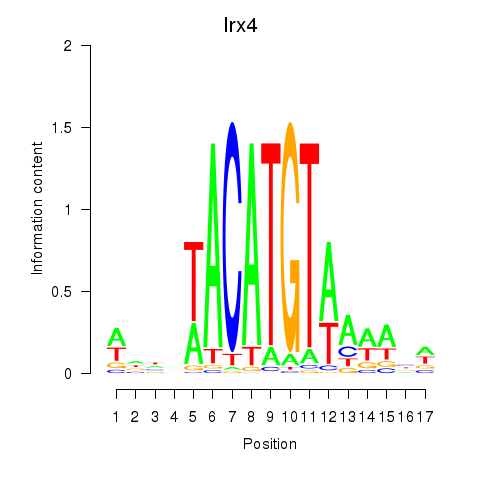
| Irx4 | 0.19 |
|
|
|
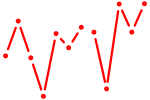
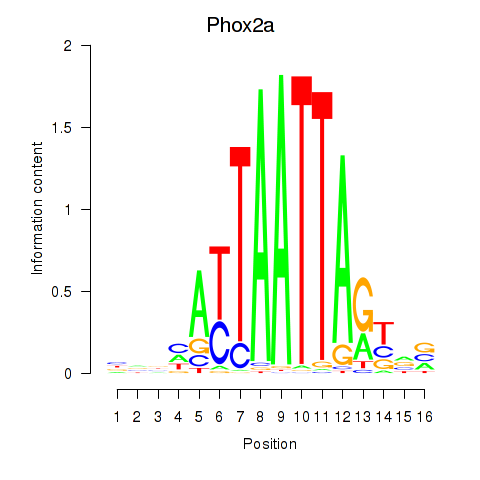
| Phox2a | 0.19 |
|
|
|
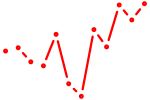
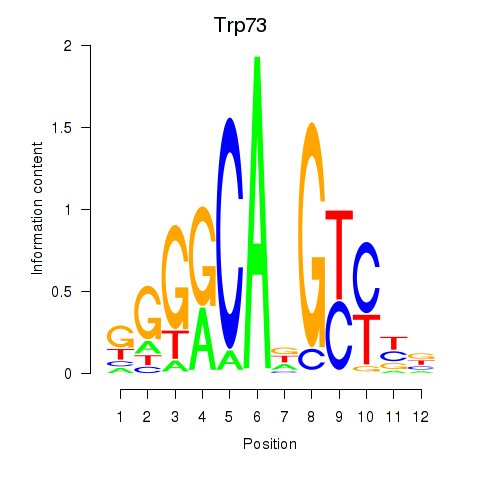
| Trp73 | 0.19 |
|
|
|
| AGUGGUU | 0.18 |
|
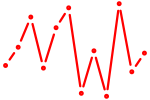
|

|
| UAAUGCU | 0.18 |
|

|

|
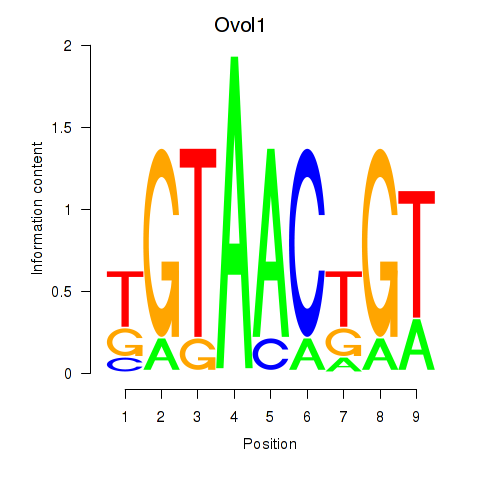
| Ovol1 | 0.18 |
|

|
|
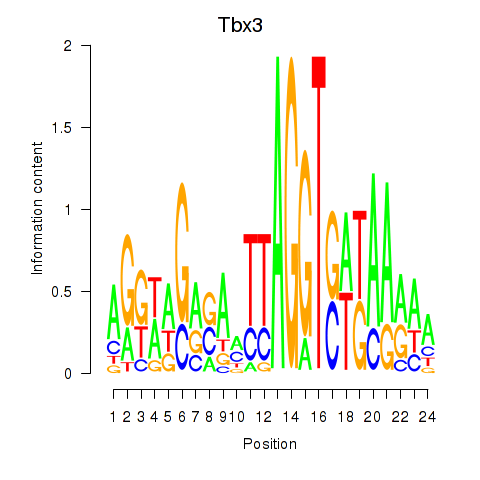
| Tbx3 | 0.18 |
|
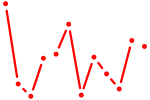
|
|
| AGGUAGU | 0.17 |
|
|

