Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
ISMARA results Epithelial-Mesenchymal Transition, human (Scheel, 2011)
ISMARA - Integrated System for Motif Actitivity Response Analysis is a free online tool that recognizes most important transcription factors that are changing their activity in a set of samples.
All motifs sorted by activity significance
| Motif name | Z-value | Associated genes | Profile | Logo |
|---|---|---|---|---|
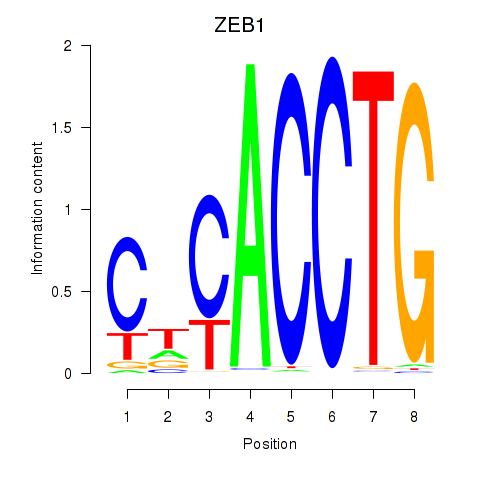
| ZEB1 | 9.62 |
|

|
|
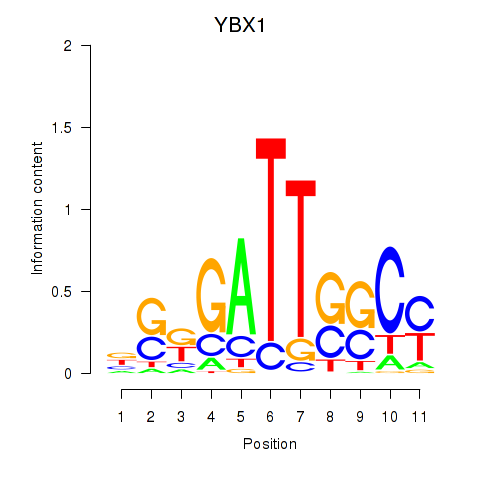
| YBX1_FOS_NFYC_NFYA_NFYB_CEBPZ | 4.79 |
|

|
|
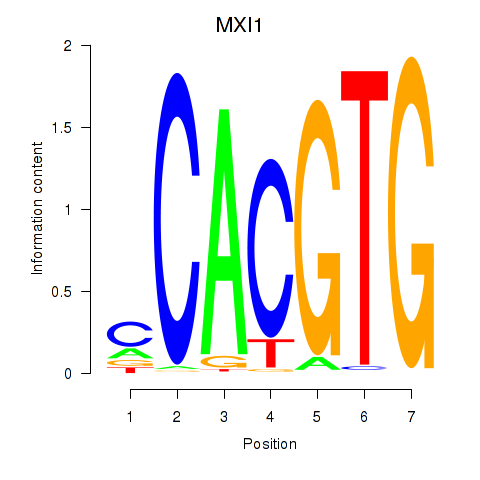
| MXI1_MYC_MYCN | 3.97 |
|

|
|
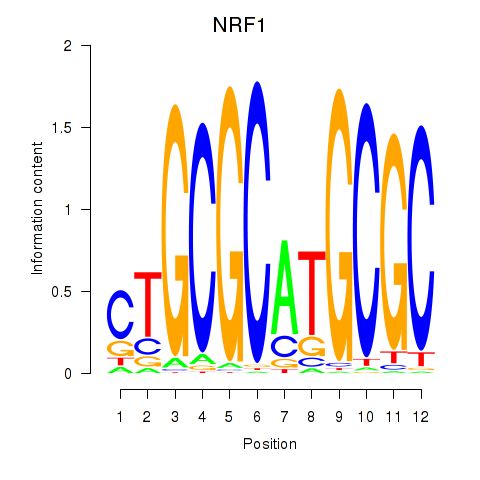
| NRF1 | 3.69 |
|
|
|
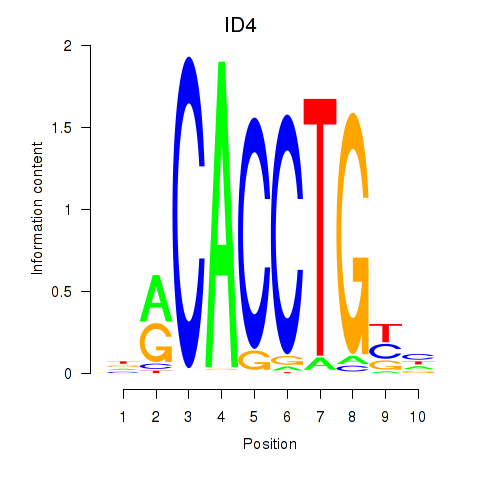
| ID4_TCF4_SNAI2 | 3.38 |
|

|
|
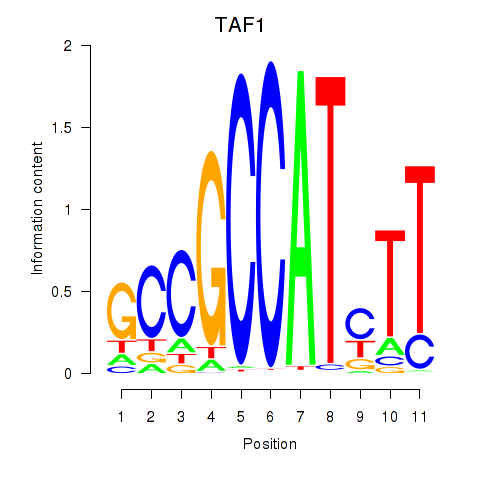
| TAF1 | 3.20 |
|

|
|
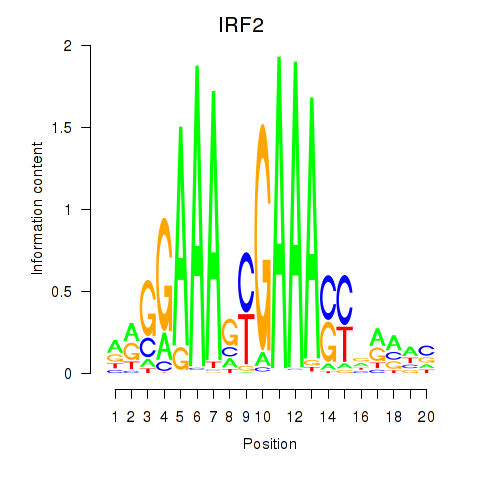
| IRF2_STAT2_IRF8_IRF1 | 3.19 |
|
|
|
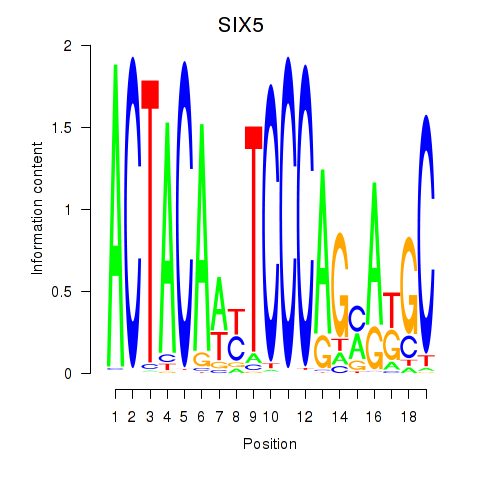
| SIX5_SMARCC2_HCFC1 | 3.04 |
|

|
|
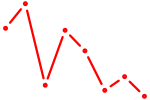
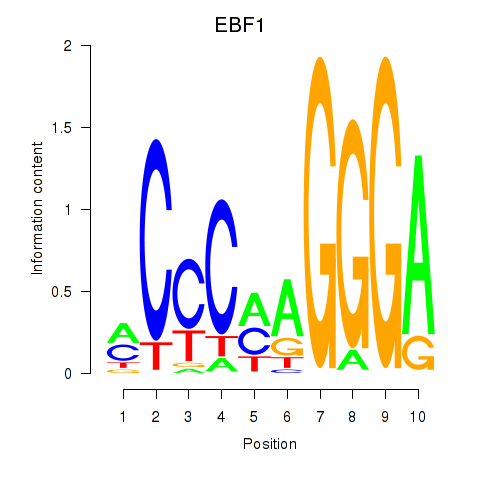
| EBF1 | 2.93 |
|
|
|
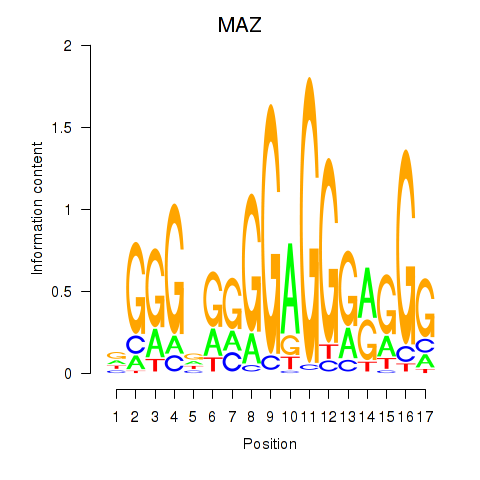
| MAZ_ZNF281_GTF2F1 | 2.73 |
|
|
|
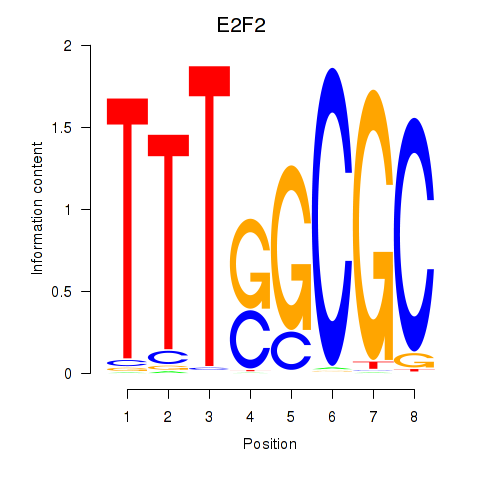
| E2F2_E2F5 | 2.72 |
|

|
|
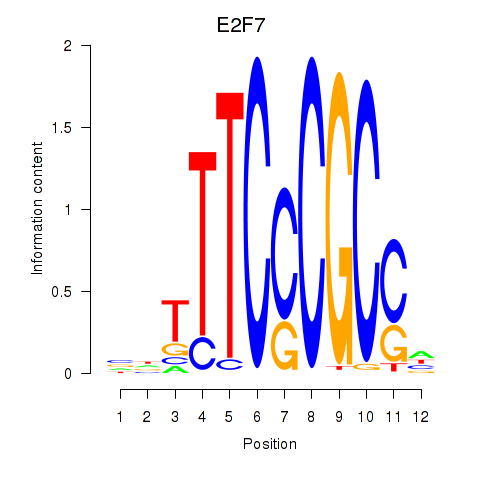
| E2F7_E2F1 | 2.71 |
|
|
|
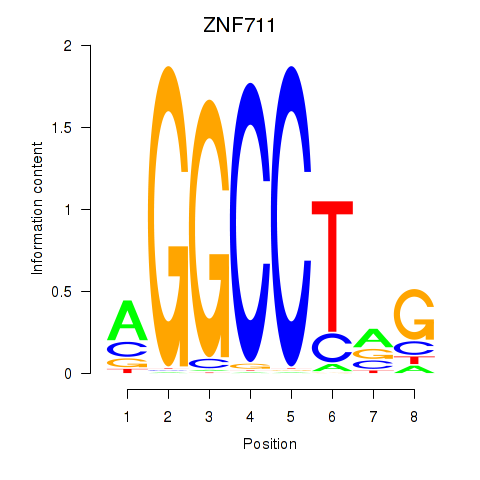
| ZNF711_TFAP2A_TFAP2D | 2.60 |
|
|
|
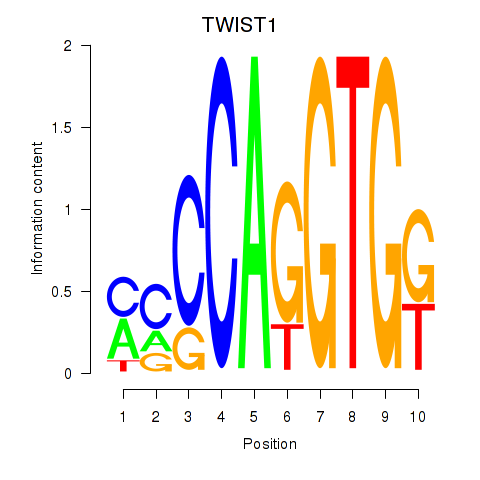
| TWIST1_SNAI1 | 2.48 |
|

|
|
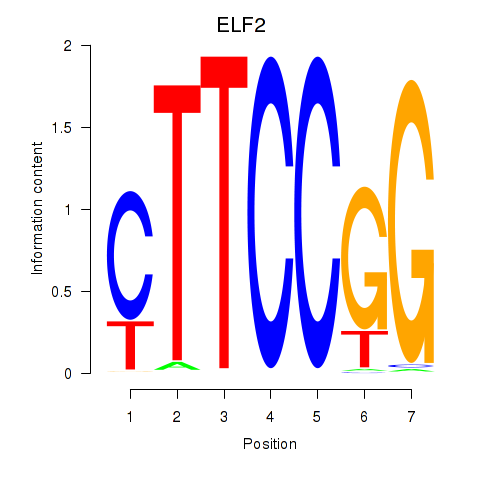
| ELF2_GABPA_ELF5 | 2.21 |
|
|
|
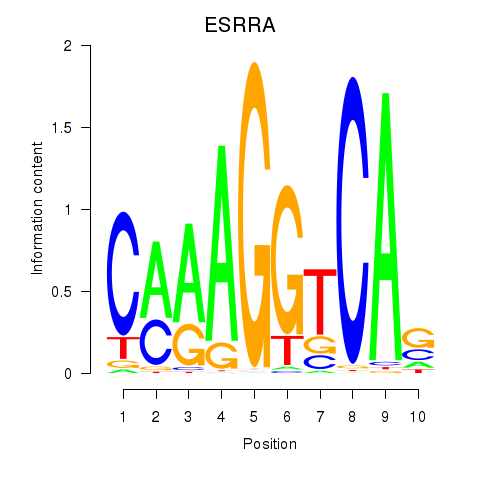
| ESRRA_ESR2 | 2.20 |
|

|
|
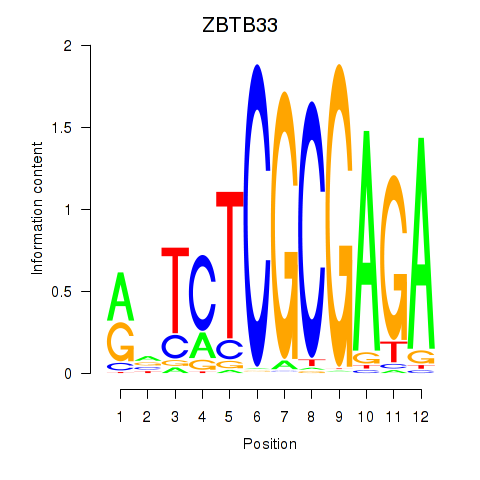
| ZBTB33_CHD2 | 2.11 |
|
|
|
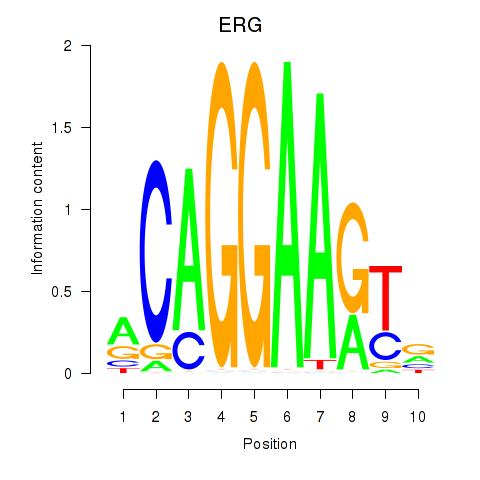
| ERG | 2.09 |
|
|
|
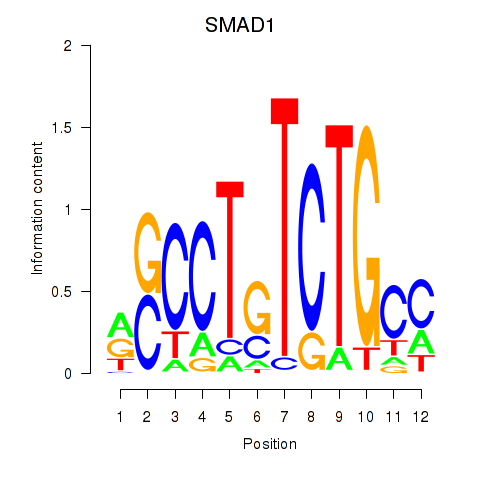
| SMAD1 | 2.04 |
|

