| WM name | Z-value | Associated genes | Logo |
|---|
|
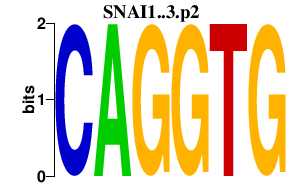
SNAI1..3.p2
|
11.175
|
SNAI2
SNAI1
SNAI3
|
|
|
FOS_FOS{B,L1}_JUN{B,D}.p2
|
9.888
|
FOS
JUNB
FOSL1
JUND
FOSB
|

|
|
HOX{A6,A7,B6,B7}.p2
|
6.877
|
HOXB6
HOXA7
HOXB7
HOXA6
|

|
|
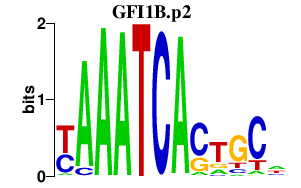
GFI1B.p2
|
5.881
|
GFI1B
|
|
|
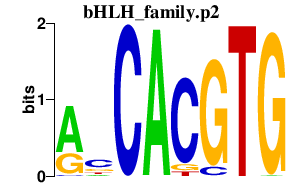
bHLH_family.p2
|
4.546
|
ARNTL
MITF
MXI1
TFE3
CLOCK
OLIG2
MXD4
HEY2
HEY1
HEYL
MLXIPL
HES6
MXD3
ARNTL2
ID1
MNT
OLIG1
NPAS2
|
|
|
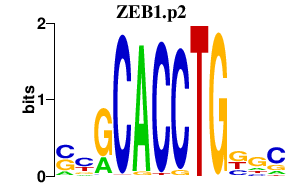
ZEB1.p2
|
4.268
|
ZEB1
|
|
|
SOX{8,9,10}.p2
|
3.523
|
SOX9
SOX8
SOX10
|
|
|
AHR_ARNT_ARNT2.p2
|
3.367
|
ARNT
AHR
ARNT2
|

|
|
GFI1.p2
|
3.278
|
GFI1
|
|
|
STAT5{A,B}.p2
|
2.983
|
STAT5B
STAT5A
|
|
|
RBPJ.p2
|
2.914
|
RBPJ
|
|
|
HLF.p2
|
2.710
|
HLF
|
|
|
NFIX.p2
|
2.539
|
NFIX
|
|
|
EP300.p2
|
2.525
|
EP300
|
|
|
MZF1.p2
|
2.516
|
MZF1
|
|
|
NKX3-2.p2
|
2.469
|
NKX3-2
|
|
|
TEAD1.p2
|
2.364
|
TEAD1
TEAD2
TEAD3
TEAD4
|
|
|
GTF2I.p2
|
2.260
|
GTF2I
|
|
|
SPZ1.p2
|
2.140
|
SPZ1
|
|
|
FOXD3.p2
|
2.053
|
FOXD3
|

|
|
HMX1.p2
|
2.019
|
HMX1
|
|
|
NFKB1_REL_RELA.p2
|
2.008
|
RELA
REL
NFKB1
|
|
|
REST.p3
|
1.914
|
REST
|
|
|
VSX1,2.p2
|
1.909
|
VSX2
VSX1
|
|
|
IKZF1.p2
|
1.876
|
IKZF1
|
|
|
HBP1_HMGB_SSRP1_UBTF.p2
|
1.462
|
HMGB2
HMGB3
SSRP1
UBTF
HBP1
|
|
|
SMAD1..7,9.p2
|
1.399
|
SMAD5
SMAD1
SMAD3
SMAD4
SMAD2
SMAD6
SMAD7
SMAD9
|

