| WM name | Z-value | Associated genes | Logo |
|---|
|
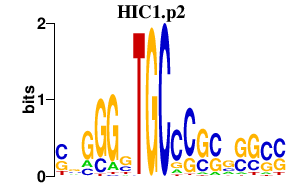
HIC1.p2
|
7.762
|
HIC1
|
|
|
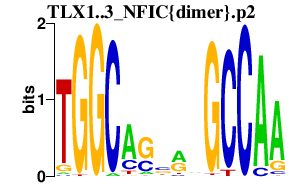
TLX1..3_NFIC{dimer}.p2
|
6.768
|
TLX1
TLX3
|
|
|
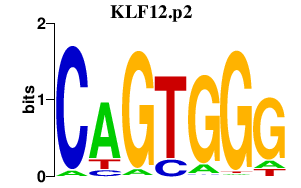
KLF12.p2
|
5.995
|
KLF12
|
|
|
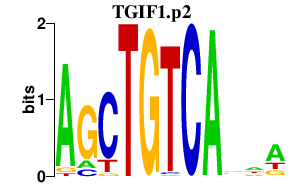
TGIF1.p2
|
5.629
|
TGIF1
|
|
|
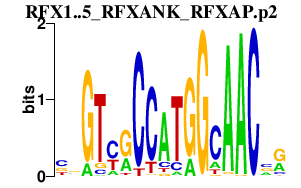
RFX1..5_RFXANK_RFXAP.p2
|
5.295
|
RFX1
RFXAP
RFX5
RFXANK
RFX4
RFX3
RFX2
|
|
|
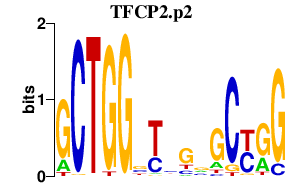
TFCP2.p2
|
4.882
|
TFCP2
|
|
|
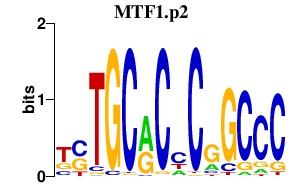
MTF1.p2
|
4.650
|
MTF1
|
|
|
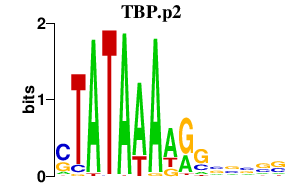
TBP.p2
|
4.630
|
TBP
|
|
|
ZIC1..3.p2
|
4.627
|
ZIC1
ZIC2
ZIC3
|
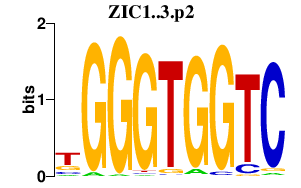
|
|
SMAD1..7,9.p2
|
4.481
|
SMAD5
SMAD1
SMAD3
SMAD4
SMAD2
SMAD6
SMAD7
SMAD9
|
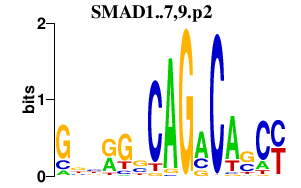
|
|
RREB1.p2
|
4.341
|
RREB1
|
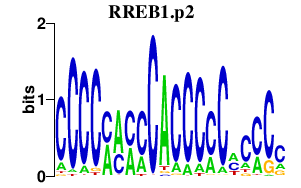
|
|
NHLH1,2.p2
|
4.299
|
NHLH1
NHLH2
|
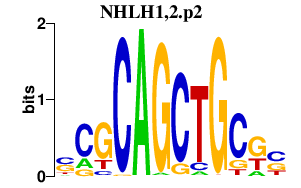
|
|
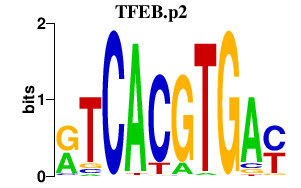
TFEB.p2
|
4.200
|
TFEB
|
|
|
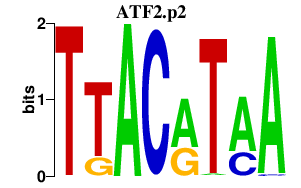
ATF2.p2
|
4.056
|
ATF2
|
|
|
AAGGCAC
|
3.882
|
|

|
|
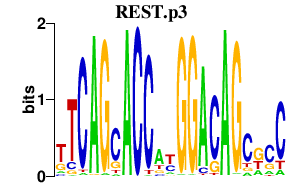
REST.p3
|
3.861
|
REST
|
|
|
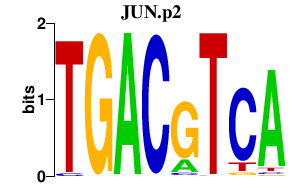
JUN.p2
|
3.842
|
JUN
JUND
JUNB
|
|
|
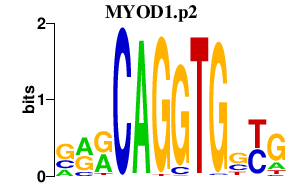
MYOD1.p2
|
3.713
|
MYOD1
|
|
|
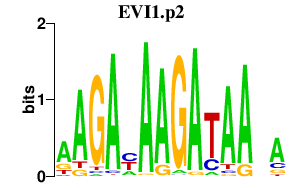
EVI1.p2
|
3.679
|
EVI1
|
|
|
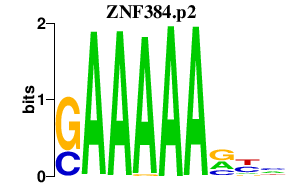
ZNF384.p2
|
3.628
|
ZNF384
|
|
|
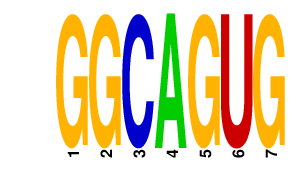
GGCAGUG
|
3.523
|
|
|
|
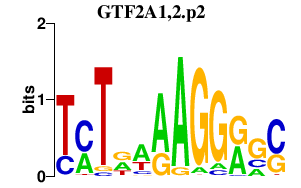
GTF2A1,2.p2
|
3.482
|
GTF2A1
GTF2A2
|
|
|
CAGCAGG
|
3.462
|
|