|
|
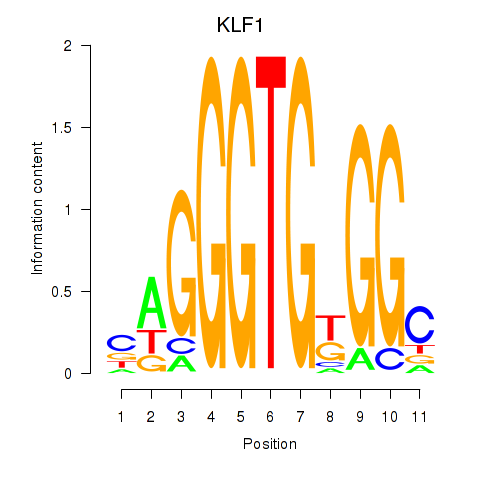
| KLF1 | 2.04 |
|

|
|
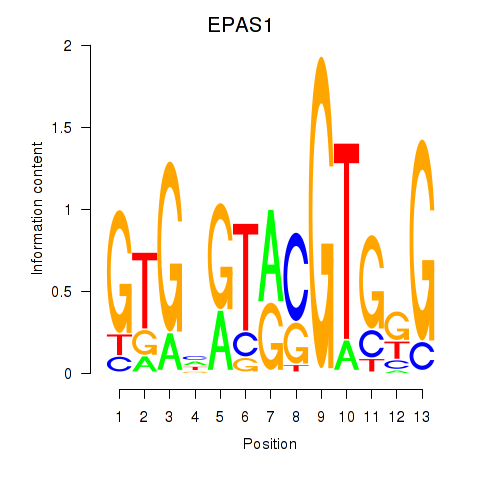
| EPAS1_BCL3 | 2.03 |
|

|
|
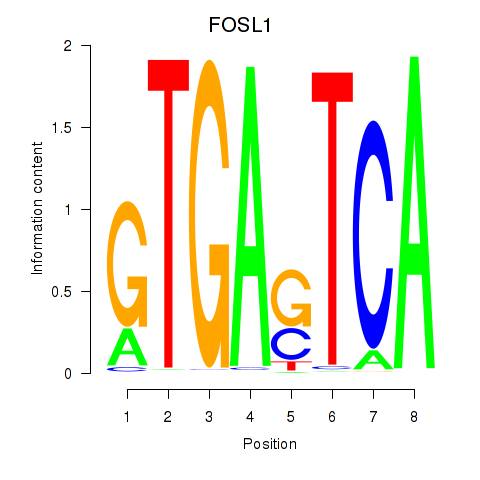
| FOSL1 | 2.03 |
|

|
|
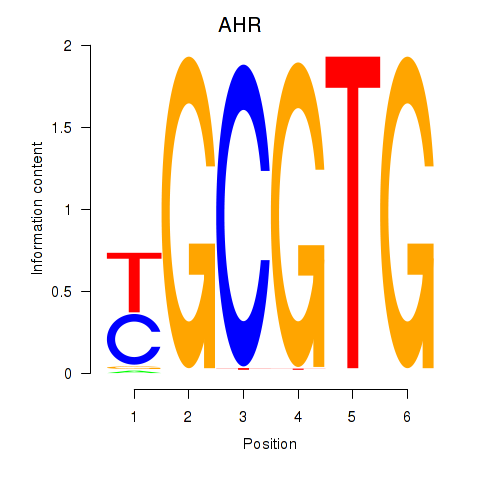
| AHR_ARNT2 | 2.01 |
|
|
|
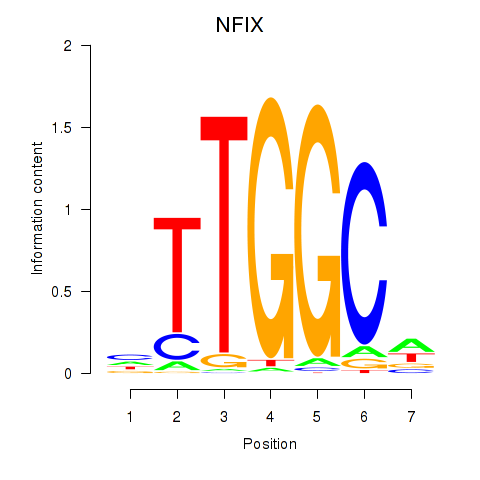
| NFIX_NFIB | 1.96 |
|
|
|
| PATZ1_KLF4 | 1.95 |
|
|
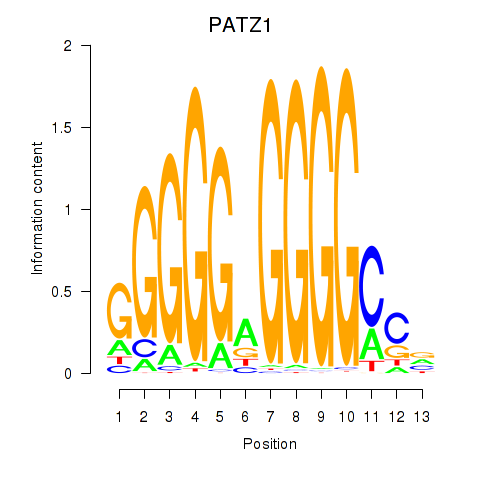
|
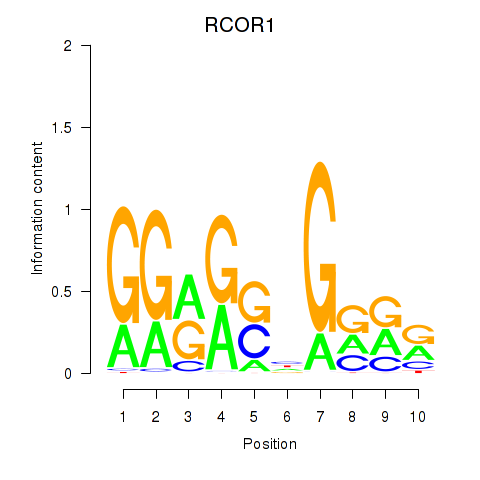
| RCOR1_MTA3 | 1.88 |
|
|
|
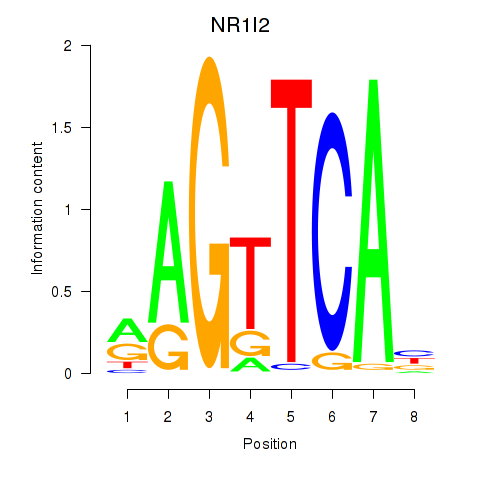
| NR1I2 | 1.81 |
|
|
|
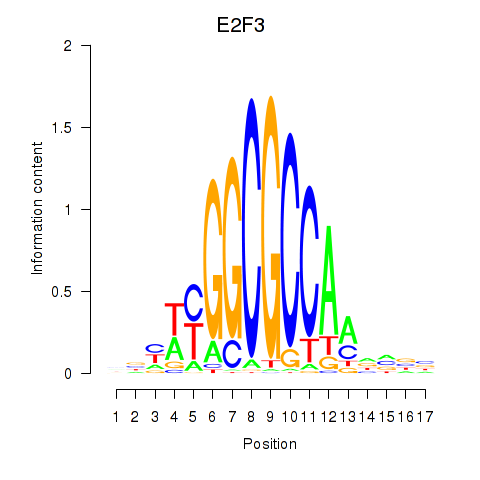
| E2F3 | 1.81 |
|
|
|
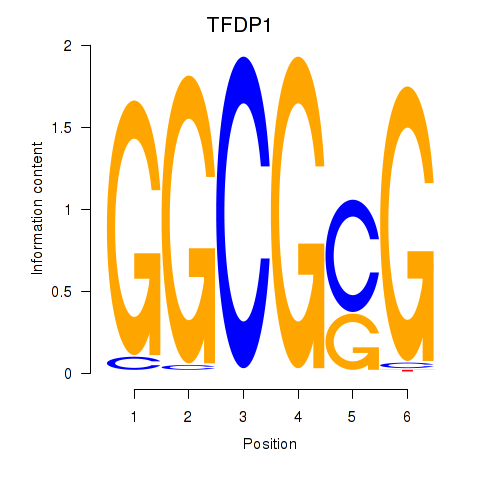
| TFDP1 | 1.75 |
|
|
|
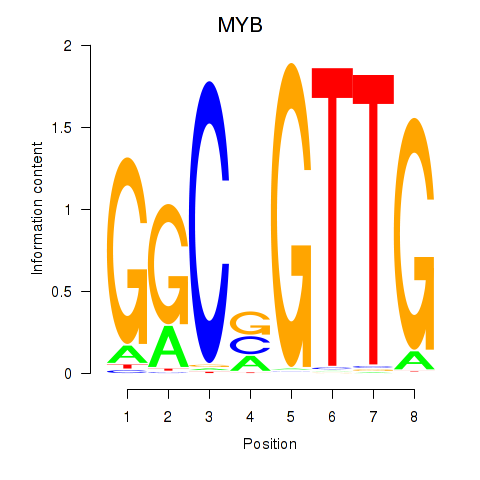
| MYB | 1.72 |
|
|
|
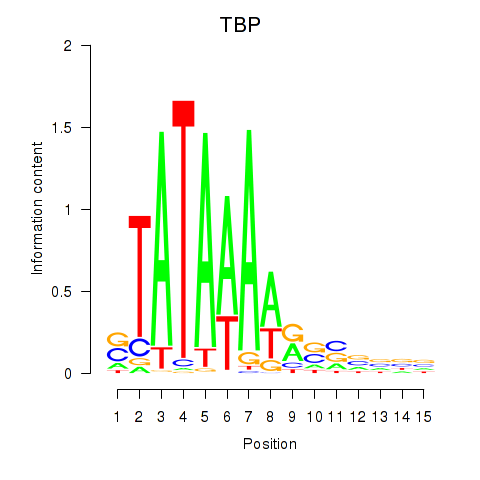
| TBP | 1.71 |
|

|
|
| IKZF1 | 1.70 |
|
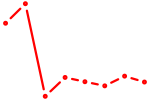
|
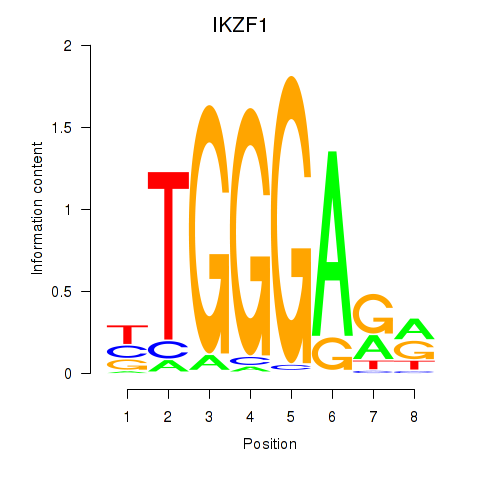
|
| NR2E3 | 1.70 |
|
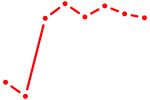
|
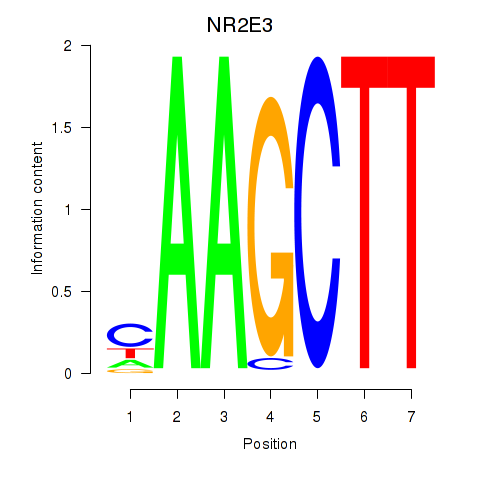
|
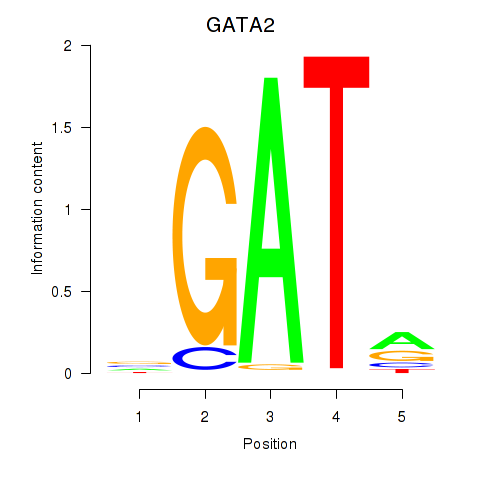
| GATA2 | 1.69 |
|

|
|
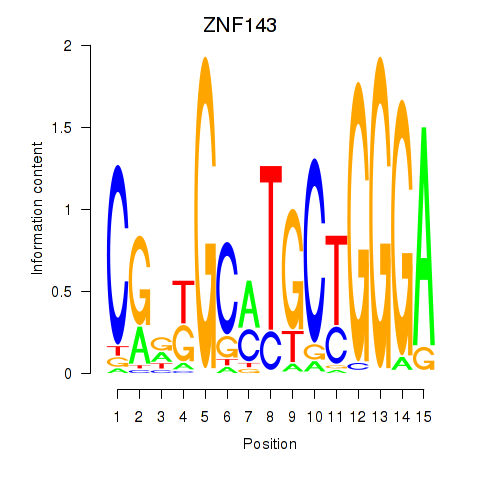
| ZNF143 | 1.69 |
|
|
|
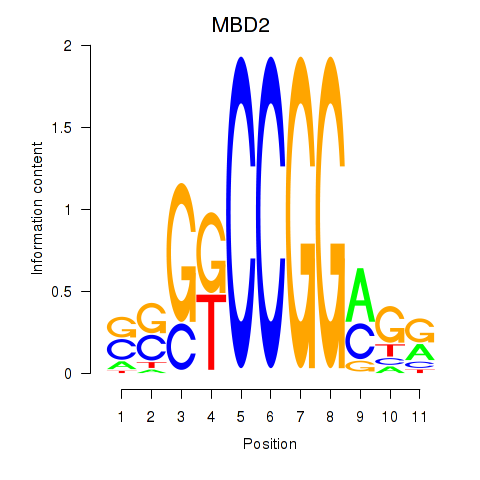
| MBD2 | 1.69 |
|

|
|
| CUX1 | 1.67 |
|

|
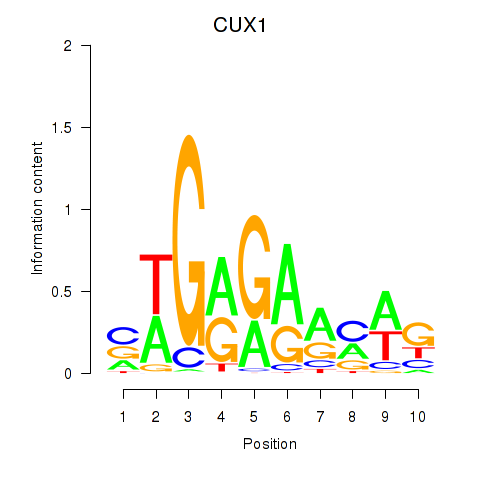
|
| ELK4_ETV5_ELK1_ELK3_ELF4 | 1.67 |
|

|
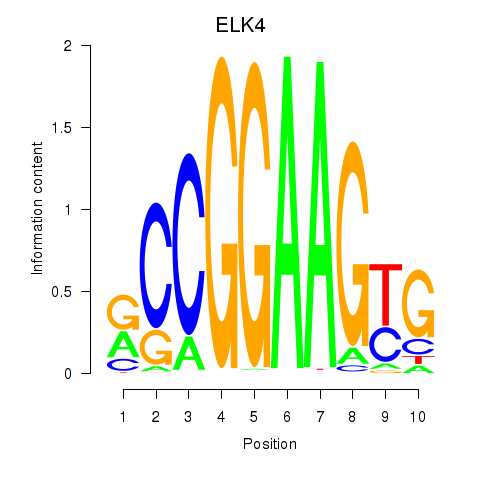
|
| MAFB | 1.66 |
|
|
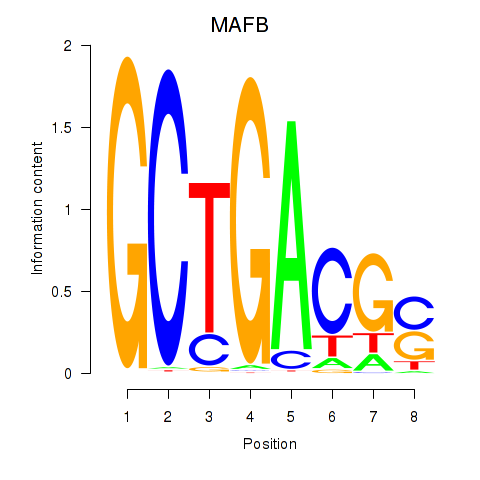
|
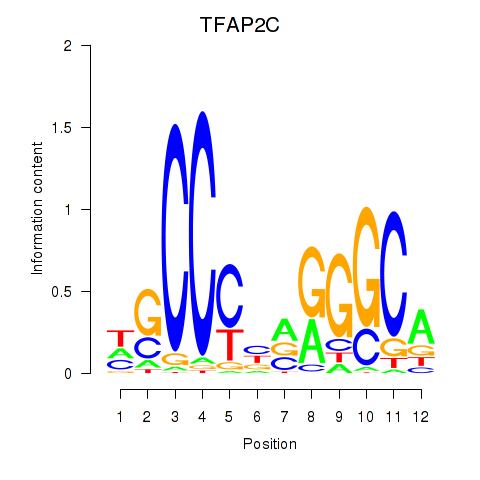
| TFAP2C | 1.66 |
|
|
|
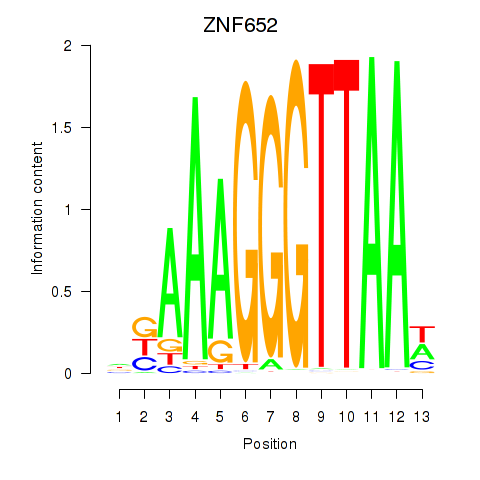
| ZNF652 | 1.65 |
|
|
|
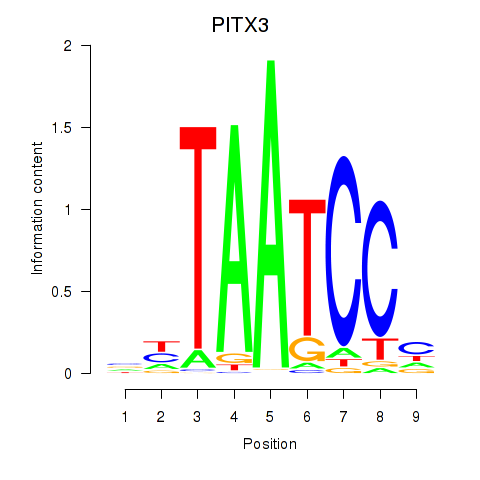
| PITX3 | 1.62 |
|