|
|
MTF1.p2
|
1.396
|
MTF1
|
|
|
FOXN1.p2
|
1.253
|
FOXN1
|
|
|
EWSR1-FLI1.p2
|
1.240
|
FLI1
EWSR1
|
|
|
EHF.p2
|
1.132
|
EHF
|
|
|
STAT2,4,6.p2
|
1.038
|
STAT2
STAT4
STAT6
|
|
|
FOX{D1,D2}.p2
|
0.917
|
FOXD2
FOXD1
|
|
|
MAFB.p2
|
0.916
|
MAFB
|
|
|
ZBTB6.p2
|
0.857
|
ZBTB6
|
|
|
TBP.p2
|
0.801
|
TBP
|
|
|
ZIC1..3.p2
|
0.703
|
ZIC1
ZIC2
ZIC3
|
|
|
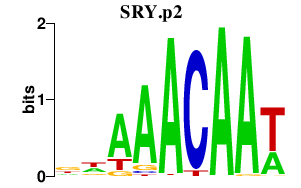
SRY.p2
|
0.673
|
SRY
|
|
|
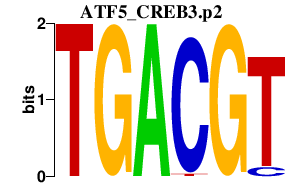
ATF5_CREB3.p2
|
0.654
|
ATF5
CREB3
|
|
|
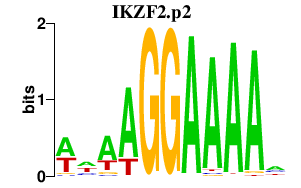
IKZF2.p2
|
0.616
|
IKZF2
|
|
|
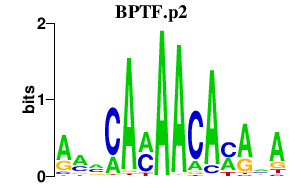
BPTF.p2
|
0.586
|
BPTF
|
|
|
FOX{I1,J2}.p2
|
0.534
|
FOXI1
FOXJ2
|
|
|
PBX1.p2
|
0.464
|
PBX1
|
|
|
EOMES.p2
|
0.393
|
EOMES
|
|
|
RFX1..5_RFXANK_RFXAP.p2
|
0.385
|
RFX1
RFXAP
RFX5
RFXANK
RFX4
RFX3
RFX2
|
|
|
GLI1..3.p2
|
0.368
|
GLI1
GLI2
GLI3
|
|
|
EBF1.p2
|
0.317
|
EBF1
|
|
|
ZNF148.p2
|
0.281
|
ZNF148
|
|
|
NKX2-3_NKX2-5.p2
|
0.252
|
NKX2-5
NKX2-3
|
|
|
KLF4.p3
|
0.252
|
KLF4
|
|
|
NHLH1,2.p2
|
0.247
|
NHLH1
NHLH2
|
|
|
HIC1.p2
|
0.219
|
HIC1
|
|
|
HSF1,2.p2
|
0.210
|
HSF1
HSF2
|
|
|
ATF4.p2
|
0.197
|
ATF4
|
|
|
HES1.p2
|
0.131
|
HES1
|
|
|
ELF1,2,4.p2
|
0.072
|
ELF2
ELF1
ELF4
|
|
|
T.p2
|
0.062
|
T
|
|
|
POU5F1.p2
|
0.034
|
POU5F1
|

|
|
ATF2.p2
|
0.020
|
ATF2
|
|
|
PRRX1,2.p2
|
0.009
|
PRRX2
PRRX1
|

|
|
NFIL3.p2
|
-0.063
|
NFIL3
|
|
|
TFAP2{A,C}.p2
|
-0.065
|
TFAP2C
TFAP2A
|
|
|
RXRA_VDR{dimer}.p2
|
-0.109
|
VDR
|

|
|
NANOG.p2
|
-0.129
|
NANOG
|

|
|
ZNF238.p2
|
-0.154
|
ZNF238
|
|
|
ESR1.p2
|
-0.155
|
ESR1
|
|
|
GZF1.p2
|
-0.211
|
GZF1
|
|
|
TFCP2.p2
|
-0.249
|
TFCP2
|
|
|
TCF4_dimer.p2
|
-0.264
|
TCF4
|
|
|
SOX5.p2
|
-0.268
|
SOX5
|