|
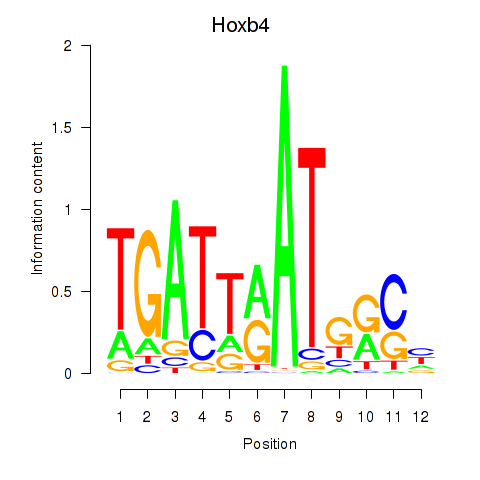
| Hoxb4 | 0.17 |
|
|
|
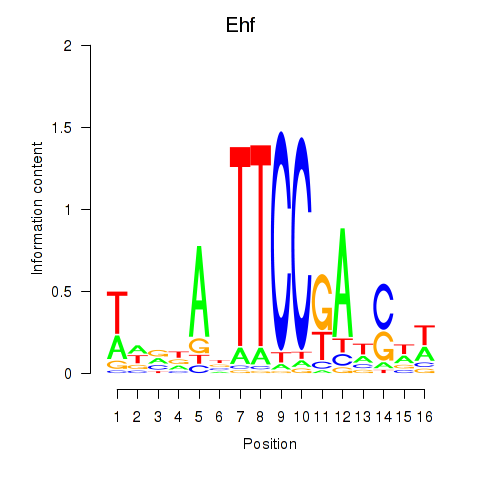
| Ehf | 0.17 |
|
|
|
| CCAGCAU | 0.16 |
|
|

|
| AAGGUGC | 0.16 |
|
|
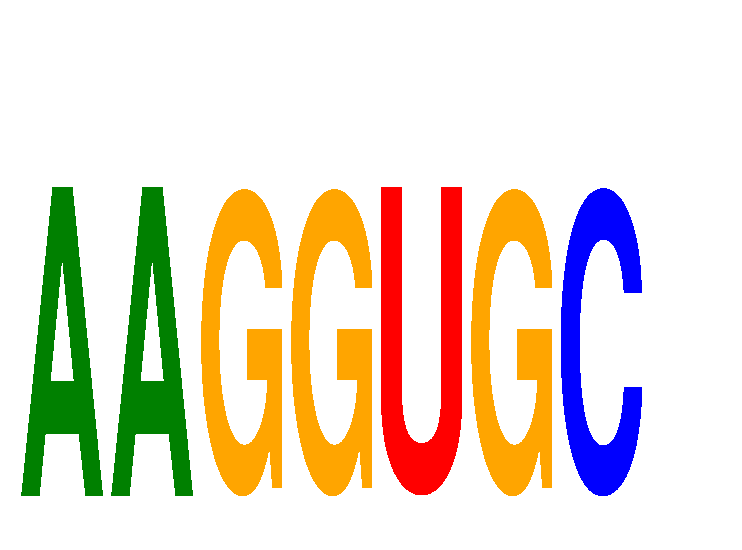
|
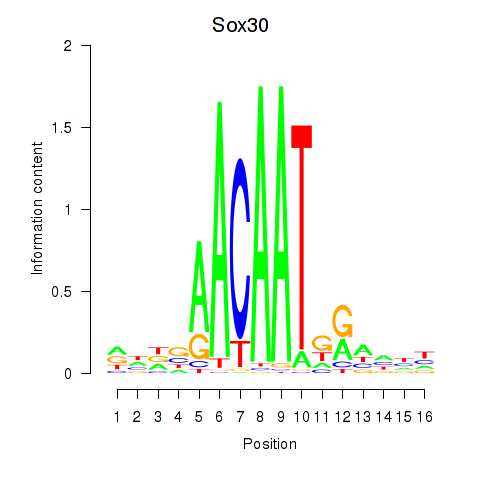
| Sox30 | 0.16 |
|
|
|
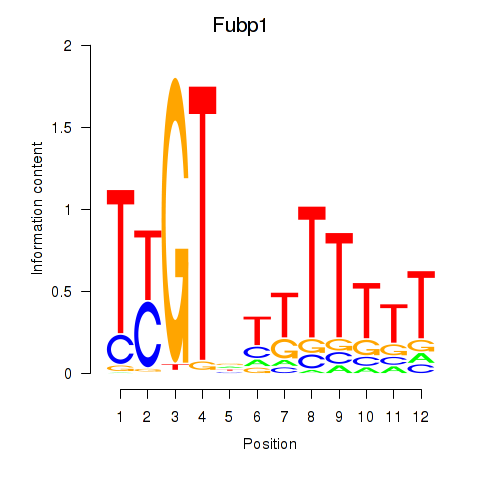
| Fubp1 | 0.16 |
|
|
|
| GUAACAG | 0.16 |
|