|
|
| AGCAGCA | 1.62 |
|

|

|
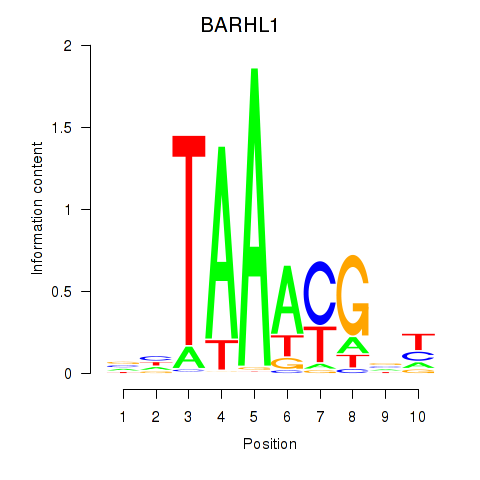
| BARHL1 | 1.60 |
|

|
|
| AGCACCA | 1.59 |
|
|

|
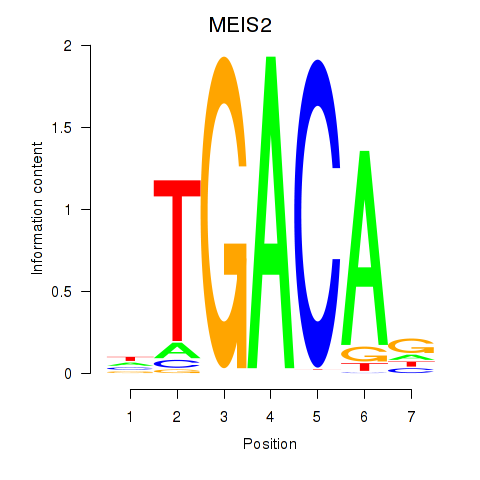
| MEIS2 | 1.56 |
|
|
|
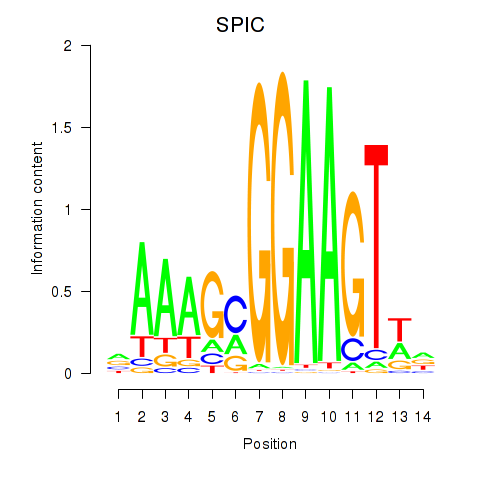
| SPIC | 1.56 |
|

|
|
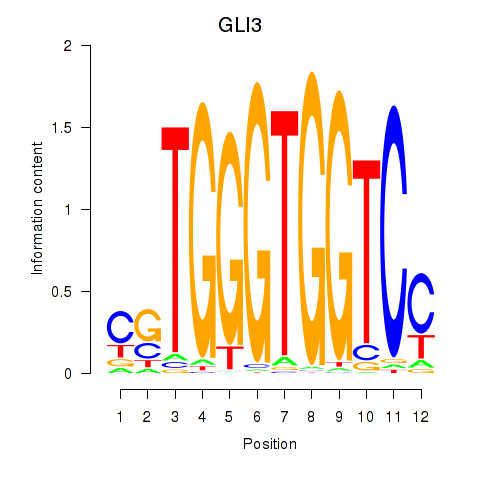
| GLI3 | 1.56 |
|
|
|
| ZBTB6 | 1.54 |
|

|
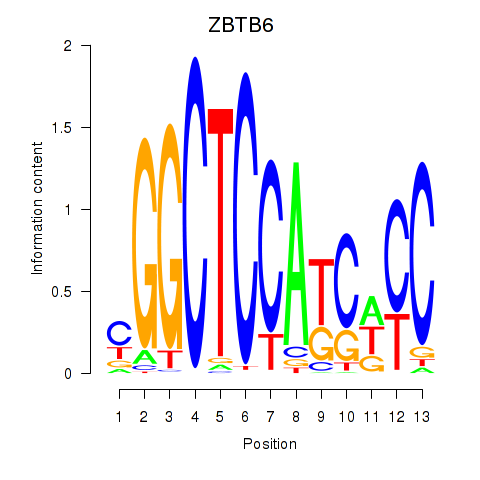
|
| MEIS1 | 1.53 |
|

|

|
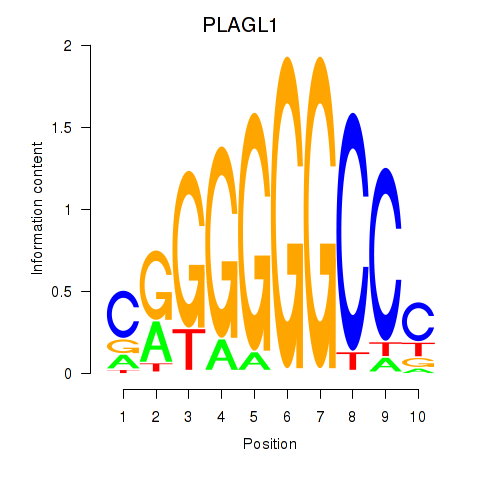
| PLAGL1 | 1.53 |
|
|
|
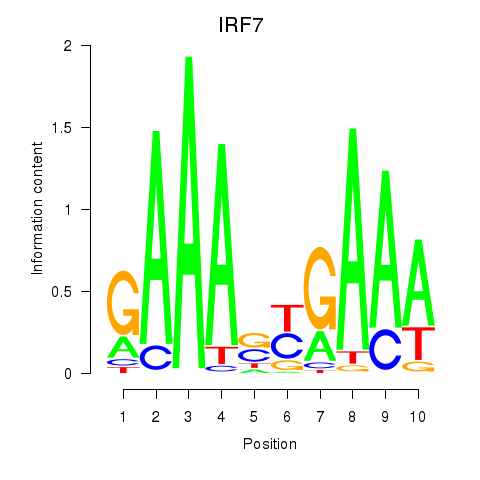
| IRF7 | 1.51 |
|
|
|
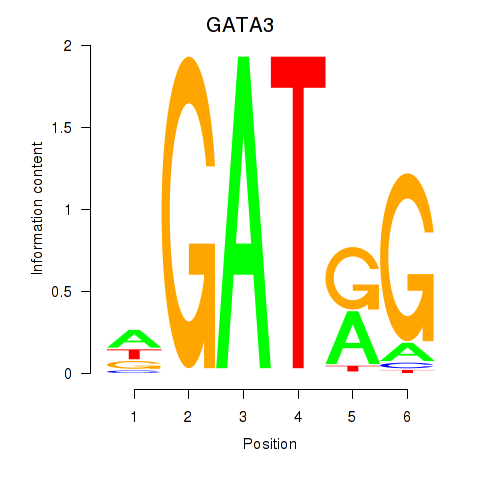
| GATA3 | 1.49 |
|

|
|
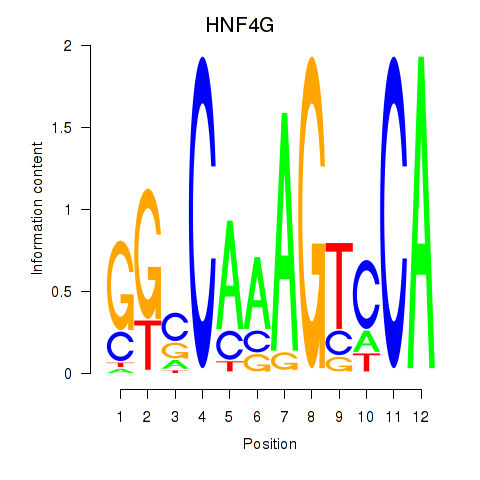
| HNF4G | 1.48 |
|

|
|
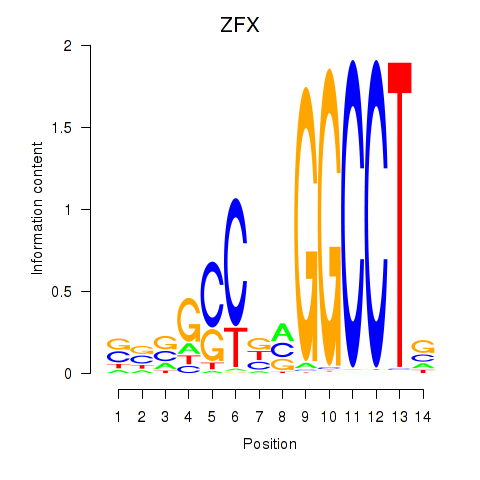
| ZFX | 1.47 |
|
|
|
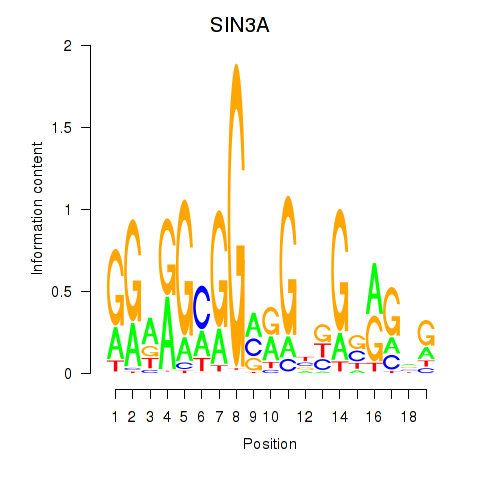
| SIN3A_CHD1 | 1.46 |
|

|
|
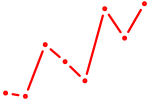
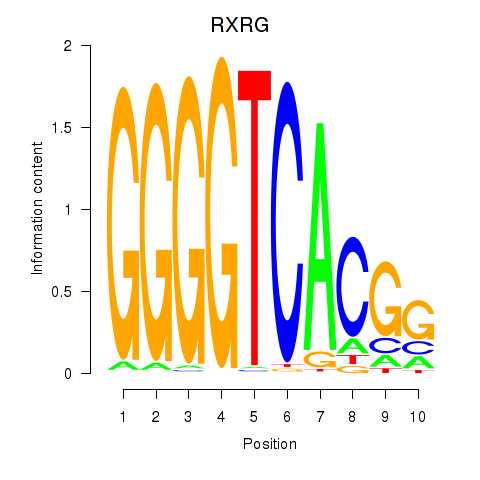
| RXRG | 1.45 |
|
|
|
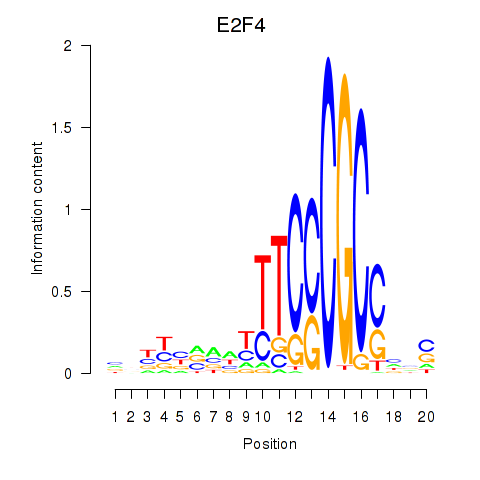
| E2F4 | 1.45 |
|
|
|
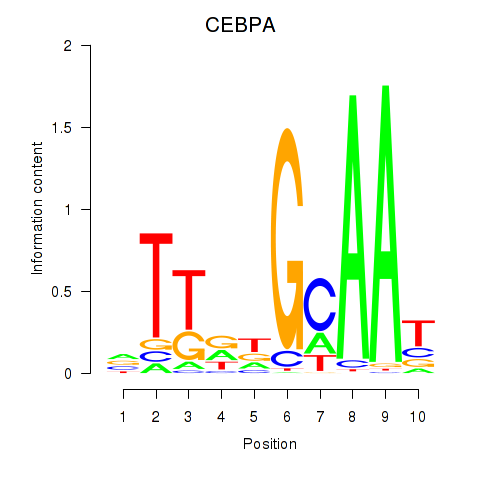
| CEBPA | 1.45 |
|

|
|
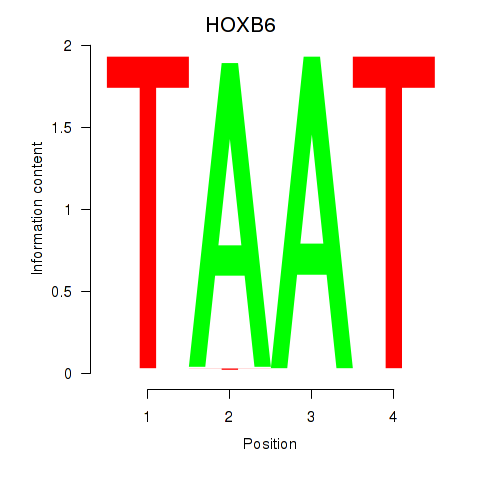
| HOXB6_PRRX2 | 1.42 |
|

|
|
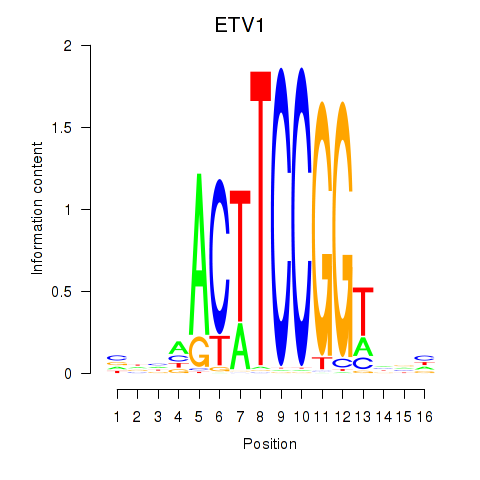
| ETV1_ERF_FEV_ELF1 | 1.42 |
|
|
|
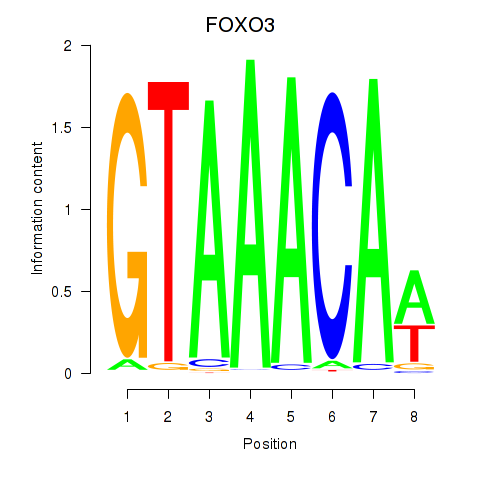
| FOXO3_FOXD2 | 1.41 |
|
|
|
| HIC2 | 1.41 |
|
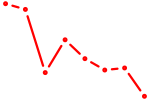
|
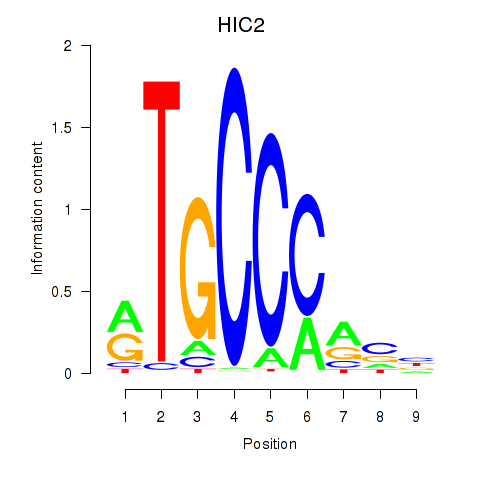
|
| NFATC4 | 1.40 |
|
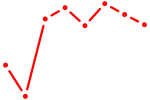
|
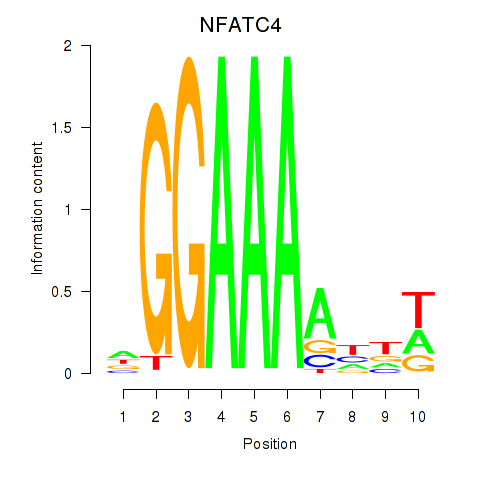
|
| ATF4 | 1.39 |
|
|
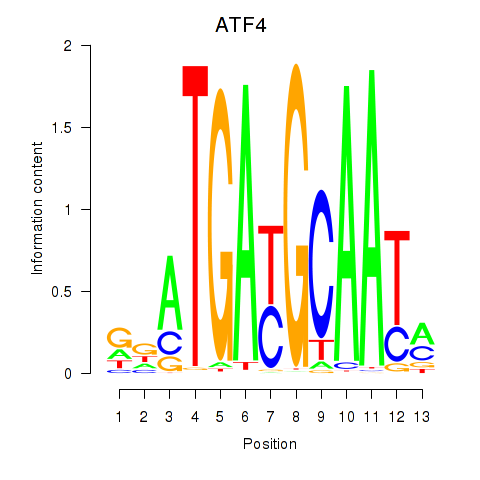
|
| ZNF384 | 1.37 |
|
|
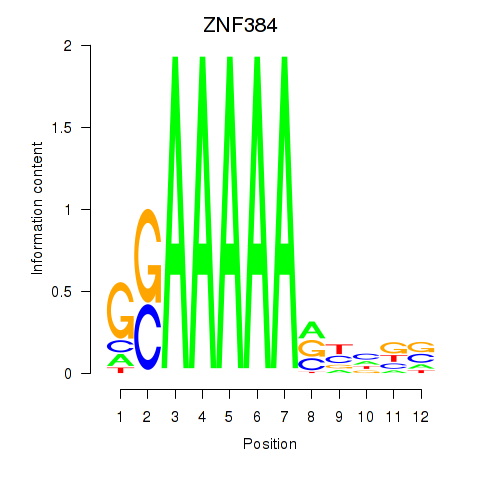
|
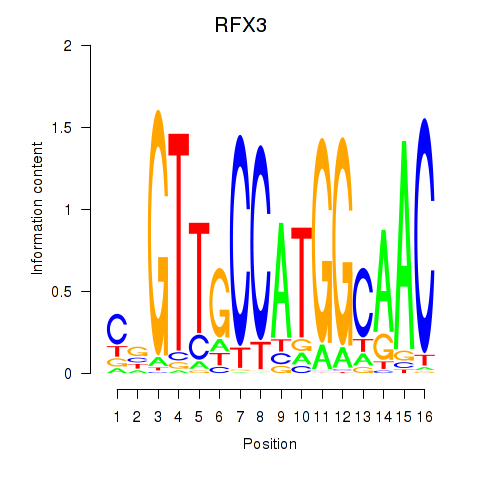
| RFX3_RFX2 | 1.37 |
|
|
|
| CCCUGAG | 1.36 |
|