|
|
RXRG_dimer.p3
|
-0.272
|
NR1H2
RXRA
PPARG
PPARA
RXRB
RXRG
|
|
|
RORA.p2
|
-0.323
|
RORA
|
|
|
PAX8.p2
|
-0.335
|
PAX8
|
|
|
E2F1..5.p2
|
-0.340
|
E2F4
E2F5
E2F2
E2F1
E2F3
|
|
|
PAX2.p2
|
-0.354
|
PAX2
|
|
|
PAX5.p2
|
-0.364
|
PAX5
|
|
|
FOSL2.p2
|
-0.364
|
FOSL2
|
|
|
ETS1,2.p2
|
-0.369
|
ETS2
ETS1
|
|
|
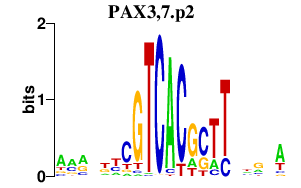
PAX3,7.p2
|
-0.379
|
PAX7
PAX3
|
|
|
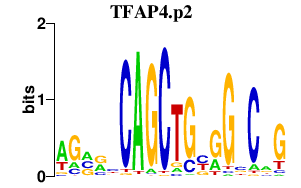
TFAP4.p2
|
-0.386
|
TFAP4
|
|
|
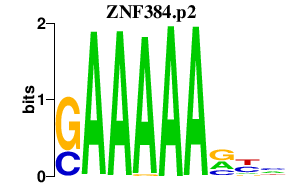
ZNF384.p2
|
-0.395
|
ZNF384
|
|
|
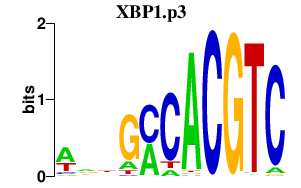
XBP1.p3
|
-0.398
|
XBP1
|
|
|
NKX6-1,2.p2
|
-0.433
|
NKX6-1
NKX6-2
|

|
|
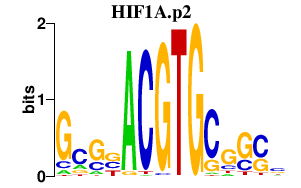
HIF1A.p2
|
-0.469
|
HIF1A
|
|
|
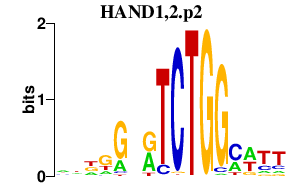
HAND1,2.p2
|
-0.489
|
HAND1
HAND2
|
|
|
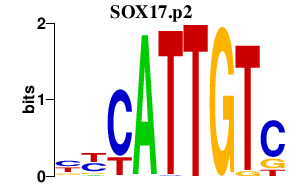
SOX17.p2
|
-0.490
|
SOX17
|
|
|
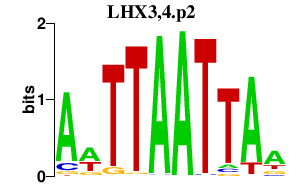
LHX3,4.p2
|
-0.549
|
LHX3
LHX4
|
|
|
PAX6.p2
|
-0.560
|
PAX6
|

|
|
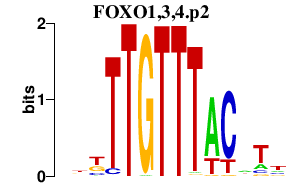
FOXO1,3,4.p2
|
-0.566
|
FOXO1
FOXO4
FOXO3
|
|
|
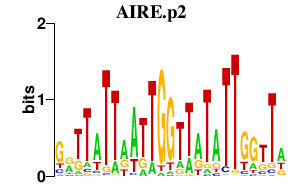
AIRE.p2
|
-0.577
|
AIRE
|
|
|
FOXA2.p3
|
-0.605
|
FOXA2
|

|
|
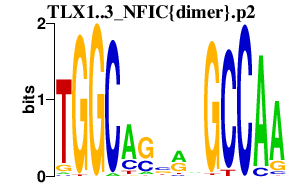
TLX1..3_NFIC{dimer}.p2
|
-0.609
|
TLX1
TLX3
|
|
|
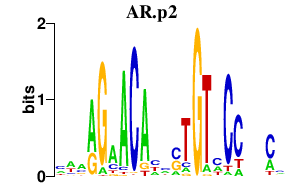
AR.p2
|
-0.619
|
AR
|
|
|
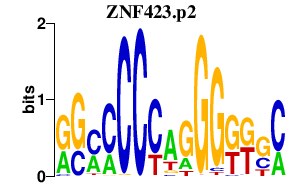
ZNF423.p2
|
-0.621
|
ZNF423
|
|
|
ATF6.p2
|
-0.627
|
ATF6
|