|
|
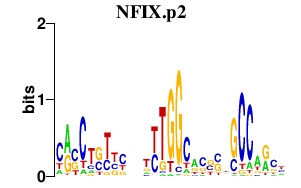
NFIX.p2
|
3.455
|
NFIX
|
|
|
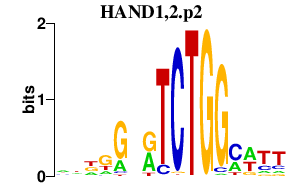
HAND1,2.p2
|
3.340
|
HAND1
HAND2
|
|
|
GUCAGUU
|
3.337
|
|

|
|
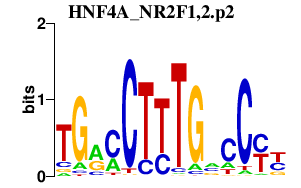
HNF4A_NR2F1,2.p2
|
3.193
|
NR2F1
NR2F2
HNF4A
|
|
|
AGUGCAA
|
3.138
|
|

|
|
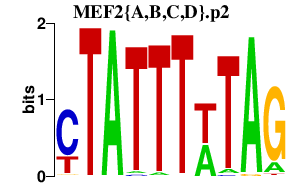
MEF2{A,B,C,D}.p2
|
3.054
|
MEF2A
MEF2C
MEF2D
MEF2BNB-MEF2B
MEF2B
|
|
|
GATA1..3.p2
|
3.053
|
GATA1
GATA2
GATA3
|

|
|
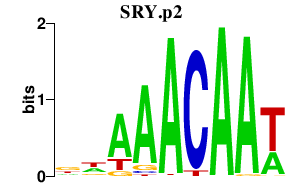
SRY.p2
|
2.951
|
SRY
|
|
|
UGCAUAG
|
2.925
|
|

|
|
GGCUCAG
|
2.845
|
|

|
|
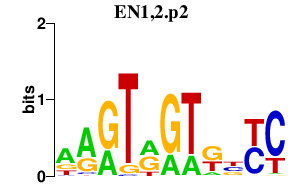
EN1,2.p2
|
2.775
|
EN1
EN2
|
|
|
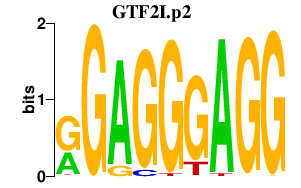
GTF2I.p2
|
2.645
|
GTF2I
|
|
|
AGCAGCA
|
2.613
|
|

|
|
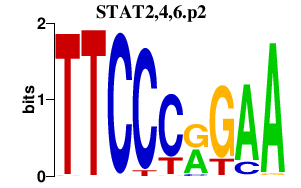
STAT2,4,6.p2
|
2.606
|
STAT2
STAT4
STAT6
|
|
|
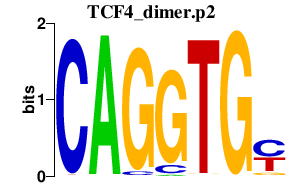
TCF4_dimer.p2
|
2.570
|
TCF4
|
|
|
NFATC1..3.p2
|
2.561
|
NFATC3
NFATC2
NFATC1
|

|
|
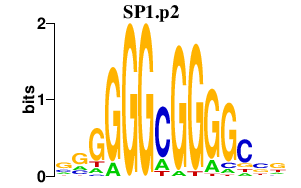
SP1.p2
|
2.549
|
SP1
|
|
|
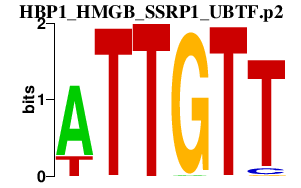
HBP1_HMGB_SSRP1_UBTF.p2
|
2.534
|
HMGB2
HMGB3
SSRP1
UBTF
HBP1
|
|
|
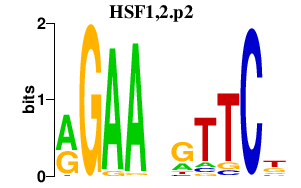
HSF1,2.p2
|
2.490
|
HSF1
HSF2
|
|
|
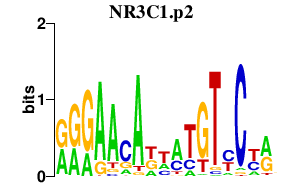
NR3C1.p2
|
2.478
|
NR3C1
|
|
|
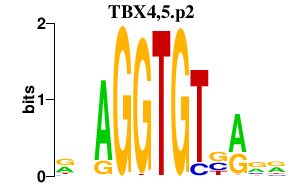
TBX4,5.p2
|
2.470
|
TBX5
TBX4
|
|
|
CACAGUG
|
2.443
|
|

|
|
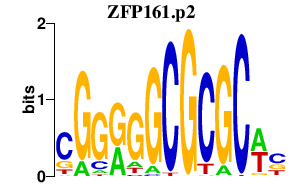
ZFP161.p2
|
2.409
|
ZFP161
|
|
|
TAL1_TCF{3,4,12}.p2
|
2.405
|
TCF3
TCF4
TAL1
TCF12
|
|
|
EBF1.p2
|
2.400
|
EBF1
|
|
|
TEAD1.p2
|
2.384
|
TEAD1
TEAD2
TEAD3
TEAD4
|
|
|
SOX2.p2
|
2.384
|
SOX2
|
|
|
MZF1.p2
|
2.331
|
MZF1
|
|
|
LMO2.p2
|
2.279
|
LMO2
|
|
|
SOX{8,9,10}.p2
|
2.259
|
SOX9
SOX8
SOX10
|
|
|
CCAGCAU
|
2.259
|
|