|

|
| ESR1 | 1.36 |
|

|
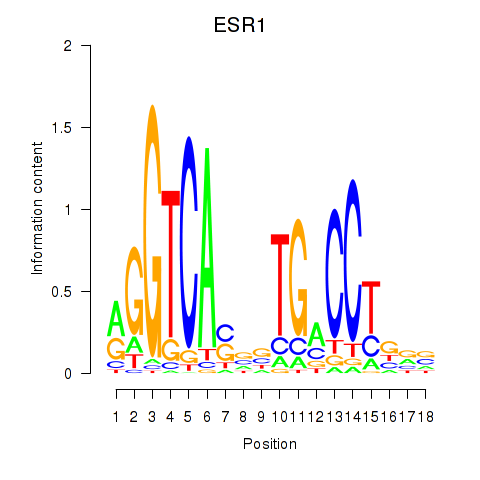
|
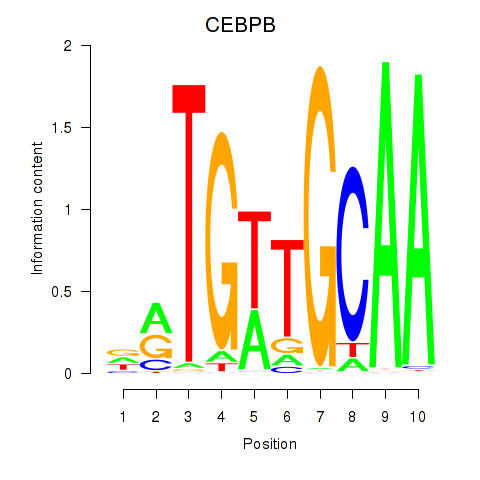
| CEBPB | 1.36 |
|
|
|
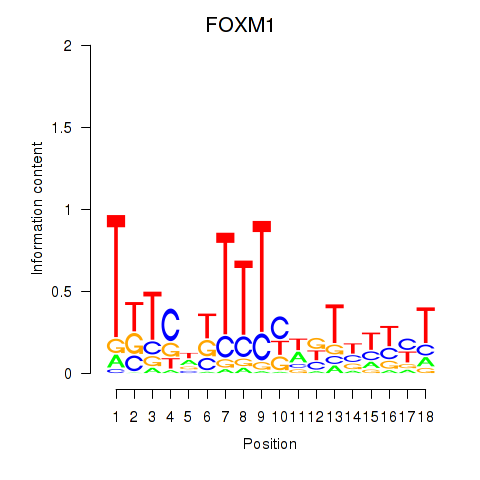
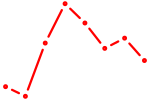
| FOXM1_TBL1XR1 | 1.36 |
|

|
|
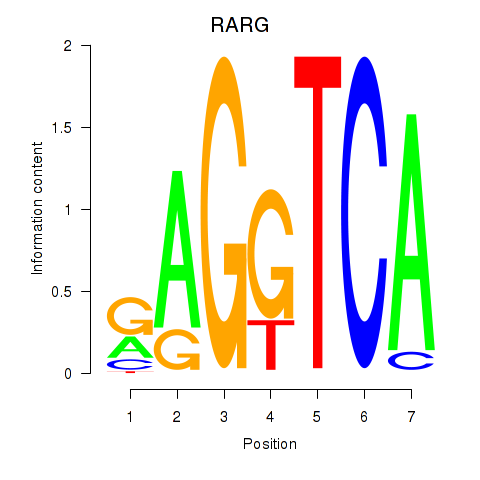
| RARG | 1.35 |
|
|
|
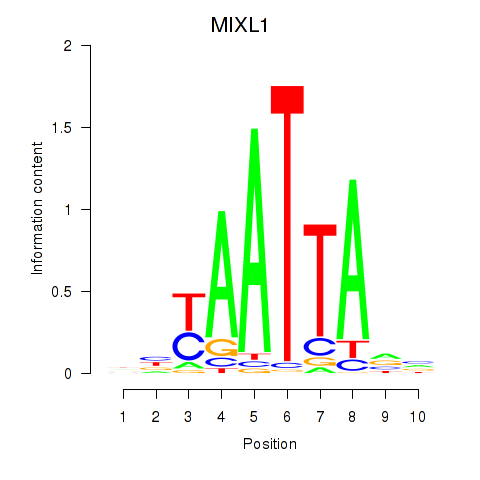
| MIXL1_GSX1_BSX_MEOX2_LHX4 | 1.35 |
|

|
|
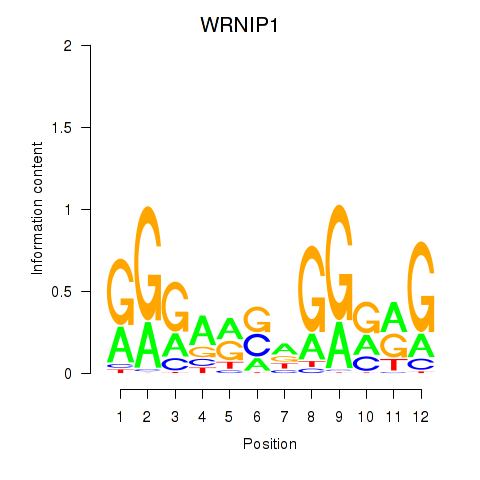
| WRNIP1 | 1.34 |
|
|
|
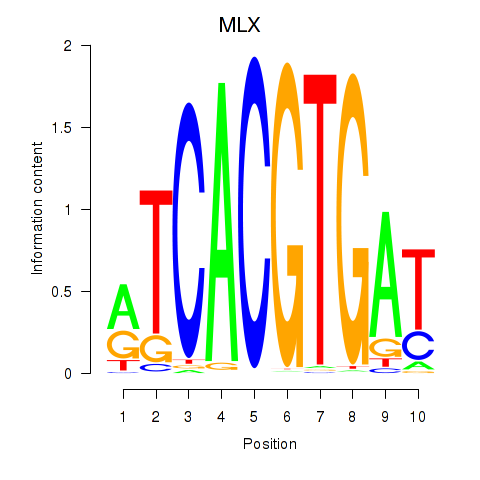
| MLX_USF2_USF1_PAX2 | 1.34 |
|

|
|
| TEAD3_TEAD1 | 1.31 |
|
|
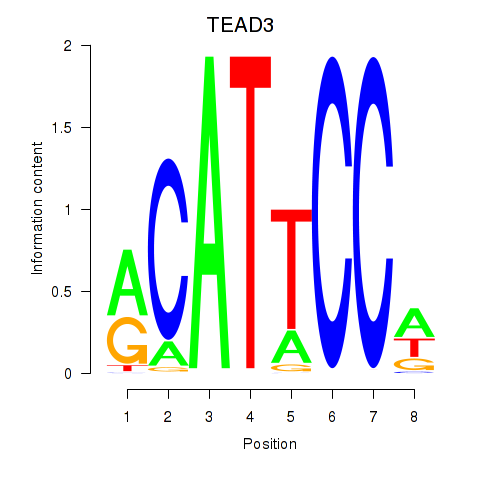
|
| SRF | 1.30 |
|
|
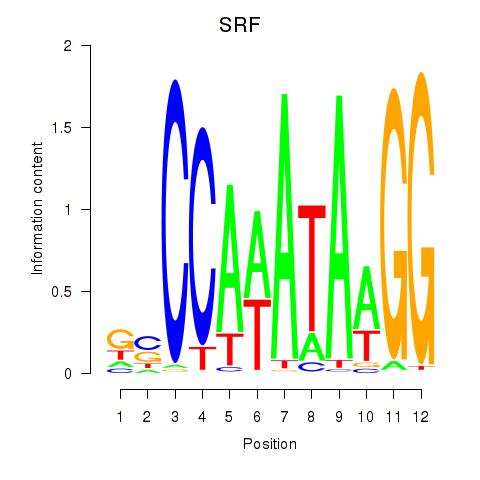
|
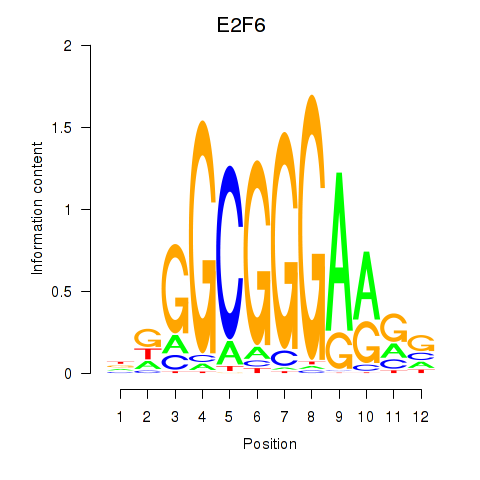
| E2F6 | 1.28 |
|
|
|
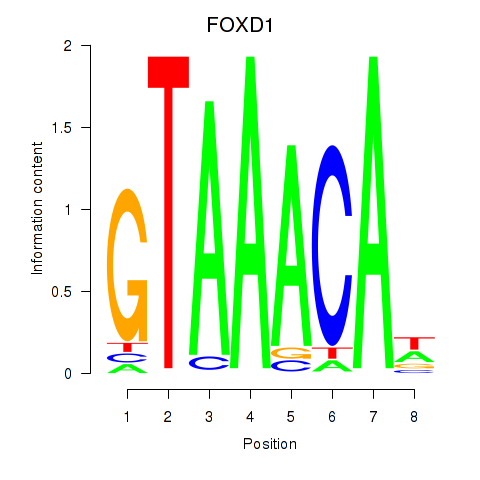
| FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 | 1.28 |
|
|
|
| NFE2L1 | 1.28 |
|

|
|
| MZF1 | 1.26 |
|

|
|
| TP63 | 1.25 |
|
|
|
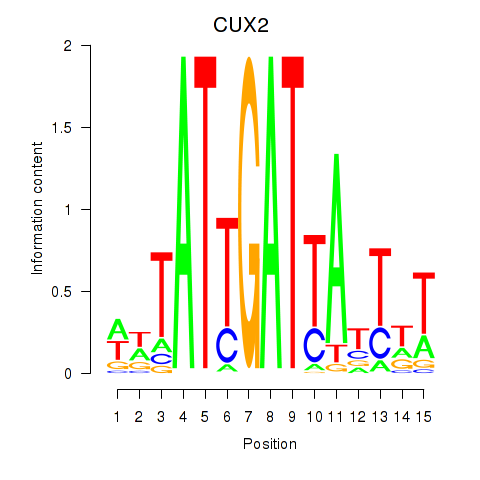
| CUX2 | 1.25 |
|
|
|
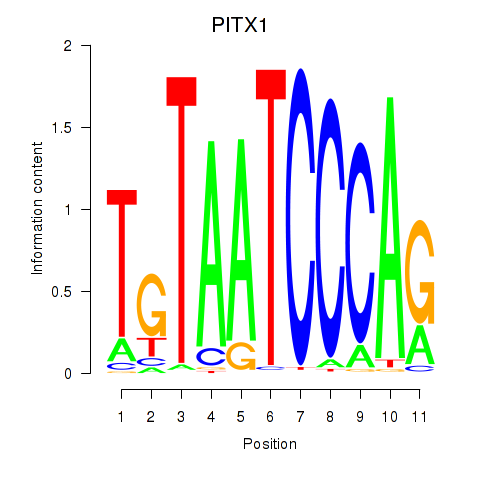
| PITX1 | 1.23 |
|
|
|
| SREBF1_TFE3 | 1.23 |
|
|
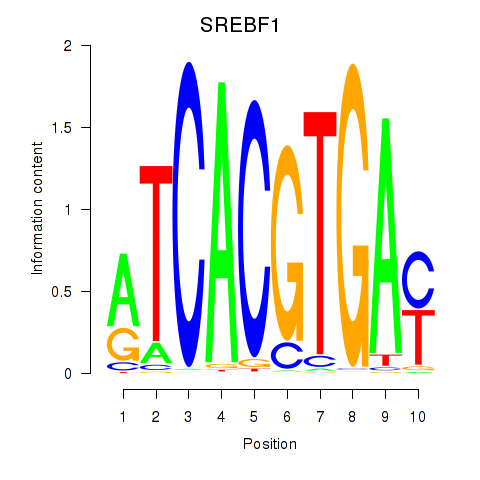
|
| SMAD4 | 1.22 |
|
|
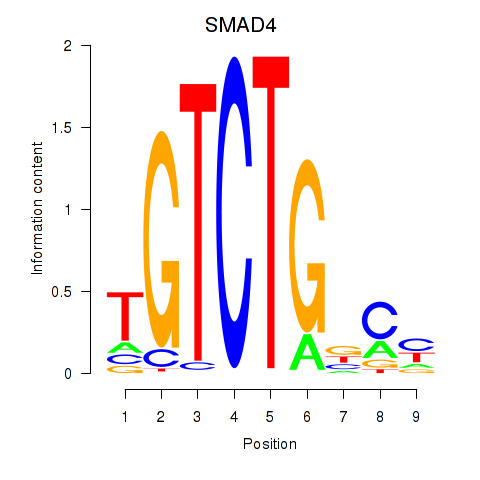
|
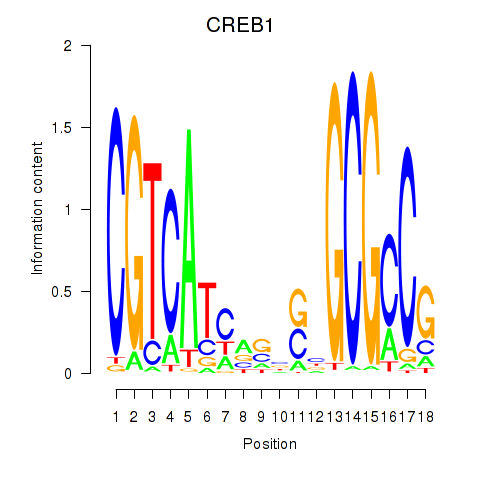
| CREB1 | 1.22 |
|

|
|
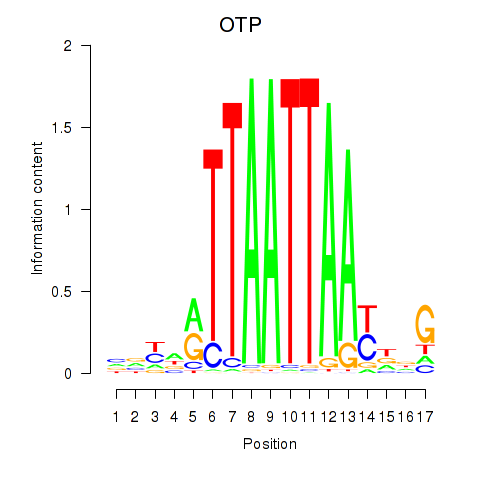
| OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 | 1.22 |
|
|
|
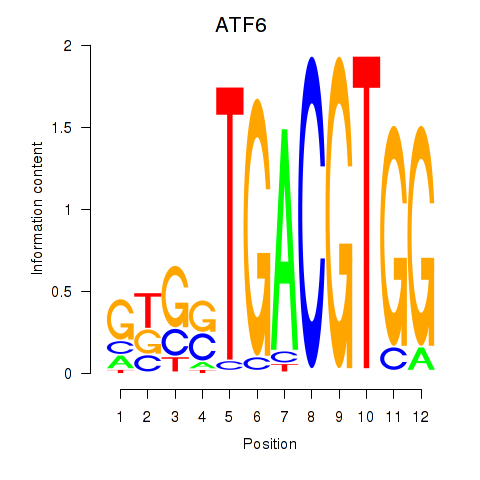
| ATF6 | 1.20 |
|
|
|
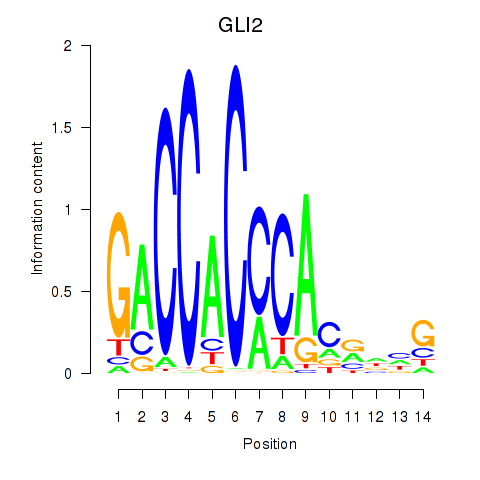
| GLI2 | 1.19 |
|
|
|
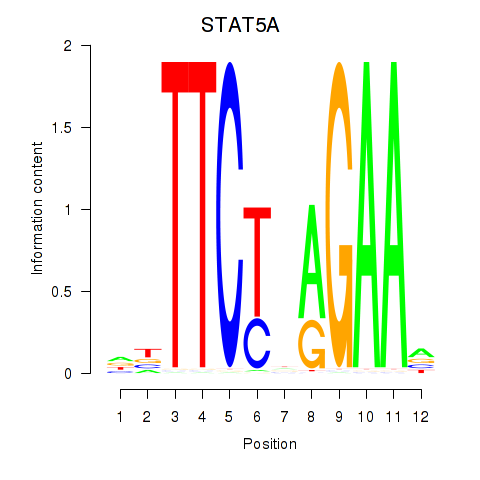
| STAT5A | 1.19 |
|