|
|
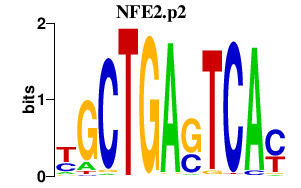
NFE2.p2
|
-0.656
|
NFE2
|
|
|
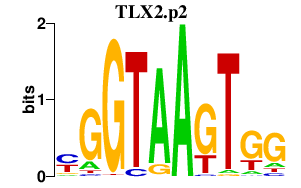
TLX2.p2
|
-0.665
|
TLX2
|
|
|
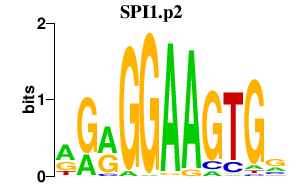
SPI1.p2
|
-0.679
|
SPI1
|
|
|
KLF12.p2
|
-0.762
|
KLF12
|
|
|
LMO2.p2
|
-0.795
|
LMO2
|
|
|
PAX4.p2
|
-0.809
|
PAX4
|
|
|
MEF2{A,B,C,D}.p2
|
-0.839
|
MEF2A
MEF2C
MEF2D
MEF2BNB-MEF2B
MEF2B
|
|
|
GATA1..3.p2
|
-0.862
|
GATA1
GATA2
GATA3
|

|
|
SREBF1,2.p2
|
-0.870
|
SREBF1
SREBF2
|
|
|
PRDM1.p3
|
-0.871
|
PRDM1
|
|
|
BACH2.p2
|
-0.874
|
BACH2
|
|
|
NKX2-2,8.p2
|
-0.881
|
NKX2-2
NKX2-8
|
|
|
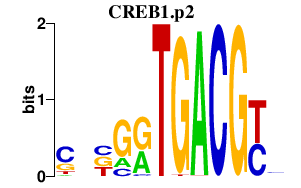
CREB1.p2
|
-0.897
|
CREB1
|
|
|
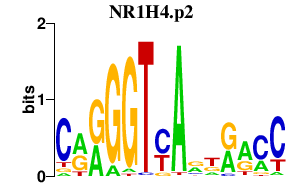
NR1H4.p2
|
-0.934
|
NR1H4
|
|
|
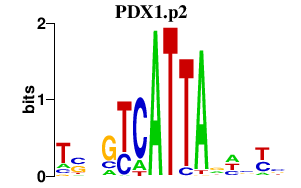
PDX1.p2
|
-0.951
|
PDX1
|
|
|
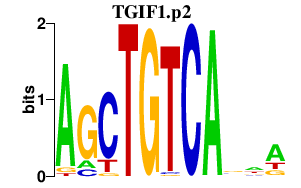
TGIF1.p2
|
-1.008
|
TGIF1
|
|
|
FOX{F1,F2,J1}.p2
|
-1.040
|
FOXF2
FOXJ1
FOXF1
|
|
|
ZFP161.p2
|
-1.041
|
ZFP161
|
|
|
YY1.p2
|
-1.044
|
YY1
|
|
|
TBX4,5.p2
|
-1.073
|
TBX5
TBX4
|
|
|
LEF1_TCF7_TCF7L1,2.p2
|
-1.081
|
TCF7
TCF7L2
LEF1
TCF7L1
|
|
|
NANOG{mouse}.p2
|
-1.090
|
NANOG
|
|
|
ELK1,4_GABP{A,B1}.p3
|
-1.120
|
GABPA
GABPB1
ELK4
ELK1
|
|
|
STAT1,3.p3
|
-1.138
|
STAT1
STAT3
|
|
|
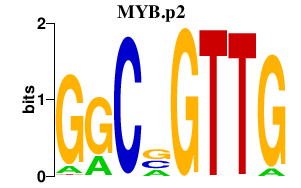
MYB.p2
|
-1.169
|
MYB
|
|
|
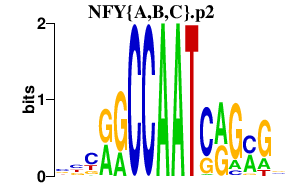
NFY{A,B,C}.p2
|
-1.221
|
NFYC
NFYB
NFYA
|
|
|
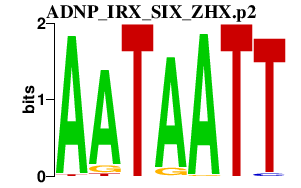
ADNP_IRX_SIX_ZHX.p2
|
-1.267
|
SIX5
ZHX1
IRX4
ADNP
ZHX3
ZHX2
IRX5
SIX2
|
|
|
NRF1.p2
|
-1.293
|
NRF1
|