|
|
NFIL3.p2
|
2.229
|
NFIL3
|
|
|
PAX4.p2
|
2.218
|
PAX4
|
|
|
UAAGACU
|
2.199
|
|

|
|
MYFfamily.p2
|
2.196
|
MYOD1
MYF6
MYF5
MYOG
|
|
|
NKX3-2.p2
|
2.164
|
NKX3-2
|
|
|
FOX{I1,J2}.p2
|
2.089
|
FOXI1
FOXJ2
|
|
|
SPZ1.p2
|
2.081
|
SPZ1
|
|
|
AUUGCAC
|
2.067
|
|

|
|
FOXP3.p2
|
2.064
|
FOXP3
|
|
|
UAAGACG
|
2.058
|
|

|
|
UCCCUUU
|
1.994
|
|
|
|
ZBTB6.p2
|
1.964
|
ZBTB6
|
|
|
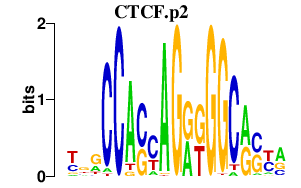
CTCF.p2
|
1.961
|
CTCF
|
|
|
UUGGCAC
|
1.961
|
|

|
|
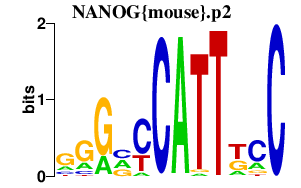
NANOG{mouse}.p2
|
1.917
|
NANOG
|
|
|
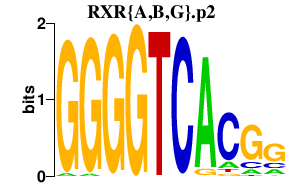
RXR{A,B,G}.p2
|
1.894
|
RXRA
RXRB
RXRG
|
|
|
AUGGCUU
|
1.890
|
|

|
|
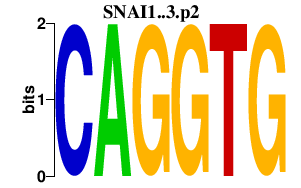
SNAI1..3.p2
|
1.863
|
SNAI2
SNAI1
SNAI3
|
|
|
POU5F1_SOX2{dimer}.p2
|
1.853
|
POU5F1
SOX2
|
|
|
POU5F1.p2
|
1.831
|
POU5F1
|

|
|
GAGAACU
|
1.816
|
|
|
|
EOMES.p2
|
1.815
|
EOMES
|
|
|
MAFB.p2
|
1.739
|
MAFB
|
|
|
AAAGUGC
|
1.703
|
|

|
|
UGUGCUU
|
1.597
|
|

|
|
FEV.p2
|
1.596
|
FEV
|
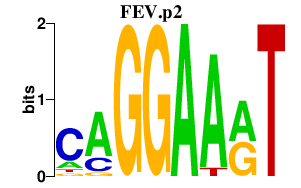
|
|
FOXO1,3,4.p2
|
1.596
|
FOXO1
FOXO4
FOXO3
|
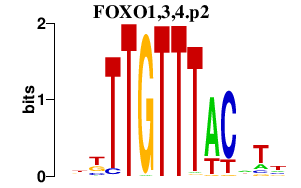
|
|
ESR1.p2
|
1.526
|
ESR1
|
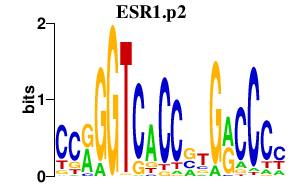
|
|
MSX1,2.p2
|
1.521
|
MSX1
MSX2
|

|
|
FOS_FOS{B,L1}_JUN{B,D}.p2
|
1.507
|
FOS
JUNB
FOSL1
JUND
FOSB
|

|
|
PAX6.p2
|
1.423
|
PAX6
|

|
|
MAZ.p2
|
1.349
|
MAZ
|
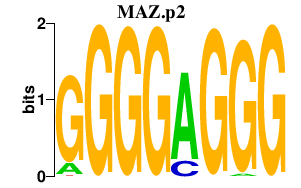
|
|
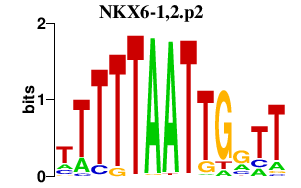
NKX6-1,2.p2
|
1.335
|
NKX6-1
NKX6-2
|
|
|
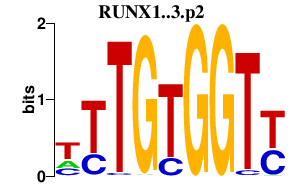
RUNX1..3.p2
|
1.333
|
RUNX1
RUNX3
RUNX2
|
|
|
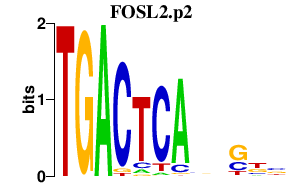
FOSL2.p2
|
1.329
|
FOSL2
|
|
|
AAUACUG
|
1.296
|
|

|
|
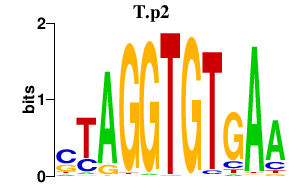
T.p2
|
1.295
|
T
|
|
|
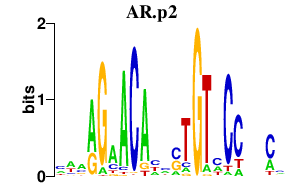
AR.p2
|
1.256
|
AR
|
|
|
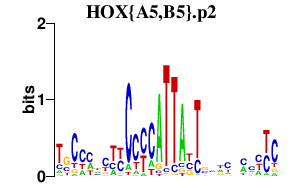
HOX{A5,B5}.p2
|
1.256
|
HOXB5
HOXA5
|
|
|
GGCAAGA
|
1.233
|
|