|
|
| EGR3_EGR2 | 1.18 |
|
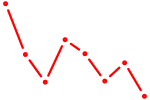
|

|
| ZBTB18 | 1.17 |
|
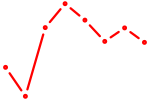
|
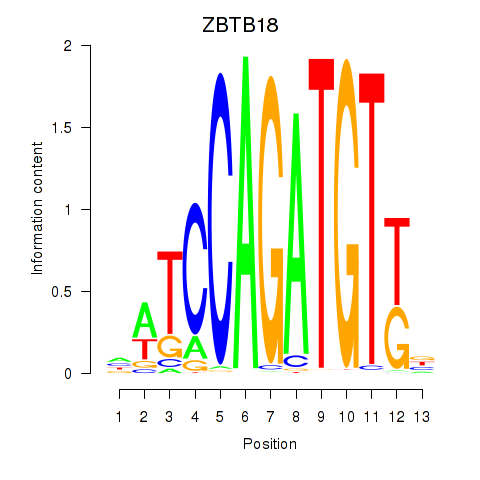
|
| SP1 | 1.17 |
|

|
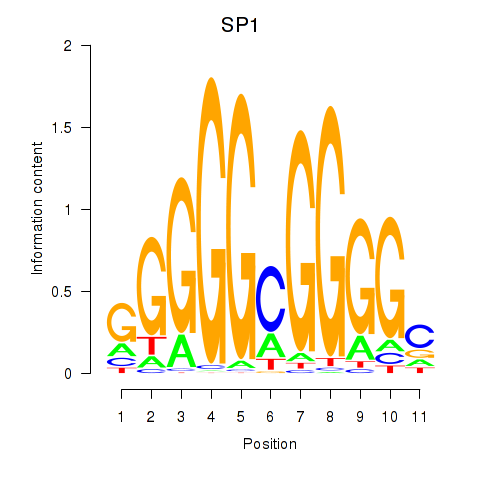
|
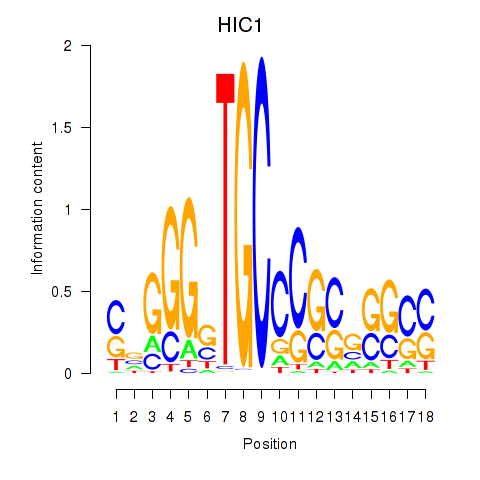
| HIC1 | 1.16 |
|
|
|
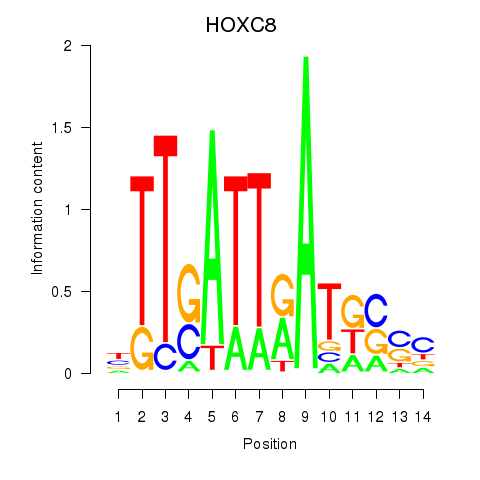
| HOXC8 | 1.16 |
|
|
|
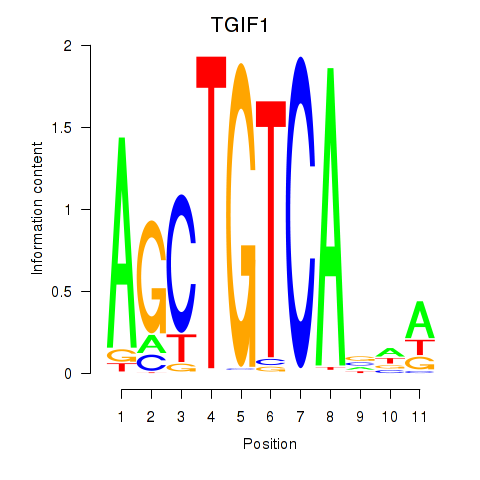
| TGIF1 | 1.16 |
|

|
|
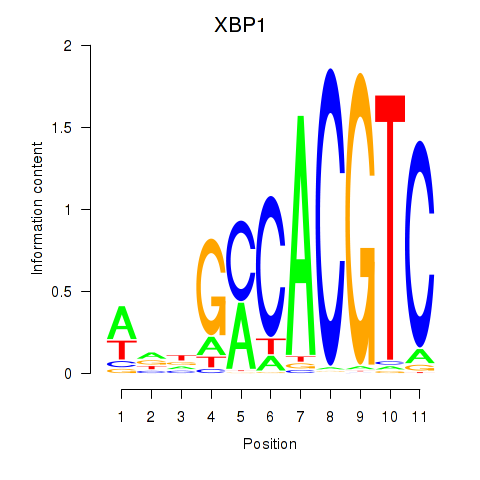
| XBP1 | 1.15 |
|
|
|
| PAX5 | 1.15 |
|
|
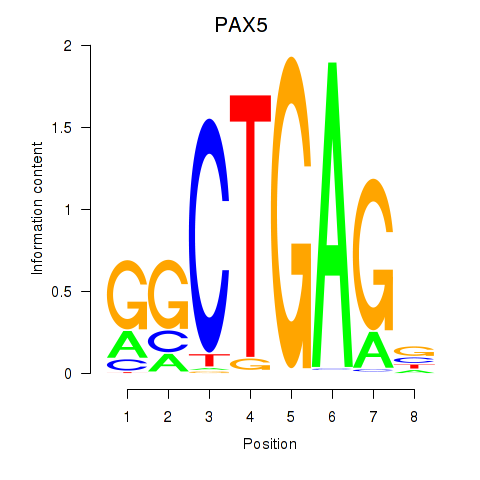
|
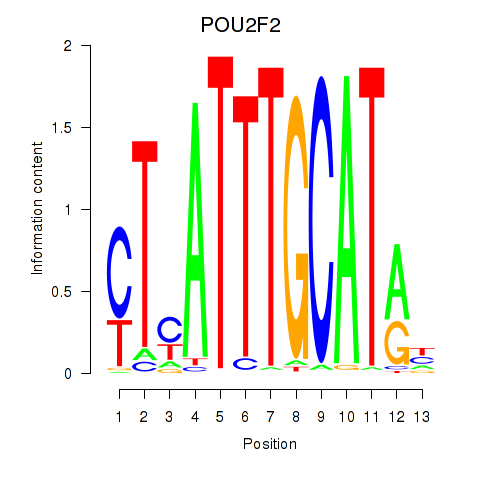
| POU2F2_POU3F1 | 1.14 |
|
|
|
| ZNF524 | 1.14 |
|
|
|
| FOXC1 | 1.14 |
|
|
|
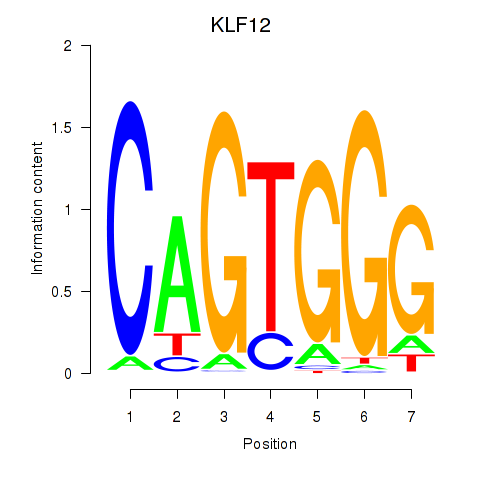
| KLF12 | 1.13 |
|
|
|
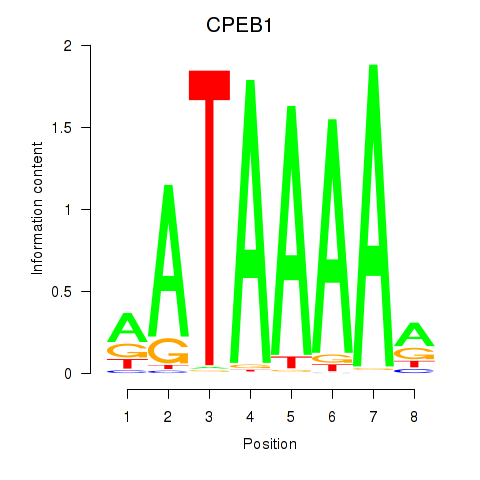
| CPEB1 | 1.13 |
|
|
|
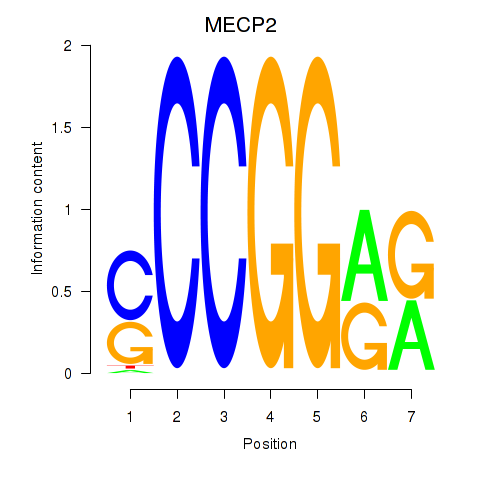
| MECP2 | 1.13 |
|
|
|
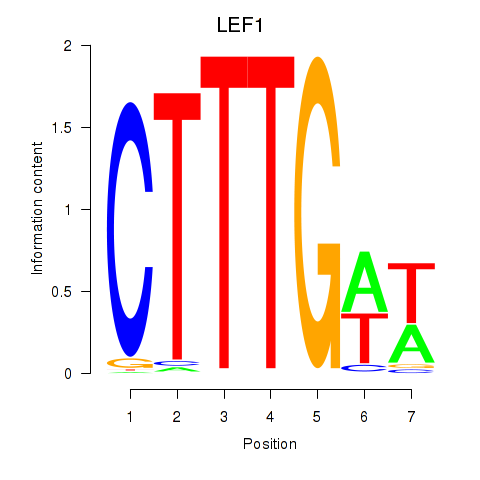
| LEF1 | 1.13 |
|

|
|
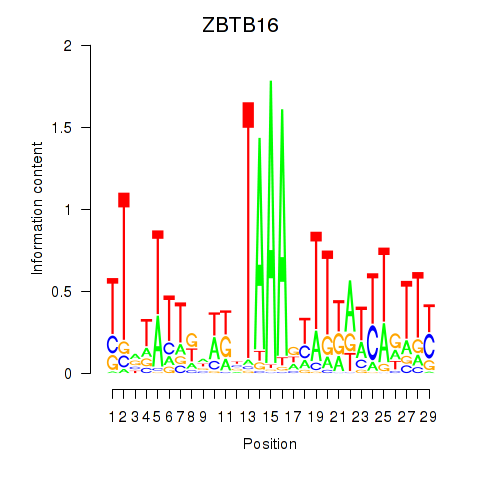
| ZBTB16 | 1.12 |
|
|
|
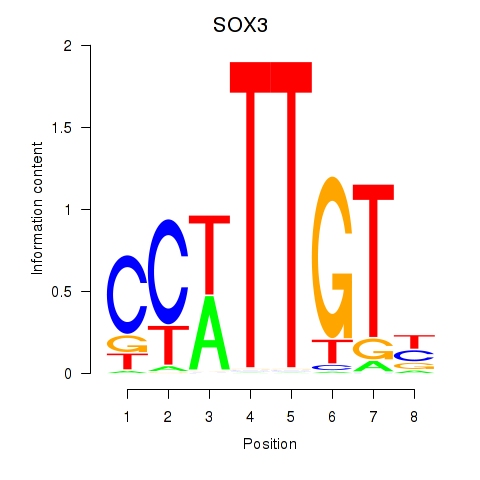
| SOX3_SOX2 | 1.11 |
|
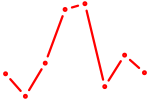
|
|
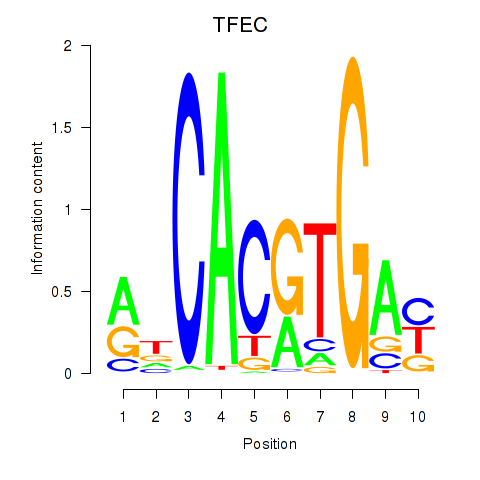
| TFEC_MITF_ARNTL_BHLHE41 | 1.11 |
|
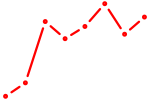
|
|
| NR0B1 | 1.11 |
|

|
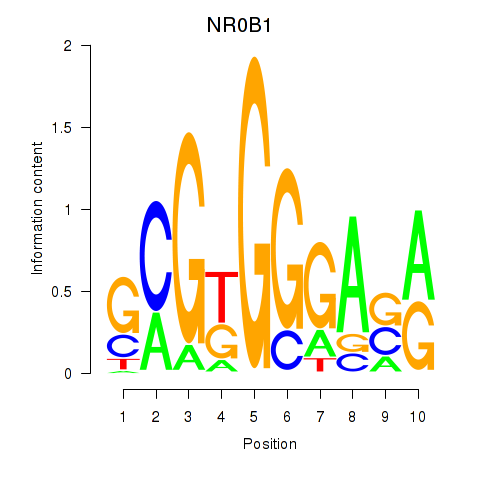
|
| GAGGUAG | 1.11 |
|

|

|
| OTX1 | 1.10 |
|
|
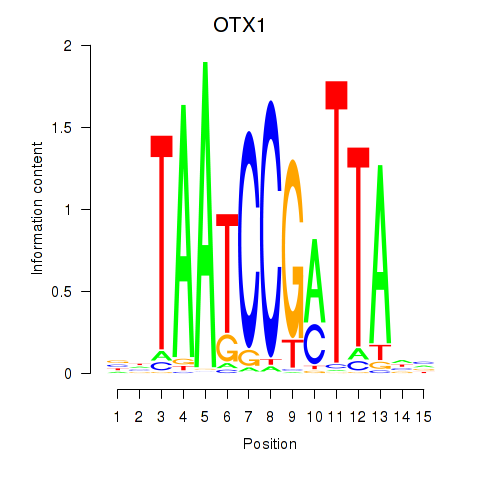
|
| DUXA | 1.09 |
|
|
|
| RELA | 1.08 |
|

|
|
| FOXL1 | 1.08 |
|

|
|
| WT1_MTF1_ZBTB7B | 1.07 |
|
|

|
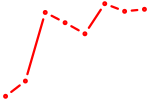
| GGAAUGU | 1.07 |
|
|

|
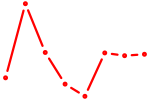
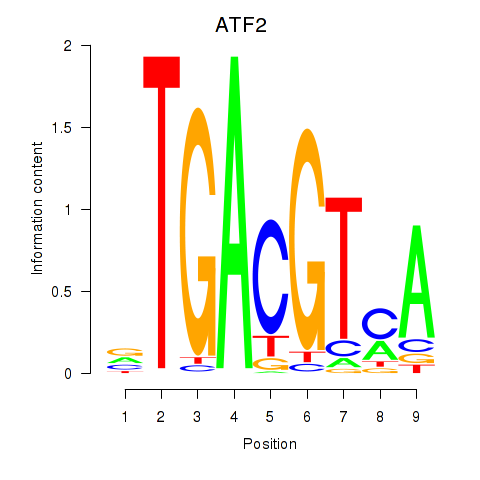
| ATF2_ATF1_ATF3 | 1.07 |
|
|
|
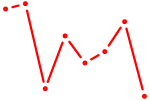
| GTF2I | 1.07 |
|
|
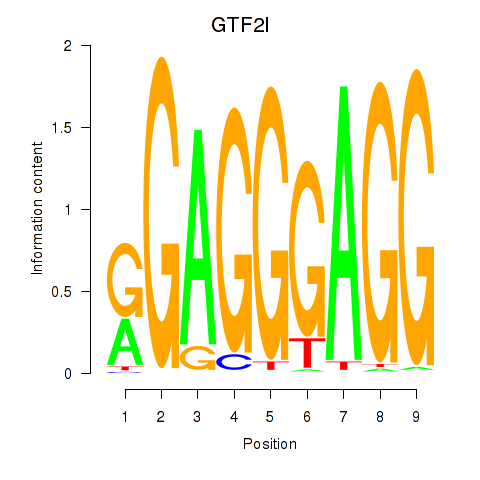
|
| YY1_YY2 | 1.07 |
|