|
|
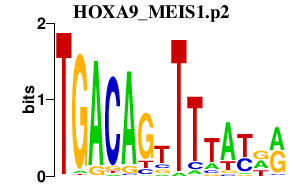
HOXA9_MEIS1.p2
|
-1.320
|
MEIS1
HOXA9
|
|
|
POU6F1.p2
|
-1.333
|
POU6F1
|
|
|
NR5A1,2.p2
|
-1.359
|
NR5A1
NR5A2
|
|
|
POU2F1..3.p2
|
-1.363
|
POU2F2
POU2F1
POU2F3
|
|
|
NFE2L1.p2
|
-1.365
|
NFE2L1
|

|
|
NR6A1.p2
|
-1.368
|
NR6A1
|
|
|
SPIB.p2
|
-1.373
|
SPIB
|
|
|
CUX2.p2
|
-1.374
|
CUX2
|
|
|
IRF1,2,7.p3
|
-1.383
|
IRF1
IRF2
IRF7
|
|
|
ARID5B.p2
|
-1.390
|
ARID5B
|
|
|
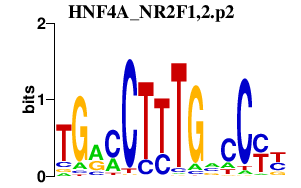
HNF4A_NR2F1,2.p2
|
-1.403
|
NR2F1
NR2F2
HNF4A
|
|
|
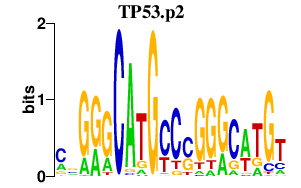
TP53.p2
|
-1.419
|
TP53
|
|
|
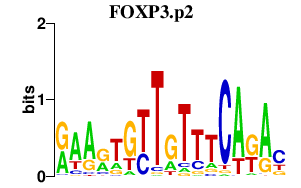
FOXP3.p2
|
-1.445
|
FOXP3
|
|
|
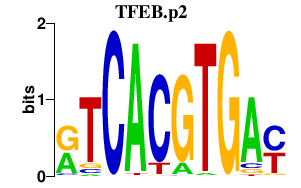
TFEB.p2
|
-1.508
|
TFEB
|
|
|
CTCF.p2
|
-1.548
|
CTCF
|
|
|
POU3F1..4.p2
|
-1.550
|
POU3F1
POU3F3
POU3F2
POU3F4
|
|
|
ARNT_ARNT2_BHLHB2_MAX_MYC_USF1.p2
|
-1.557
|
ARNT
MAX
MYC
USF1
BHLHB2
ARNT2
|
|
|
NKX3-1.p2
|
-1.592
|
NKX3-1
|
|
|
TFAP2B.p2
|
-1.604
|
TFAP2B
|
|
|
HMGA1,2.p2
|
-1.605
|
HMGA1
HMGA2
|
|
|
DMAP1_NCOR{1,2}_SMARC.p2
|
-1.624
|
SMARCA1
SMARCC2
SMARCA5
NCOR1
NCOR2
DMAP1
|
|
|
ZNF143.p2
|
-1.655
|
ZNF143
|
|
|
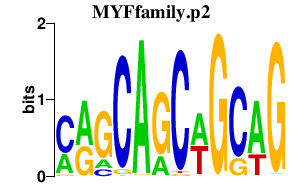
MYFfamily.p2
|
-1.768
|
MYOD1
MYF6
MYF5
MYOG
|
|
|
NFATC1..3.p2
|
-1.862
|
NFATC3
NFATC2
NFATC1
|