|
|
ESRRA.p2
|
1.186
|
ESRRA
|

|
|
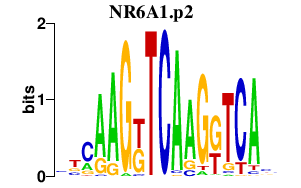
NR6A1.p2
|
1.141
|
NR6A1
|
|
|
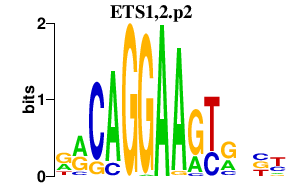
ETS1,2.p2
|
1.125
|
ETS2
ETS1
|
|
|
GAUCAGA
|
1.124
|
|

|
|
UUGGUCC
|
1.106
|
|

|
|
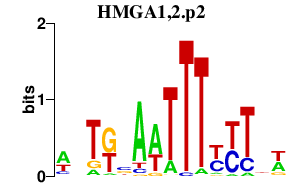
HMGA1,2.p2
|
1.097
|
HMGA1
HMGA2
|
|
|
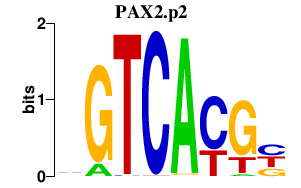
PAX2.p2
|
1.097
|
PAX2
|
|
|
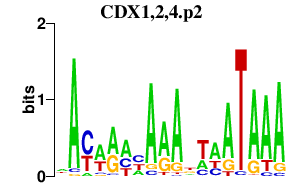
CDX1,2,4.p2
|
1.062
|
CDX1
CDX2
CDX4
|
|
|
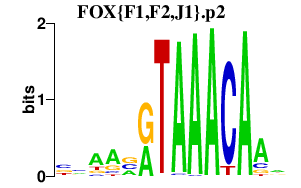
FOX{F1,F2,J1}.p2
|
1.052
|
FOXF2
FOXJ1
FOXF1
|
|
|
CGUGUCU
|
1.045
|
|

|
|
AAGUGCU
|
1.032
|
|
|
|
STAT1,3.p3
|
1.011
|
STAT1
STAT3
|
|
|
EWSR1-FLI1.p2
|
0.984
|
FLI1
EWSR1
|
|
|
NR5A1,2.p2
|
0.968
|
NR5A1
NR5A2
|
|
|
GUGCAAA
|
0.934
|
|

|
|
AGCUGCC
|
0.912
|
|

|
|
PRRX1,2.p2
|
0.882
|
PRRX2
PRRX1
|

|
|
HOX{A6,A7,B6,B7}.p2
|
0.851
|
HOXB6
HOXA7
HOXB7
HOXA6
|

|
|
RXRA_VDR{dimer}.p2
|
0.850
|
VDR
|

|
|
GGAAGAC
|
0.836
|
|

|
|
SRF.p3
|
0.829
|
SRF
|
|
|
GLI1..3.p2
|
0.825
|
GLI1
GLI2
GLI3
|
|
|
CEBPA,B_DDIT3.p2
|
0.810
|
DDIT3
CEBPA
CEBPB
|
|
|
NR4A2.p2
|
0.802
|
NR4A2
|

|
|
MYBL2.p2
|
0.796
|
MYBL2
|
|
|
GAGAUGA
|
0.791
|
|

|
|
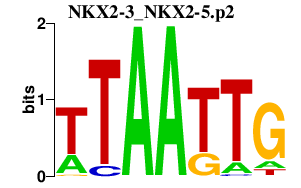
NKX2-3_NKX2-5.p2
|
0.785
|
NKX2-5
NKX2-3
|
|
|
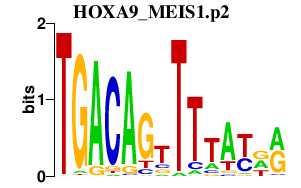
HOXA9_MEIS1.p2
|
0.756
|
MEIS1
HOXA9
|
|
|
UGCAUUG
|
0.733
|
|

|
|
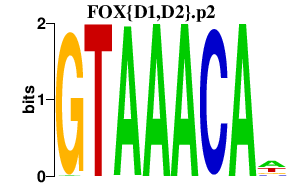
FOX{D1,D2}.p2
|
0.723
|
FOXD2
FOXD1
|
|
|
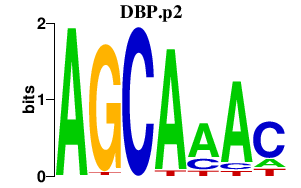
DBP.p2
|
0.697
|
DBP
|
|
|
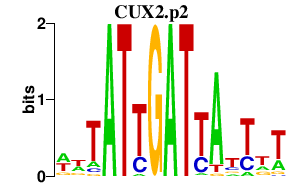
CUX2.p2
|
0.695
|
CUX2
|
|
|
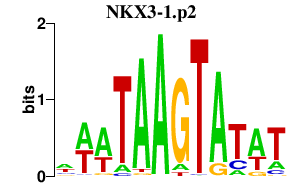
NKX3-1.p2
|
0.643
|
NKX3-1
|
|
|
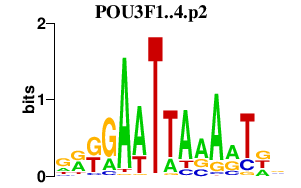
POU3F1..4.p2
|
0.572
|
POU3F1
POU3F3
POU3F2
POU3F4
|
|
|
CUACAGU
|
0.536
|
|

|
|
AACACUG
|
0.510
|
|

|
|
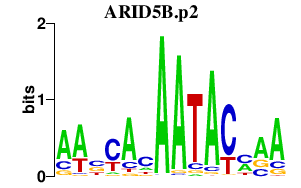
ARID5B.p2
|
0.496
|
ARID5B
|
|
|
CCAGUGU
|
0.465
|
|