|
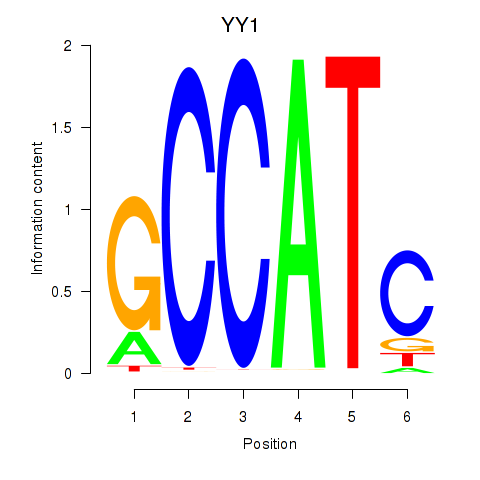
|
| OLIG3_NEUROD2_NEUROG2 | 1.07 |
|
|
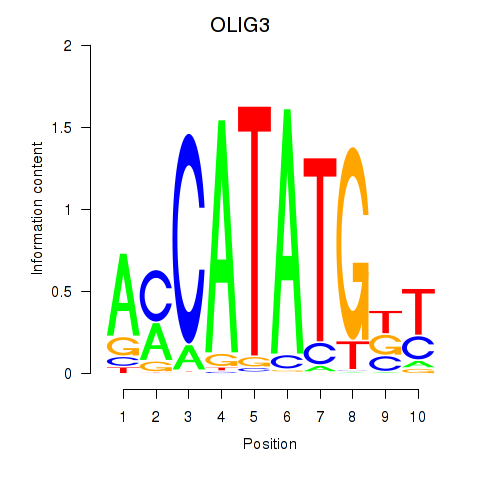
|
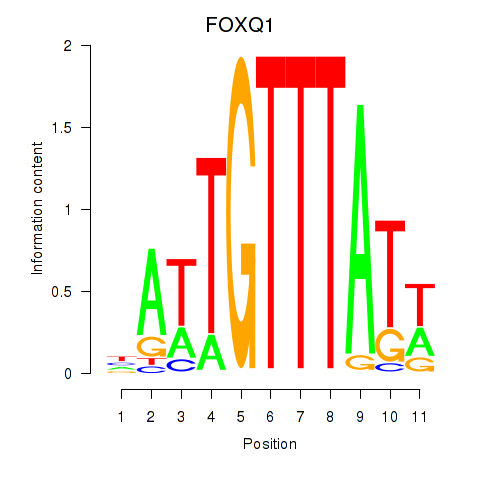
| FOXQ1 | 1.07 |
|
|
|
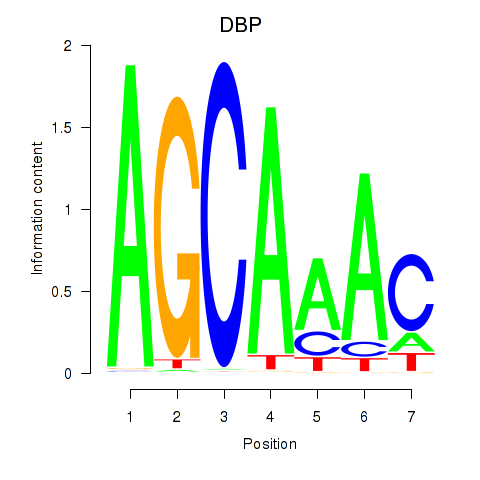
| DBP | 1.06 |
|
|
|
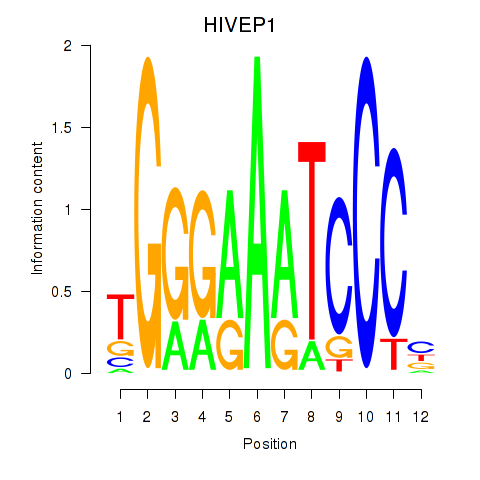
| HIVEP1 | 1.06 |
|
|
|
| NKX2-2 | 1.06 |
|

|
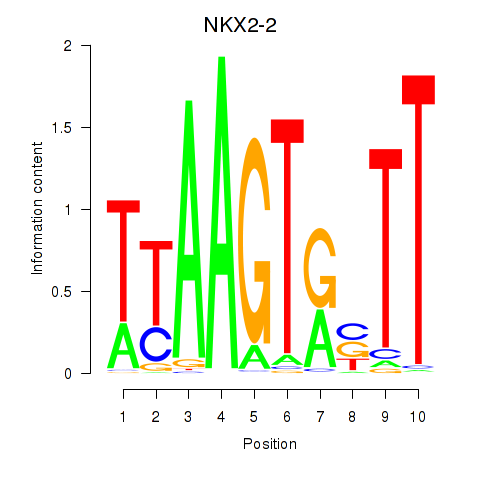
|
| FOSB | 1.05 |
|

|
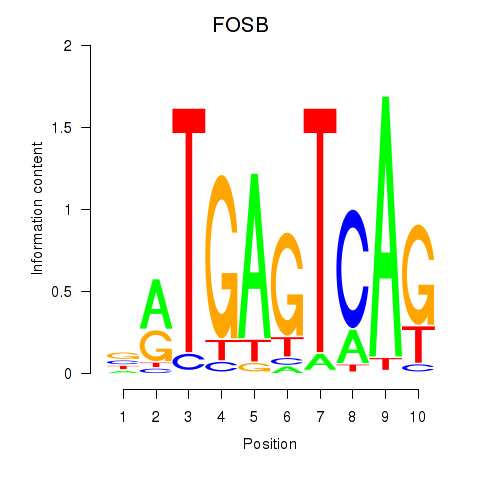
|
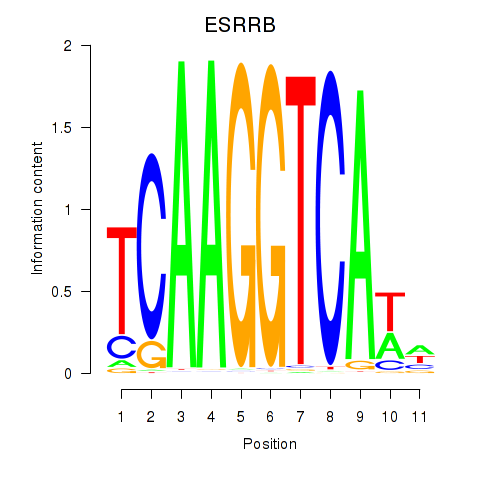
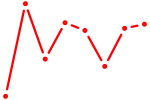
| ESRRB_ESRRG | 1.05 |
|

|
|
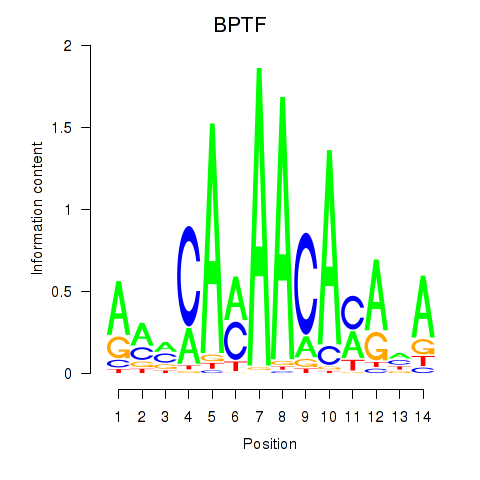
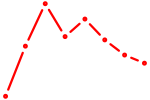
| BPTF | 1.04 |
|
|
|
| PPARG | 1.04 |
|
|
|
| CDC5L | 1.03 |
|

|
|
| ALX3 | 1.02 |
|

|
|
| FIGLA | 1.02 |
|

|
|
| TBX2 | 1.01 |
|

|
|
| RHOXF1 | 1.01 |
|

|
|
| FOXD3_FOXI1_FOXF1 | 1.01 |
|
|
|
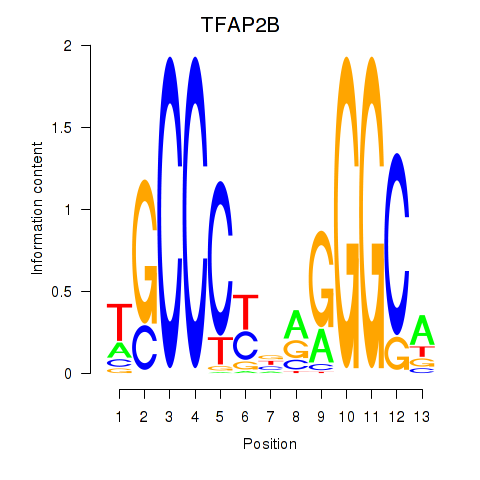
| TFAP2B | 1.01 |
|
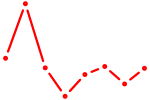
|
|
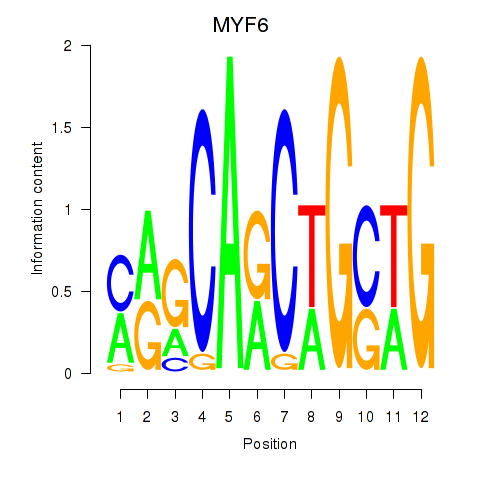
| MYF6 | 1.01 |
|

|
|
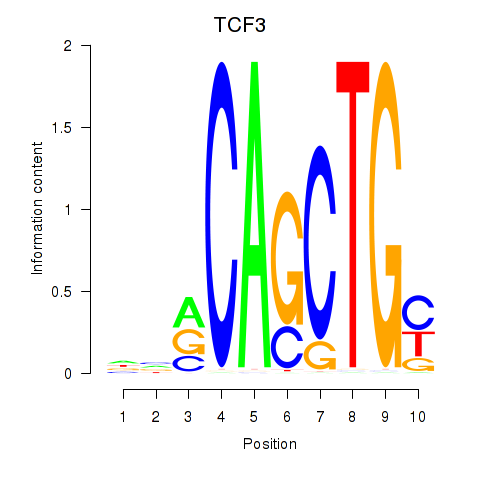
| TCF3_MYOG | 1.01 |
|
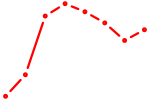
|
|
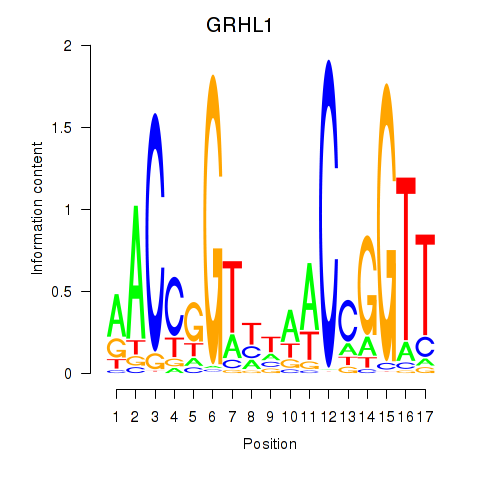
| GRHL1 | 1.01 |
|
|
|
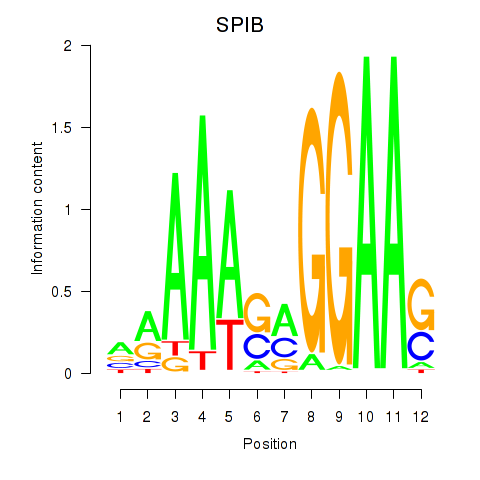
| SPIB | 1.01 |
|

|
|
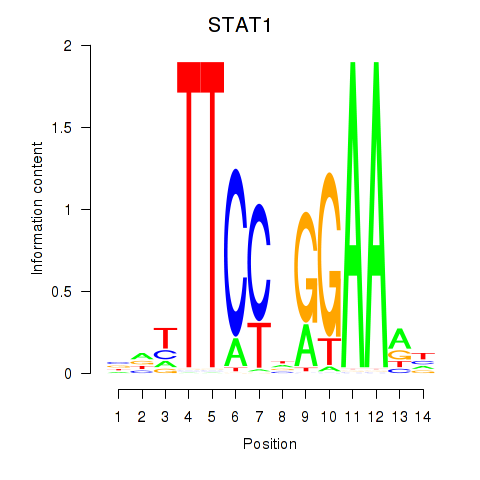
| STAT1_STAT3_BCL6 | 1.01 |
|
|
|
| MYBL2 | 1.00 |
|

|
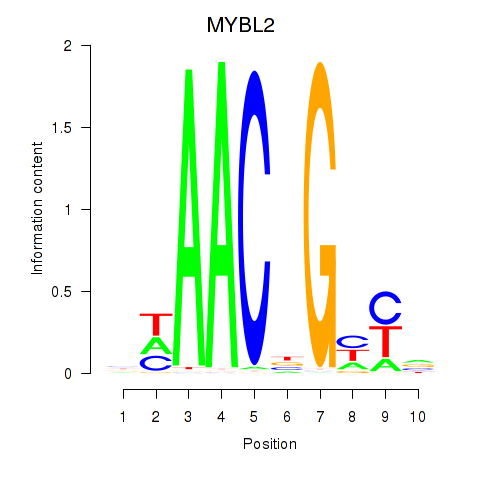
|
| SHOX | 0.99 |
|
|
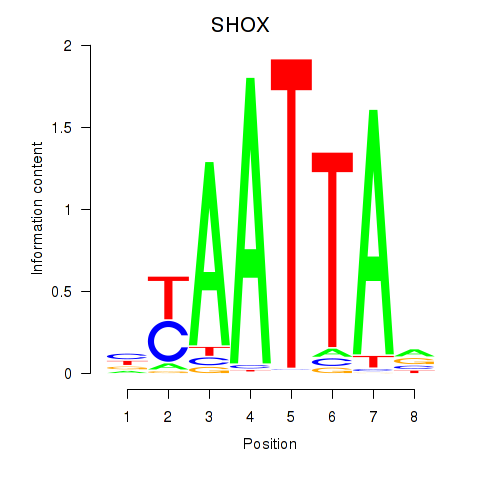
|
| GMEB2 | 0.99 |
|

|
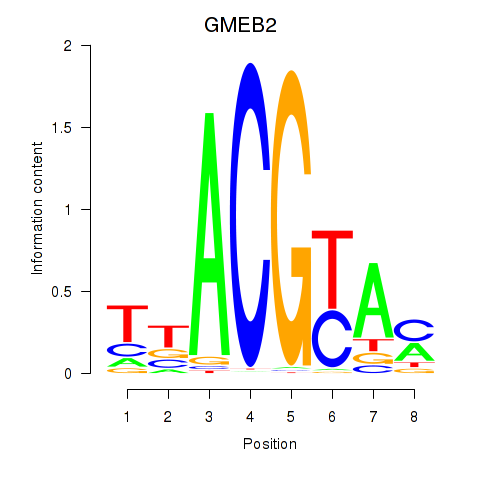
|
| KLF16_SP2 | 0.99 |
|

|
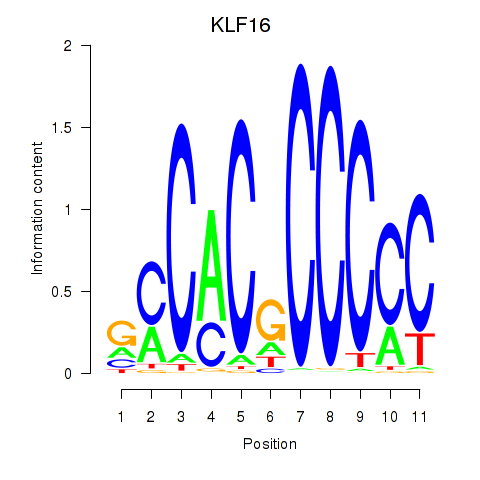
|
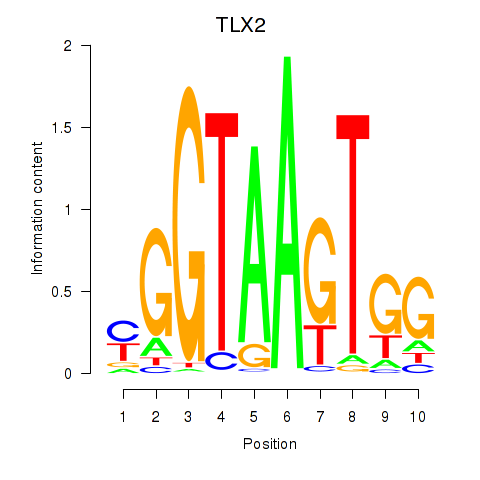
| TLX2 | 0.98 |
|
|
|
| UUGGUCC | 0.98 |
|
|