|
|
PPARG.p2
|
-1.891
|
PPARG
|

|
|
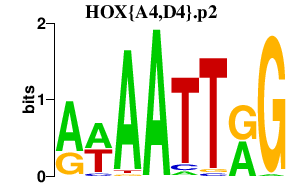
HOX{A4,D4}.p2
|
-1.919
|
HOXA4
HOXD4
|
|
|
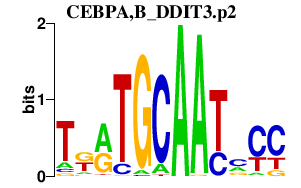
CEBPA,B_DDIT3.p2
|
-1.974
|
DDIT3
CEBPA
CEBPB
|
|
|
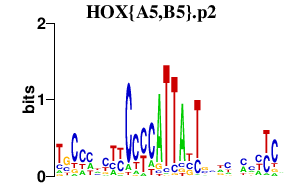
HOX{A5,B5}.p2
|
-1.980
|
HOXB5
HOXA5
|
|
|
PITX1..3.p2
|
-1.996
|
PITX2
PITX3
PITX1
|
|
|
MYBL2.p2
|
-2.008
|
MYBL2
|
|
|
CDC5L.p2
|
-2.008
|
CDC5L
|
|
|
EN1,2.p2
|
-2.009
|
EN1
EN2
|
|
|
ALX1.p2
|
-2.098
|
ALX1
|
|
|
SP1.p2
|
-2.127
|
SP1
|
|
|
FOX{C1,C2}.p2
|
-2.129
|
FOXC1
FOXC2
|
|
|
NR3C1.p2
|
-2.255
|
NR3C1
|
|
|
POU5F1_SOX2{dimer}.p2
|
-2.259
|
POU5F1
SOX2
|
|
|
CDX1,2,4.p2
|
-2.275
|
CDX1
CDX2
CDX4
|
|
|
EGR1..3.p2
|
-2.280
|
EGR3
EGR1
EGR2
|
|
|
PATZ1.p2
|
-2.421
|
PATZ1
|
|
|
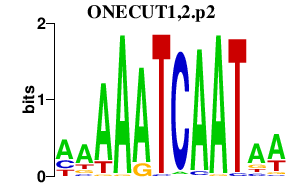
ONECUT1,2.p2
|
-2.464
|
ONECUT2
ONECUT1
|
|
|
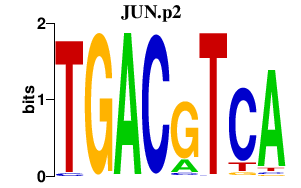
JUN.p2
|
-2.478
|
JUN
JUND
JUNB
|
|
|
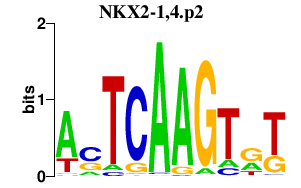
NKX2-1,4.p2
|
-2.674
|
NKX2-1
NKX2-4
|
|
|
ESRRA.p2
|
-2.838
|
ESRRA
|

|
|
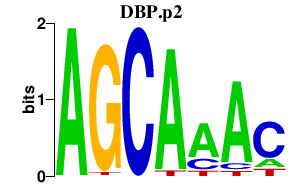
DBP.p2
|
-2.851
|
DBP
|
|
|
GTF2A1,2.p2
|
-2.863
|
GTF2A1
GTF2A2
|
|
|
CRX.p2
|
-2.971
|
CRX
|
|
|
FOXL1.p2
|
-3.148
|
FOXL1
|
|
|
TAL1_TCF{3,4,12}.p2
|
-3.153
|
TCF3
TCF4
TAL1
TCF12
|
|
|
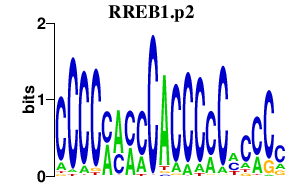
RREB1.p2
|
-3.215
|
RREB1
|
|
|
MSX1,2.p2
|
-3.308
|
MSX1
MSX2
|