|
|
AUGACAC
|
0.460
|
|

|
|
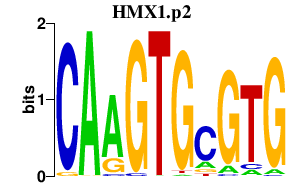
HMX1.p2
|
0.447
|
HMX1
|
|
|
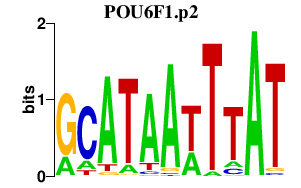
POU6F1.p2
|
0.435
|
POU6F1
|
|
|
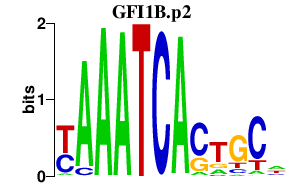
GFI1B.p2
|
0.382
|
GFI1B
|
|
|
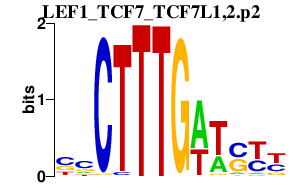
LEF1_TCF7_TCF7L1,2.p2
|
0.360
|
TCF7
TCF7L2
LEF1
TCF7L1
|
|
|
GUAGUGU
|
0.317
|
|

|
|
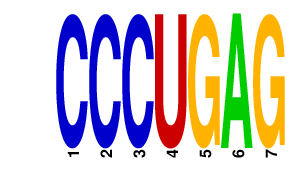
CCCUGAG
|
0.300
|
|
|
|
UGUGCGU
|
0.263
|
|

|
|
AAGGUGC
|
0.262
|
|

|
|
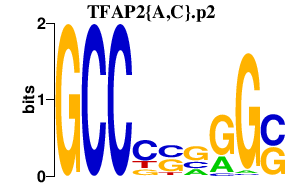
TFAP2{A,C}.p2
|
0.226
|
TFAP2C
TFAP2A
|
|
|
AGCACCA
|
0.218
|
|

|
|
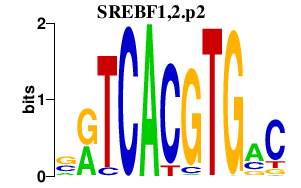
SREBF1,2.p2
|
0.217
|
SREBF1
SREBF2
|
|
|
AGUGGUU
|
0.191
|
|

|
|
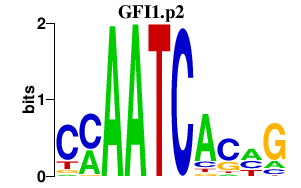
GFI1.p2
|
0.175
|
GFI1
|
|
|
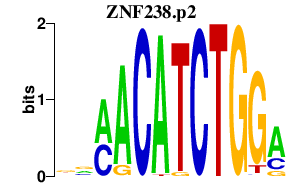
ZNF238.p2
|
0.170
|
ZNF238
|
|
|
GCUACAU
|
0.149
|
|

|
|
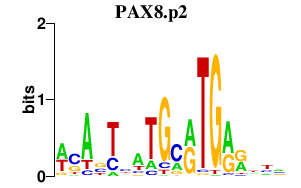
PAX8.p2
|
0.129
|
PAX8
|
|
|
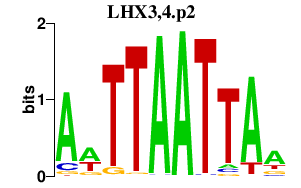
LHX3,4.p2
|
0.126
|
LHX3
LHX4
|
|
|
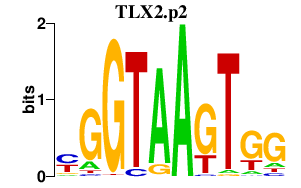
TLX2.p2
|
0.094
|
TLX2
|
|
|
NKX2-1,4.p2
|
0.089
|
NKX2-1
NKX2-4
|
|
|
FOXL1.p2
|
0.080
|
FOXL1
|
|
|
PAX3,7.p2
|
0.076
|
PAX7
PAX3
|
|
|
ALX1.p2
|
0.018
|
ALX1
|
|
|
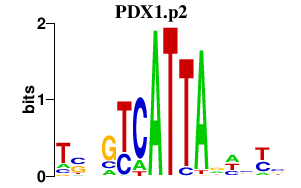
PDX1.p2
|
0.015
|
PDX1
|
|
|
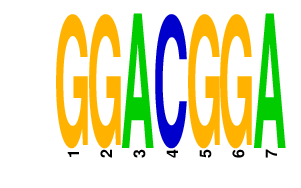
GGACGGA
|
0.012
|
|
|
|
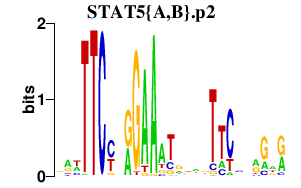
STAT5{A,B}.p2
|
0.001
|
STAT5B
STAT5A
|
|
|
GCUGGUG
|
-0.018
|
|

|
|
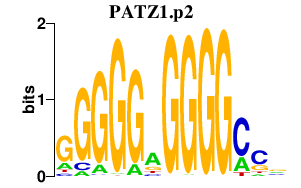
PATZ1.p2
|
-0.061
|
PATZ1
|
|
|
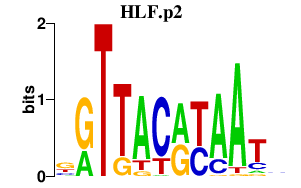
HLF.p2
|
-0.065
|
HLF
|
|
|
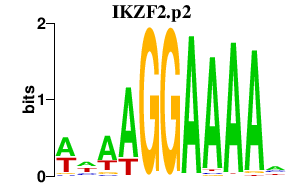
IKZF2.p2
|
-0.067
|
IKZF2
|
|
|
UGACCUA
|
-0.103
|
|