|
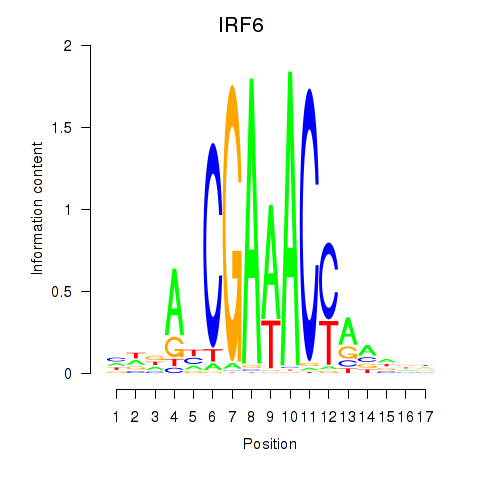
| IRF6_IRF4_IRF5 | 0.98 |
|

|
|
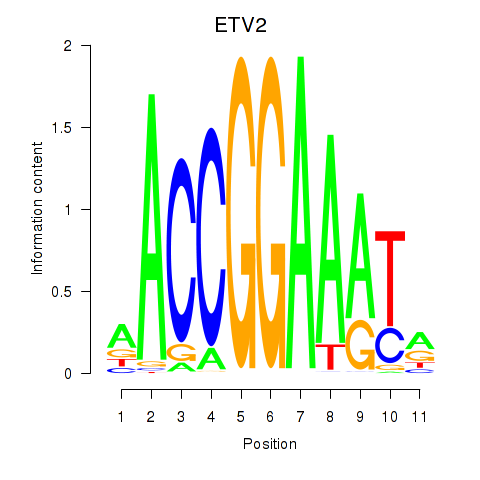
| ETV2 | 0.98 |
|

|
|
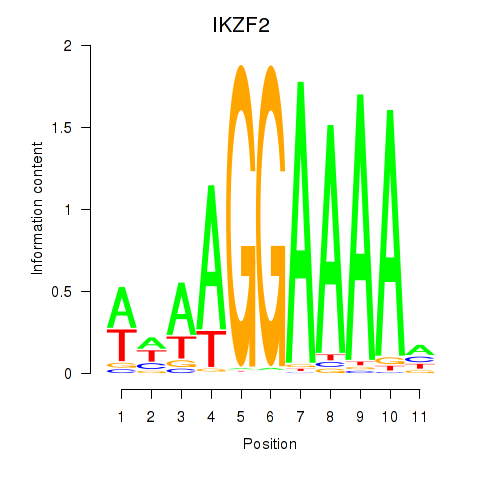
| IKZF2 | 0.98 |
|
|
|
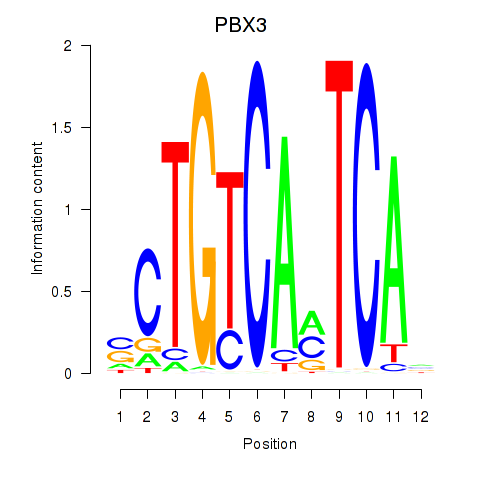
| PBX3 | 0.97 |
|
|
|
| HES1 | 0.97 |
|
|
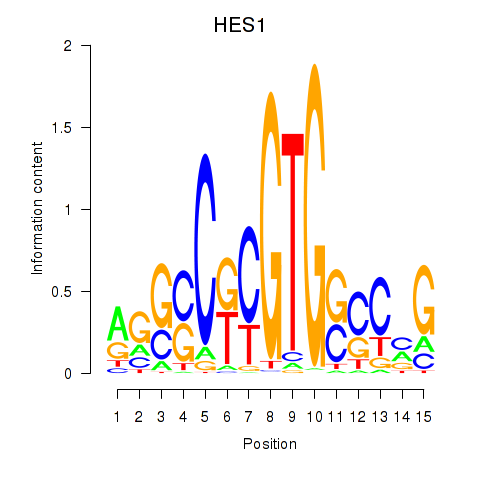
|
| AAUACUG | 0.97 |
|
|

|
| KLF3 | 0.96 |
|

|
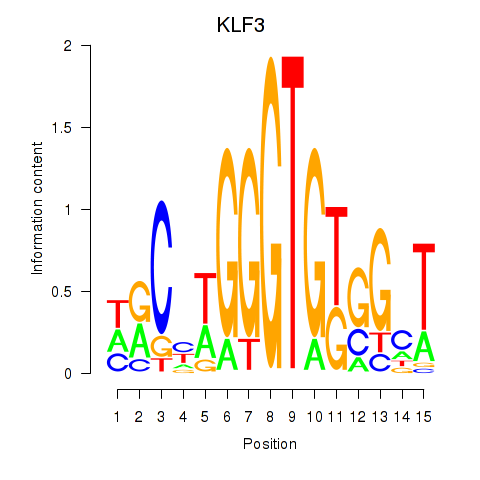
|
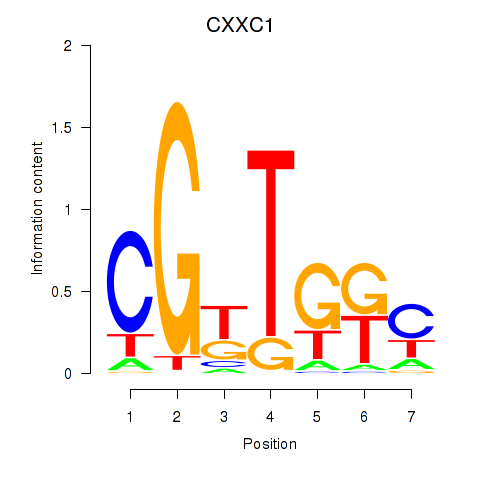
| CXXC1 | 0.96 |
|
|
|
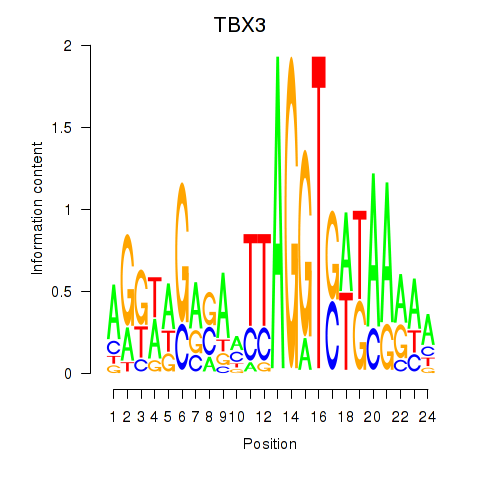
| TBX3 | 0.96 |
|
|
|
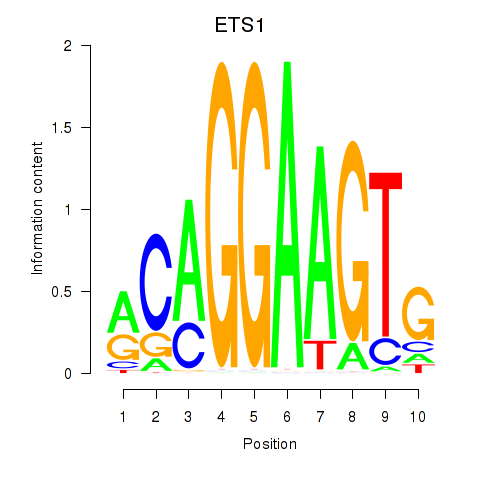
| ETS1 | 0.96 |
|
|
|
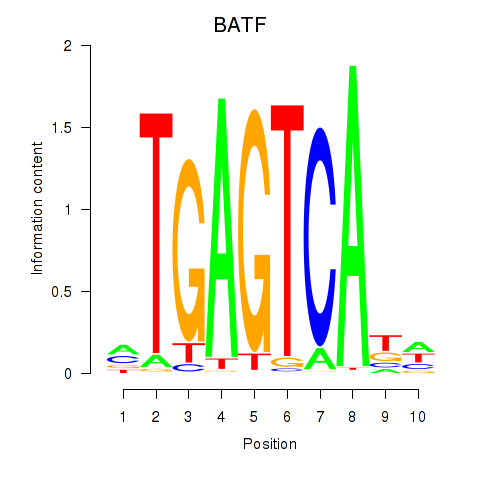
| BATF | 0.95 |
|

|
|
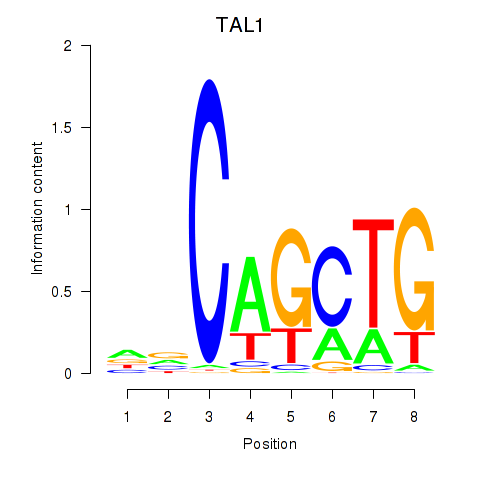
| TAL1 | 0.95 |
|

|
|
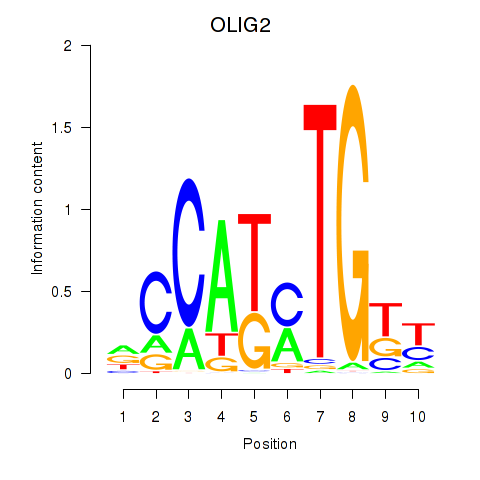
| OLIG2_NEUROD1_ATOH1 | 0.95 |
|

|
|
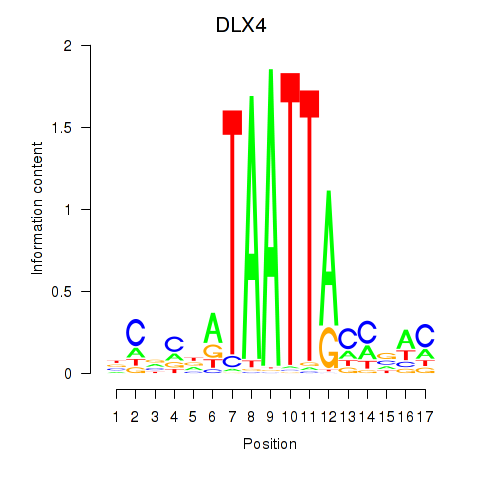
| DLX4_HOXD8 | 0.95 |
|
|
|
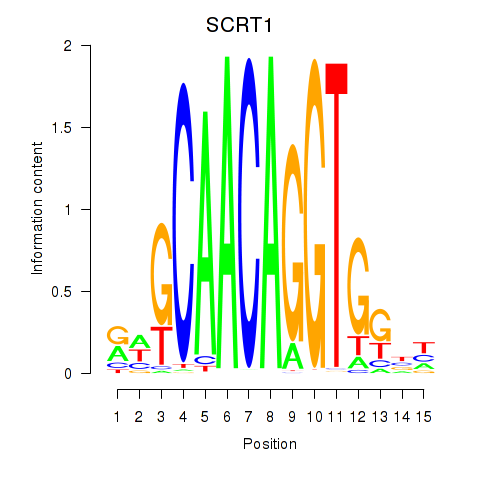
| SCRT1_SCRT2 | 0.94 |
|
|
|
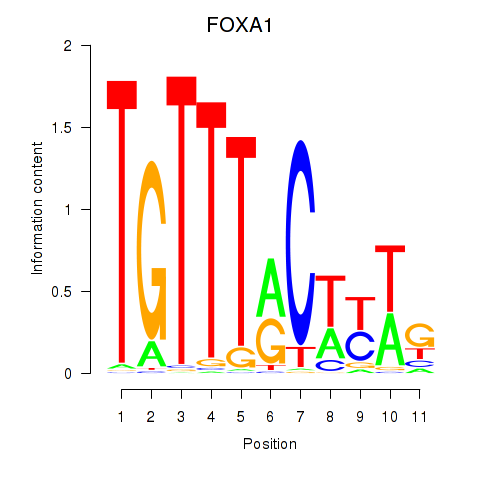
| FOXA1 | 0.93 |
|
|
|
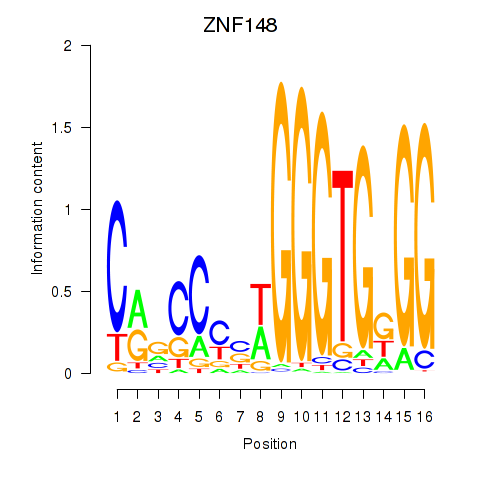
| ZNF148 | 0.93 |
|
|
|
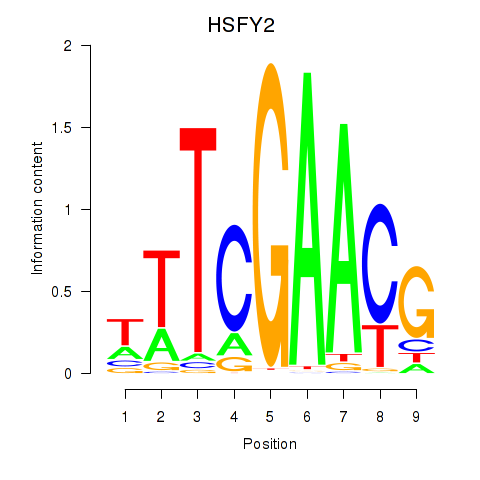
| HSFY2 | 0.93 |
|
|
|
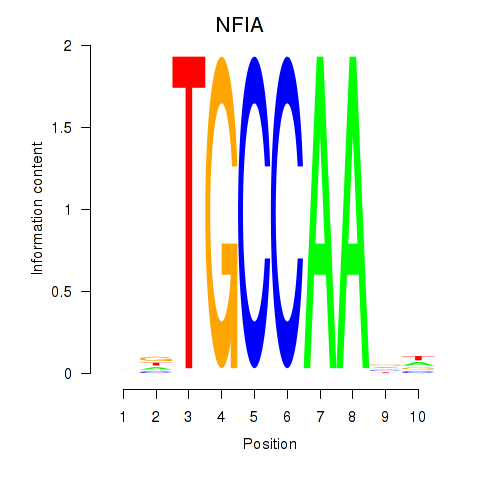
| NFIA | 0.92 |
|
|
|
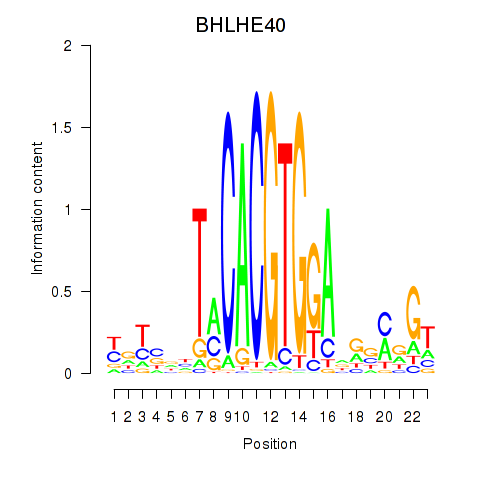
| BHLHE40 | 0.92 |
|

|
|
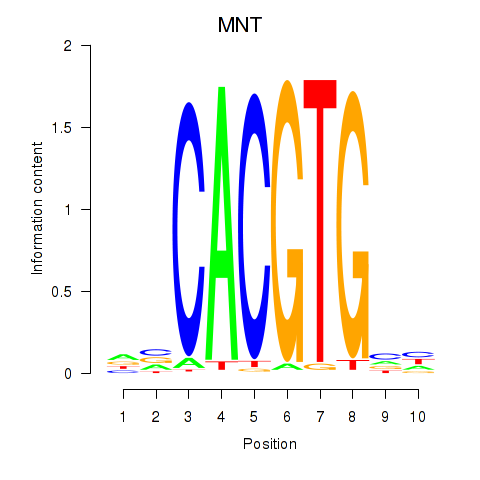
| MNT_HEY1_HEY2 | 0.92 |
|
|
|
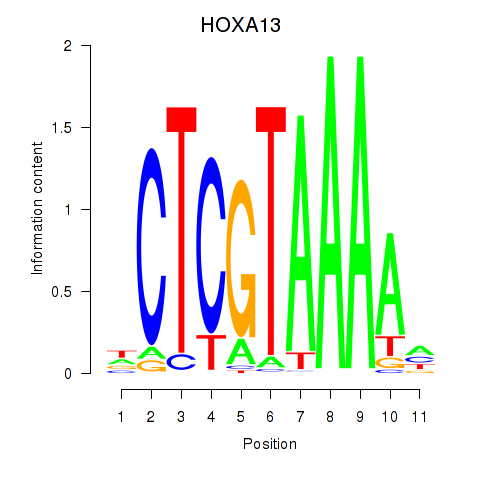
| HOXA13 | 0.91 |
|

|
|
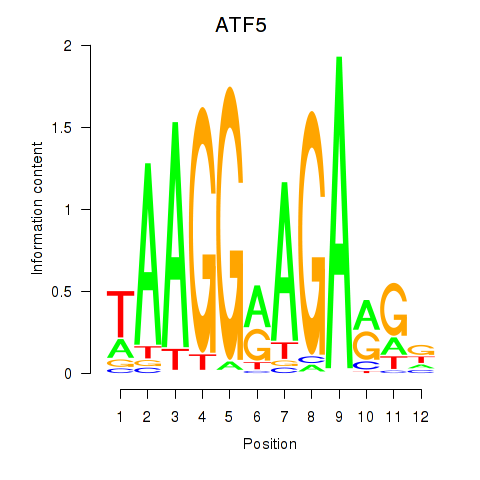
| ATF5 | 0.91 |
|
|
|
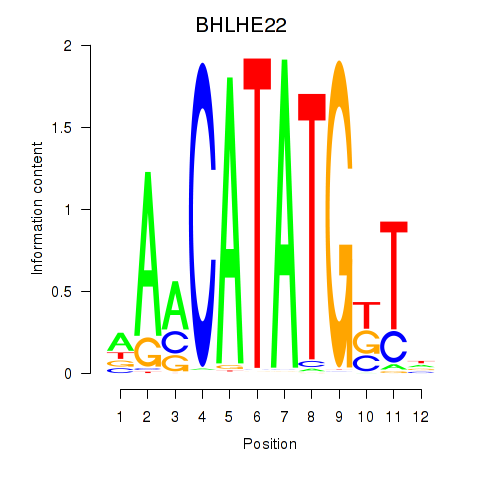
| BHLHE22_BHLHA15_BHLHE23 | 0.91 |
|
|
|
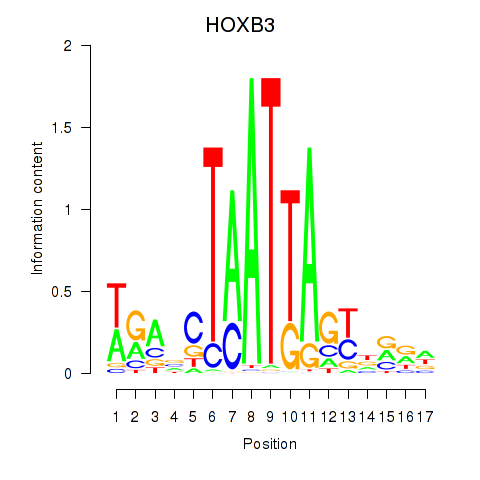
| HOXB3 | 0.91 |
|