|
|
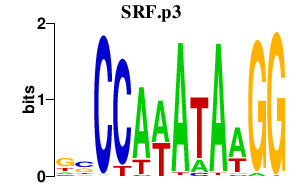
SRF.p3
|
-3.462
|
SRF
|
|
|
TFDP1.p2
|
-3.570
|
TFDP1
|

|
|
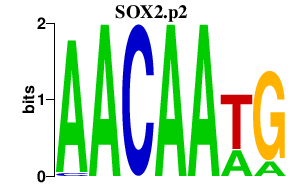
SOX2.p2
|
-3.650
|
SOX2
|
|
|
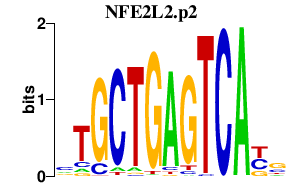
NFE2L2.p2
|
-3.682
|
NFE2L2
|
|
|
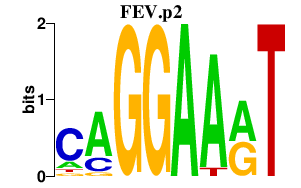
FEV.p2
|
-3.743
|
FEV
|
|
|
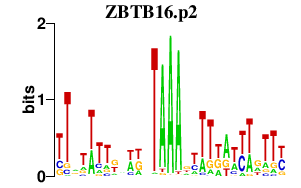
ZBTB16.p2
|
-3.885
|
ZBTB16
|
|
|
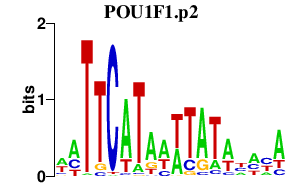
POU1F1.p2
|
-3.919
|
POU1F1
|
|
|
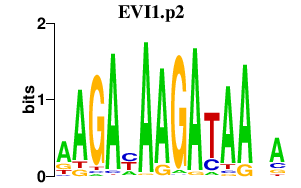
EVI1.p2
|
-3.923
|
EVI1
|
|
|
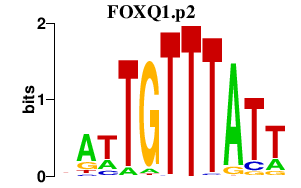
FOXQ1.p2
|
-3.930
|
FOXQ1
|
|
|
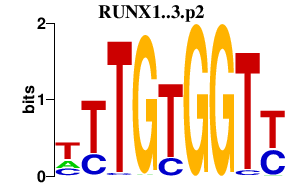
RUNX1..3.p2
|
-4.400
|
RUNX1
RUNX3
RUNX2
|
|
|
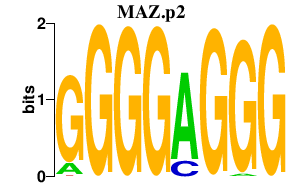
MAZ.p2
|
-4.667
|
MAZ
|
|
|
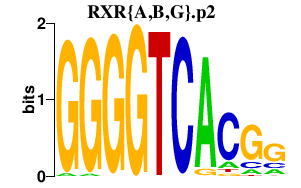
RXR{A,B,G}.p2
|
-4.692
|
RXRA
RXRB
RXRG
|
|
|
MYOD1.p2
|
-5.546
|
MYOD1
|
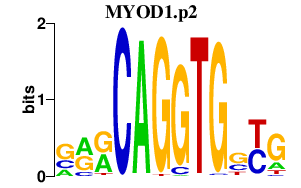
|
|
HNF1A.p2
|
-5.909
|
HNF1A
|
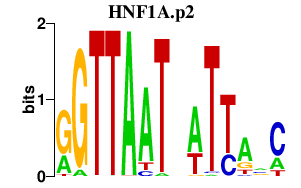
|
|
GATA6.p2
|
-6.178
|
GATA6
|
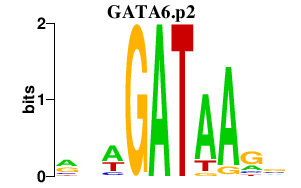
|