|
|
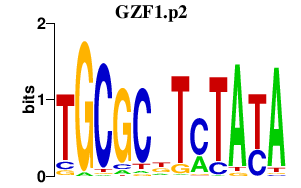
GZF1.p2
|
-0.109
|
GZF1
|
|
|
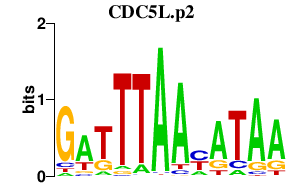
CDC5L.p2
|
-0.112
|
CDC5L
|
|
|
GUAACAG
|
-0.118
|
|

|
|
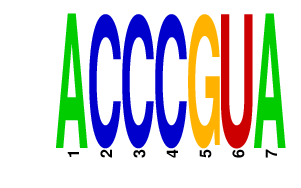
ACCCGUA
|
-0.124
|
|
|
|
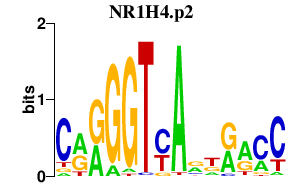
NR1H4.p2
|
-0.125
|
NR1H4
|
|
|
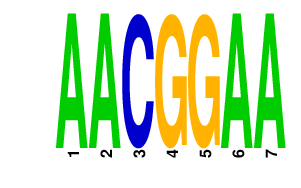
AACGGAA
|
-0.130
|
|
|
|
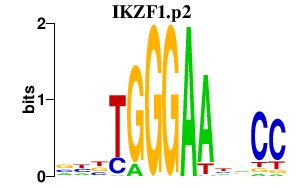
IKZF1.p2
|
-0.173
|
IKZF1
|
|
|
UCACAGU
|
-0.184
|
|

|
|
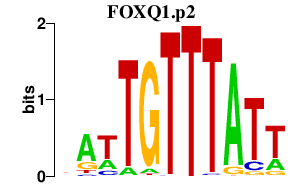
FOXQ1.p2
|
-0.211
|
FOXQ1
|
|
|
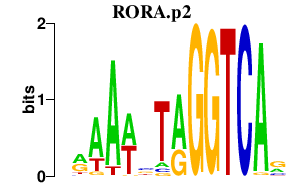
RORA.p2
|
-0.228
|
RORA
|
|
|
CCUUCAU
|
-0.281
|
|

|
|
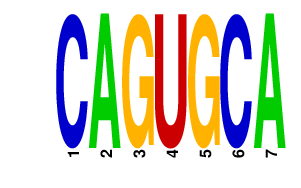
NANOG.p2
|
-0.314
|
NANOG
|

|
|
CAGUGCA
|
-0.319
|
|
|
|
CGUACCG
|
-0.357
|
|

|
|
AAUCUCA
|
-0.364
|
|

|
|
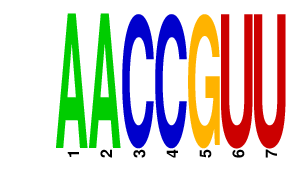
AACCGUU
|
-0.391
|
|
|
|
CUCCCAA
|
-0.400
|
|

|
|
AAUCUCU
|
-0.408
|
|

|
|
UAUUGCU
|
-0.417
|
|

|
|
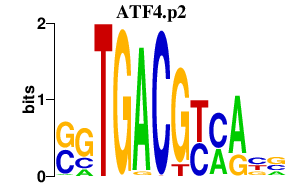
ATF4.p2
|
-0.428
|
ATF4
|
|
|
UCCAGUU
|
-0.439
|
|

|
|
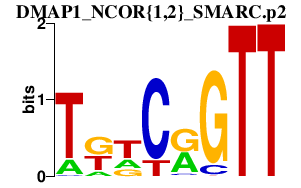
DMAP1_NCOR{1,2}_SMARC.p2
|
-0.445
|
SMARCA1
SMARCC2
SMARCA5
NCOR1
NCOR2
DMAP1
|
|
|
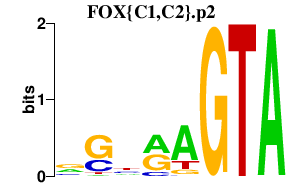
FOX{C1,C2}.p2
|
-0.454
|
FOXC1
FOXC2
|
|
|
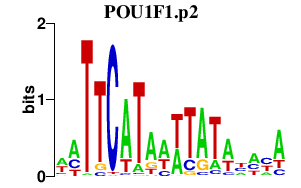
POU1F1.p2
|
-0.463
|
POU1F1
|
|
|
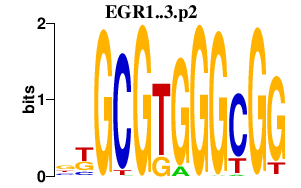
EGR1..3.p2
|
-0.465
|
EGR3
EGR1
EGR2
|
|
|
AGCAGCG
|
-0.475
|
|

|
|
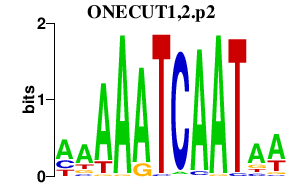
ONECUT1,2.p2
|
-0.521
|
ONECUT2
ONECUT1
|
|
|
AACAGUC
|
-0.537
|
|

|
|
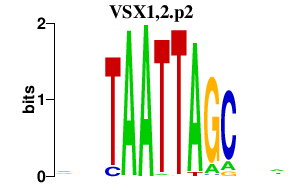
VSX1,2.p2
|
-0.578
|
VSX2
VSX1
|
|
|
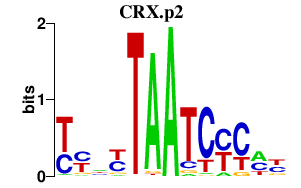
CRX.p2
|
-0.584
|
CRX
|
|
|
AIRE.p2
|
-0.607
|
AIRE
|
|
|
NFE2L1.p2
|
-0.615
|
NFE2L1
|