|

|
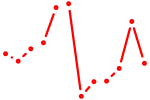
| UCCCUUU | 0.15 |
|
|
|
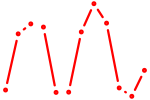
| UGUGCGU | 0.15 |
|
|

|
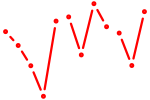
| GAGAUGA | 0.14 |
|
|

|
| GGAGUGU | 0.13 |
|
|

|
| AUGUGCC | 0.12 |
|
|

|
| Hoxa3 | 0.12 |
|

|
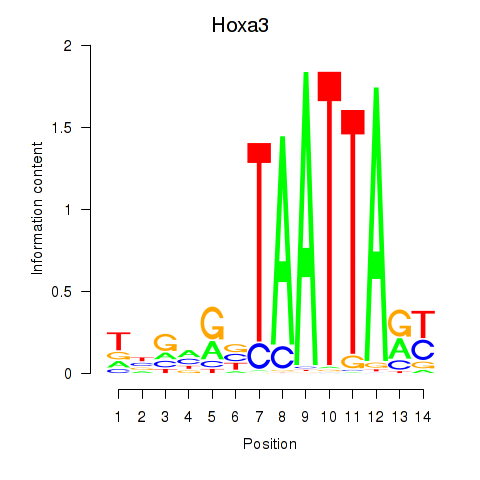
|
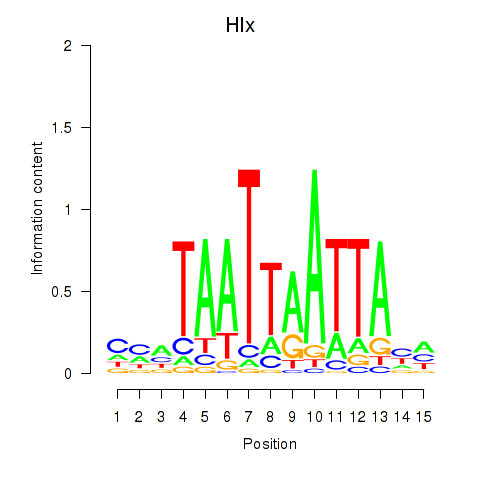
| Hlx | 0.12 |
|
|
|
| UGCAUUG | 0.12 |
|
|

|
| AACGGAA | 0.11 |
|
|
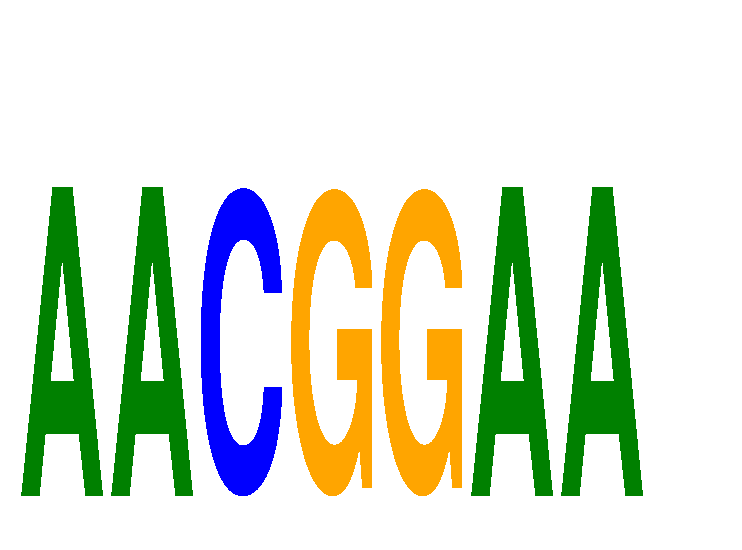
|
| GGCAAGA | 0.09 |
|
|