|
|
UAAUGCU
|
-0.620
|
|

|
|
GGAGUGU
|
-0.627
|
|
|
|
PBX1.p2
|
-0.676
|
PBX1
|
|
|
NKX2-2,8.p2
|
-0.738
|
NKX2-2
NKX2-8
|
|
|
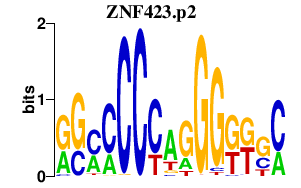
ZNF423.p2
|
-0.770
|
ZNF423
|
|
|
ACCCUGU
|
-0.808
|
|

|
|
ACUGCAU
|
-0.822
|
|

|
|
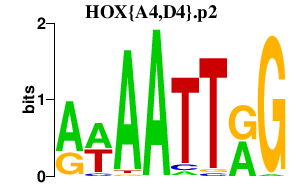
HOX{A4,D4}.p2
|
-0.829
|
HOXA4
HOXD4
|
|
|
GAGGUAG
|
-0.843
|
|

|
|
UUGUUCG
|
-0.860
|
|

|
|
ACUGGCC
|
-0.886
|
|

|
|
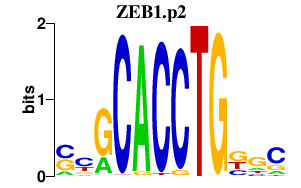
ZEB1.p2
|
-0.900
|
ZEB1
|
|
|
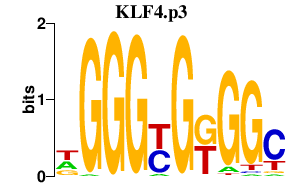
KLF4.p3
|
-0.955
|
KLF4
|
|
|
AACCUGG
|
-0.974
|
|

|
|
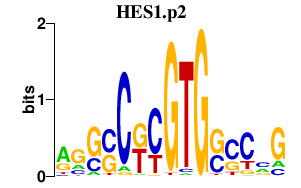
HES1.p2
|
-1.041
|
HES1
|
|
|
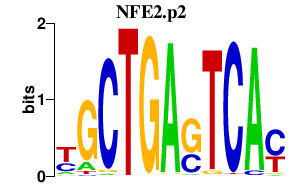
NFE2.p2
|
-1.056
|
NFE2
|
|
|
AGGUAGU
|
-1.061
|
|

|
|
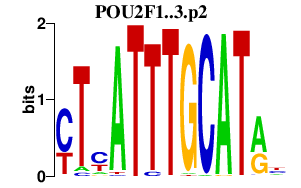
POU2F1..3.p2
|
-1.069
|
POU2F2
POU2F1
POU2F3
|
|
|
ACAUUCA
|
-1.104
|
|

|
|
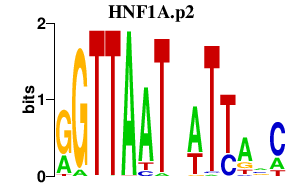
HNF1A.p2
|
-1.113
|
HNF1A
|
|
|
ATF6.p2
|
-1.178
|
ATF6
|

|
|
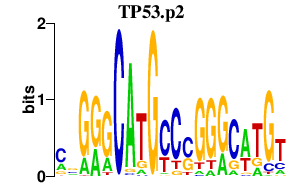
TP53.p2
|
-1.273
|
TP53
|
|
|
GAUAUGU
|
-1.315
|
|

|
|
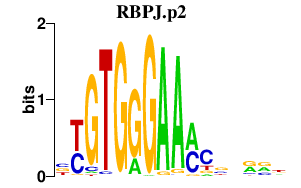
RBPJ.p2
|
-1.317
|
RBPJ
|
|
|
AUGGCAC
|
-1.344
|
|

|
|
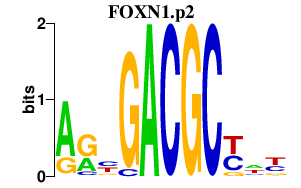
FOXN1.p2
|
-1.346
|
FOXN1
|
|
|
AGCUUAU
|
-1.373
|
|

|
|
AAUGCCC
|
-1.443
|
|

|
|
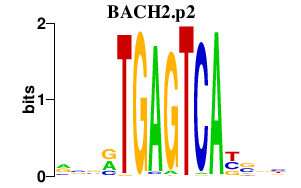
BACH2.p2
|
-1.449
|
BACH2
|
|
|
FOXA2.p3
|
-1.519
|
FOXA2
|

|
|
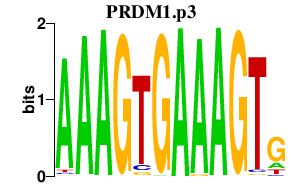
PRDM1.p3
|
-1.528
|
PRDM1
|
|
|
UCAAGUA
|
-1.552
|
|

|
|
ACAGUAU
|
-1.608
|
|

|
|
GCAGCAU
|
-1.698
|
|

|
|
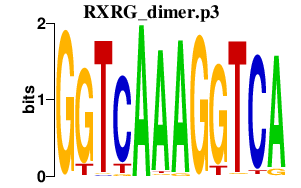
RXRG_dimer.p3
|
-1.702
|
NR1H2
RXRA
PPARG
PPARA
RXRB
RXRG
|
|
|
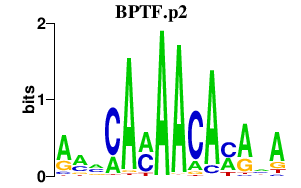
BPTF.p2
|
-1.708
|
BPTF
|
|
|
AUGUGCC
|
-1.712
|
|

|
|
UCACAUU
|
-1.783
|
|

|
|
ACAGUAC
|
-1.798
|
|

|
|
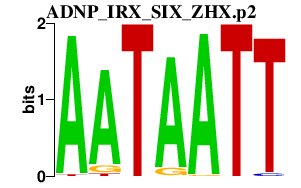
ADNP_IRX_SIX_ZHX.p2
|
-1.825
|
SIX5
ZHX1
IRX4
ADNP
ZHX3
ZHX2
IRX5
SIX2
|
|
|
NFE2L2.p2
|
-1.865
|
NFE2L2
|
|
|
TFAP2B.p2
|
-1.868
|
TFAP2B
|
|
|
UGAAAUG
|
-1.981
|
|

|
|
UUGGCAA
|
-2.004
|
|

|
|
GUAAACA
|
-2.055
|
|
|
|
PPARG.p2
|
-2.103
|
PPARG
|
|
|
PAX5.p2
|
-2.153
|
PAX5
|
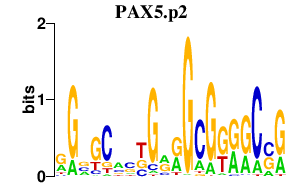
|
|
TFAP4.p2
|
-2.215
|
TFAP4
|
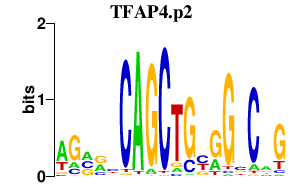
|
|
CREB1.p2
|
-2.230
|
CREB1
|
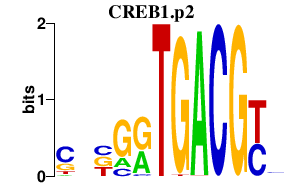
|
|
HIF1A.p2
|
-2.285
|
HIF1A
|
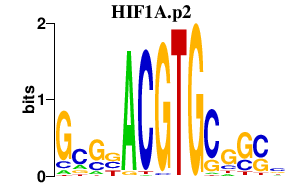
|
|
CUUUGGU
|
-2.303
|
|

|
|
ZNF148.p2
|
-2.333
|
ZNF148
|
|
|
SOX17.p2
|
-2.391
|
SOX17
|
|
|
GATA6.p2
|
-2.452
|
GATA6
|
|
|
GGAAUGU
|
-2.869
|
|

|
|
SPIB.p2
|
-2.903
|
SPIB
|
|
|
NFKB1_REL_RELA.p2
|
-2.924
|
RELA
REL
NFKB1
|
|
|
UUUUUGC
|
-3.020
|
|

|
|
IRF1,2,7.p3
|
-3.059
|
IRF1
IRF2
IRF7
|
|
|
ZBTB16.p2
|
-3.221
|
ZBTB16
|
|
|
SPI1.p2
|
-3.474
|
SPI1
|
|
|
SOX5.p2
|
-3.585
|
SOX5
|

|
|
XBP1.p3
|
-3.614
|
XBP1
|
|
|
FOXD3.p2
|
-3.635
|
FOXD3
|

|
|
EP300.p2
|
-3.707
|
EP300
|
|
|
bHLH_family.p2
|
-3.751
|
ARNTL
MITF
MXI1
TFE3
CLOCK
OLIG2
MXD4
HEY2
HEY1
HEYL
MLXIPL
HES6
MXD3
ARNTL2
ID1
MNT
OLIG1
NPAS2
|
|
|
PITX1..3.p2
|
-4.165
|
PITX2
PITX3
PITX1
|
|
|
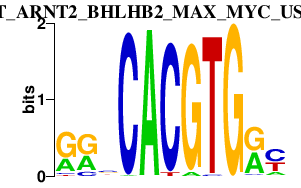
ARNT_ARNT2_BHLHB2_MAX_MYC_USF1.p2
|
-4.371
|
ARNT
MAX
MYC
USF1
BHLHB2
ARNT2
|
|
|
AHR_ARNT_ARNT2.p2
|
-4.682
|
ARNT
AHR
ARNT2
|

|
|
GAUUGUC
|
-4.957
|
|

|
|
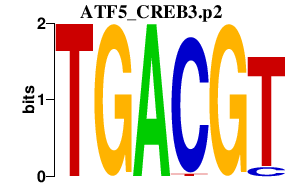
ATF5_CREB3.p2
|
-5.193
|
ATF5
CREB3
|
|
|
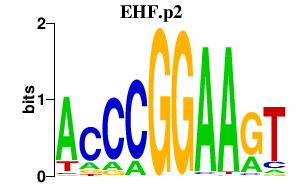
EHF.p2
|
-5.262
|
EHF
|
|
|
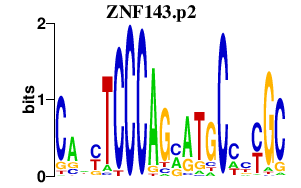
ZNF143.p2
|
-5.983
|
ZNF143
|
|
|
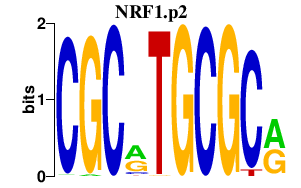
NRF1.p2
|
-6.728
|
NRF1
|
|
|
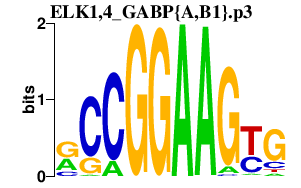
ELK1,4_GABP{A,B1}.p3
|
-7.048
|
GABPA
GABPB1
ELK4
ELK1
|
|
|
TFDP1.p2
|
-7.105
|
TFDP1
|

|
|
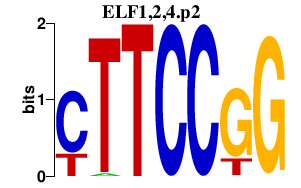
ELF1,2,4.p2
|
-7.516
|
ELF2
ELF1
ELF4
|
|
|
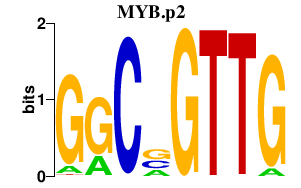
MYB.p2
|
-8.395
|
MYB
|
|
|
NFY{A,B,C}.p2
|
-8.813
|
NFYC
NFYB
NFYA
|
|
|
E2F1..5.p2
|
-9.665
|
E2F4
E2F5
E2F2
E2F1
E2F3
|
|
|
YY1.p2
|
-10.669
|
YY1
|
|