|
| GAGAACU | 0.09 |
|
|

|
| AACCUGG | 0.09 |
|

|
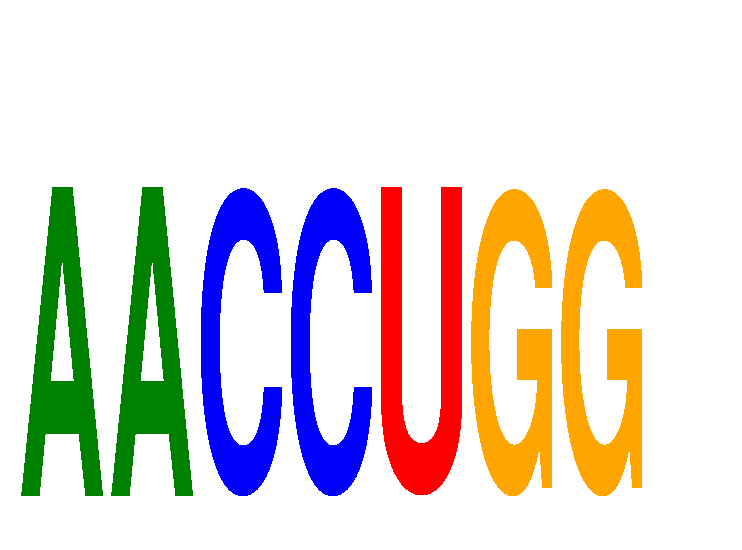
|
| UAAGACU | 0.09 |
|
|

|
| GAUAUGU | 0.08 |
|
|

|
| UGACCUA | 0.06 |
|
|

|
| Glis1 | 0.05 |
|
|
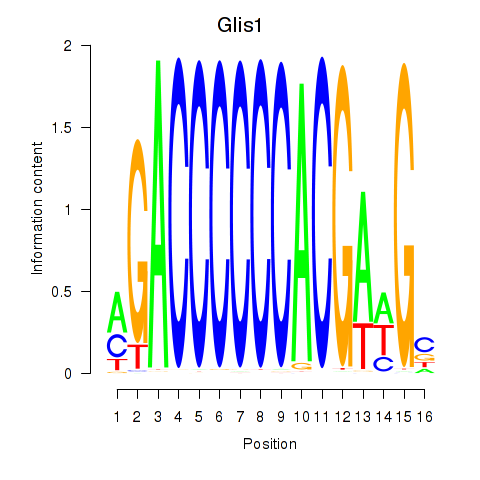
|
| AUGACAC | 0.05 |
|
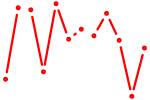
|

|
| AAUCUCU | 0.05 |
|
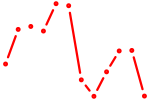
|

|
| AAUCUCA | 0.04 |
|
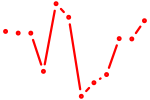
|

|
| CUACAGU | 0.03 |
|

|

|
| AACCGUU | 0.03 |
|
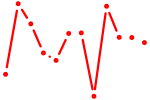
|
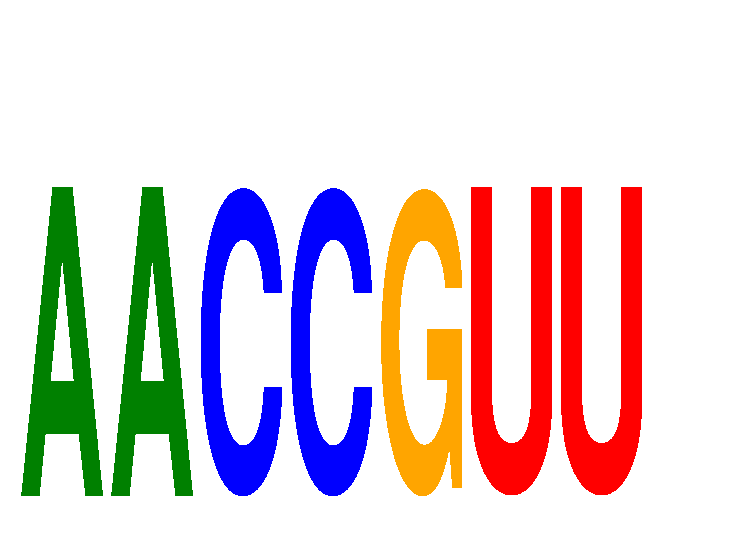
|
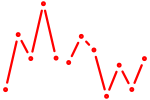
| CGUGUCU | 0.02 |
|
|

|
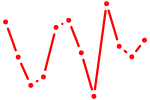
| CUCCCAA | 0.01 |
|
|

|
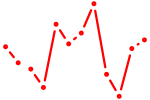
| GGACGGA | 0.00 |
|
|

|