|
|
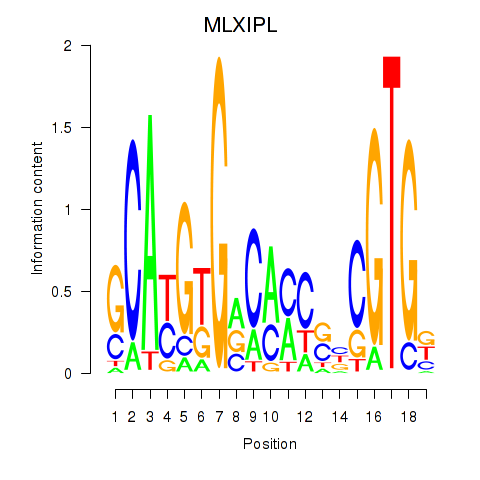
| MLXIPL | 0.91 |
|

|
|
| TP53 | 0.91 |
|
|

|
| FOXK1_FOXP2_FOXB1_FOXP3 | 0.89 |
|

|
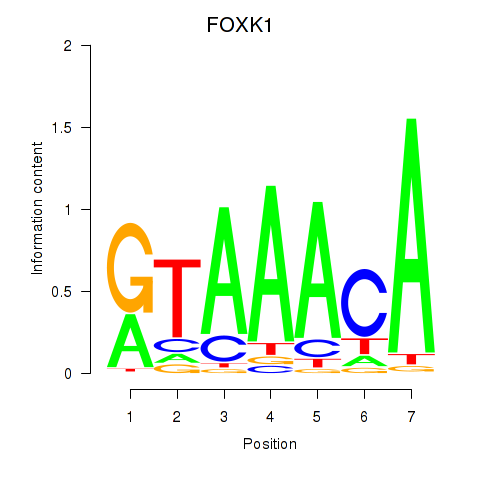
|
| IRX2 | 0.87 |
|

|
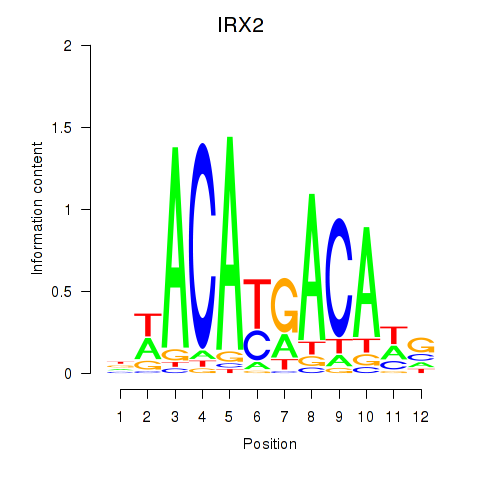
|
| HOXB8 | 0.86 |
|

|
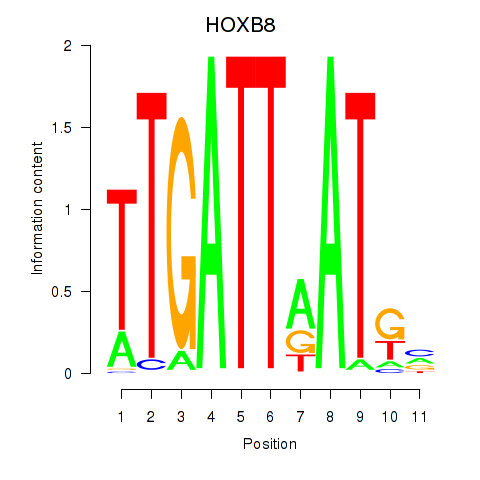
|
| AUUGCAC | 0.86 |
|
|

|
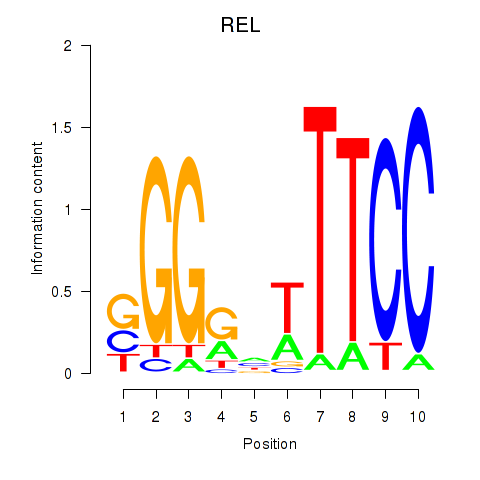
| REL | 0.86 |
|
|
|
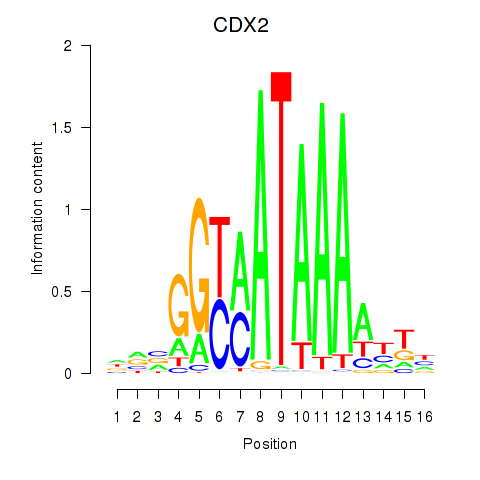
| CDX2 | 0.86 |
|

|
|
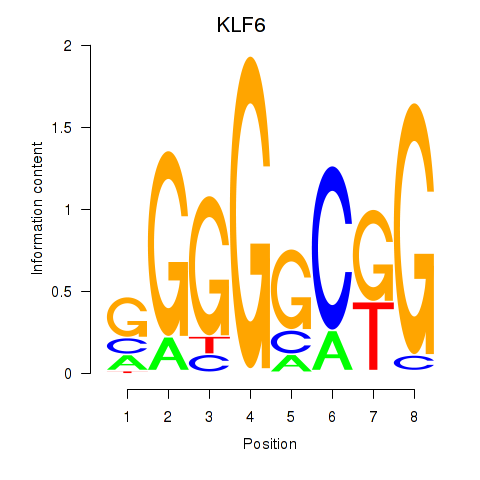
| KLF6 | 0.85 |
|
|
|
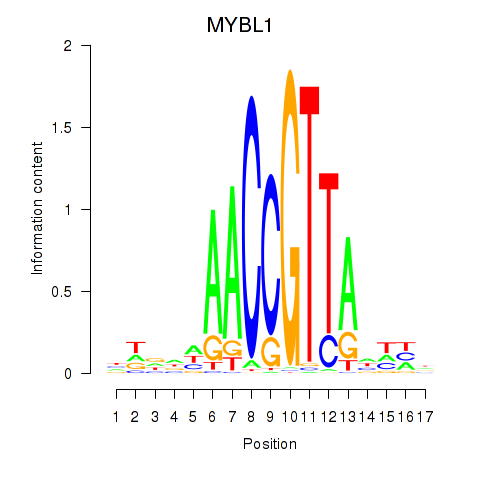
| MYBL1 | 0.85 |
|

|
|
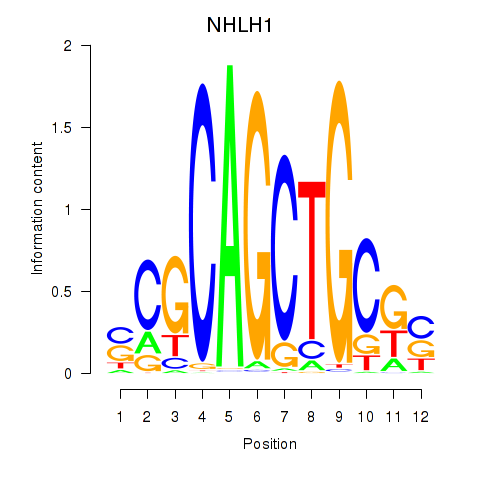
| NHLH1 | 0.85 |
|
|
|
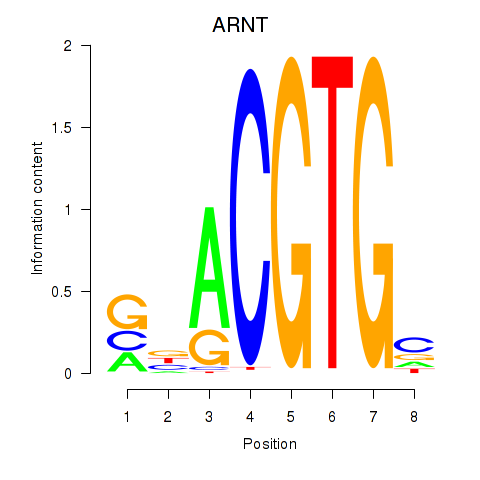
| ARNT | 0.85 |
|
|
|
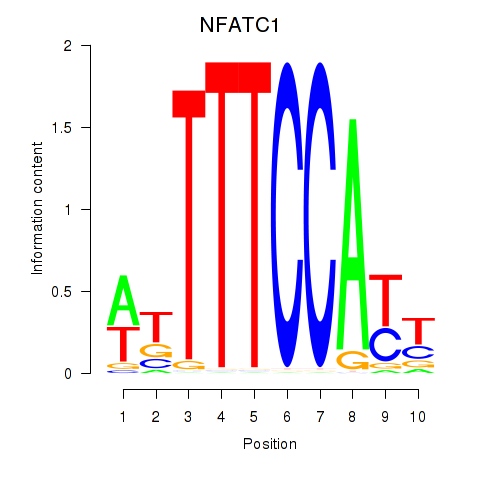
| NFATC1 | 0.84 |
|

|
|
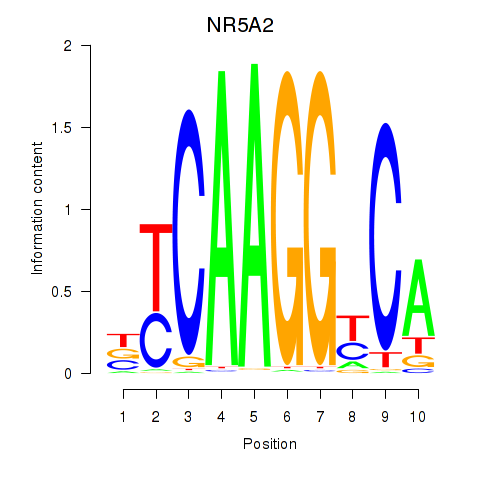
| NR5A2 | 0.84 |
|
|
|
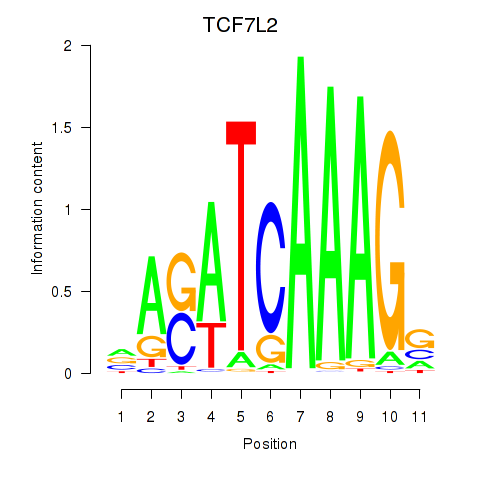
| TCF7L2 | 0.84 |
|
|
|
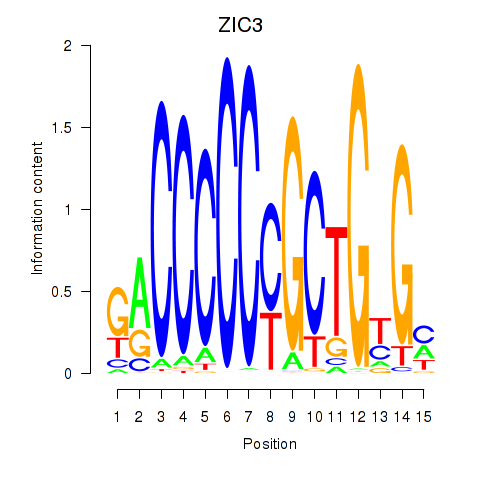
| ZIC3_ZIC4 | 0.84 |
|
|
|
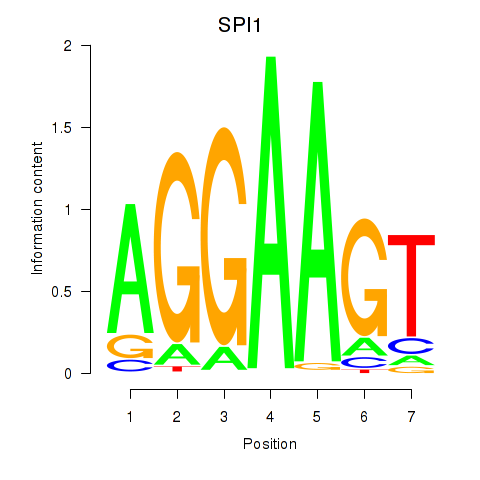
| SPI1 | 0.83 |
|

|
|
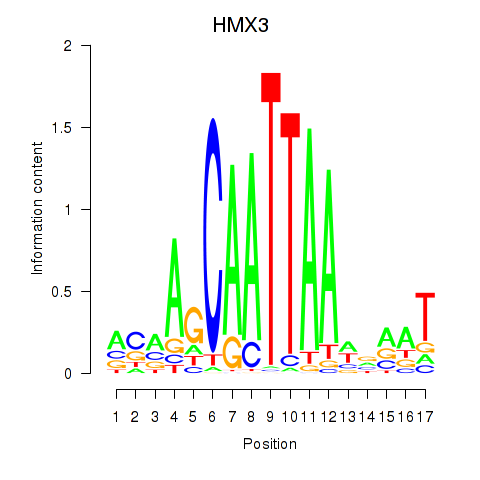
| HMX3 | 0.83 |
|
|
|
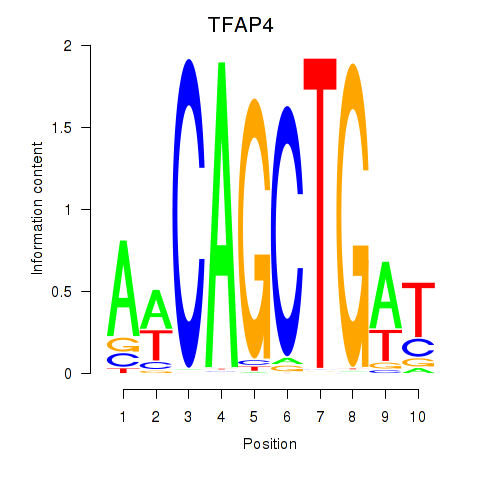
| TFAP4_MSC | 0.82 |
|
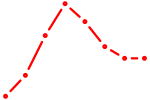
|
|
| GMEB1 | 0.82 |
|

|
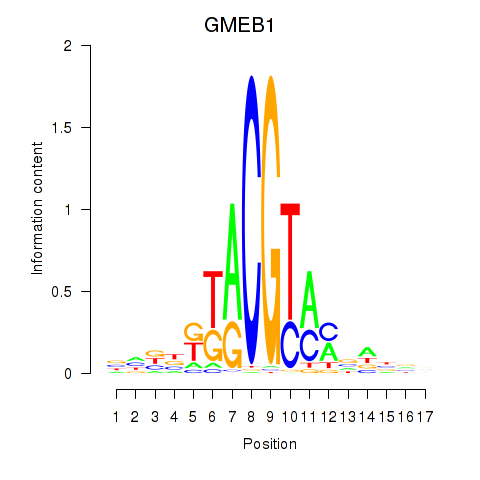
|
| ZNF740_ZNF219 | 0.82 |
|

|
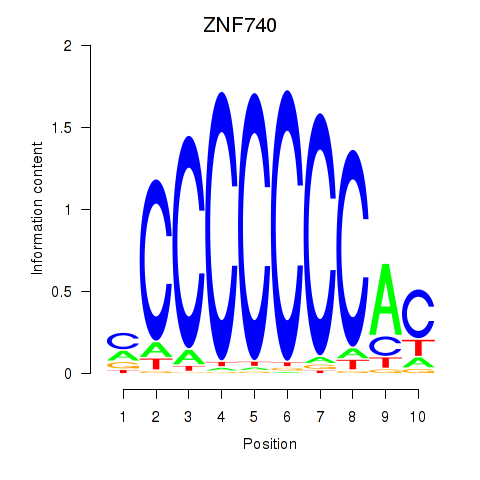
|
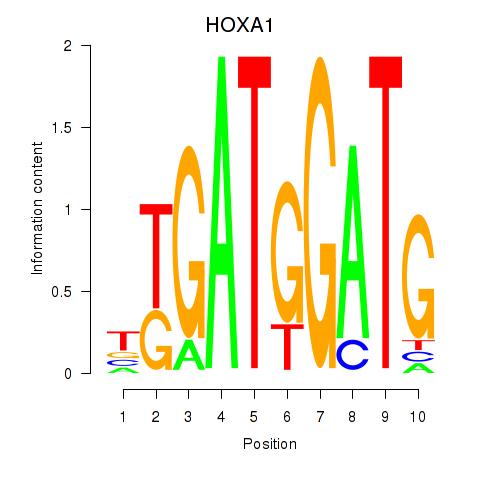
| HOXA1 | 0.81 |
|

|
|
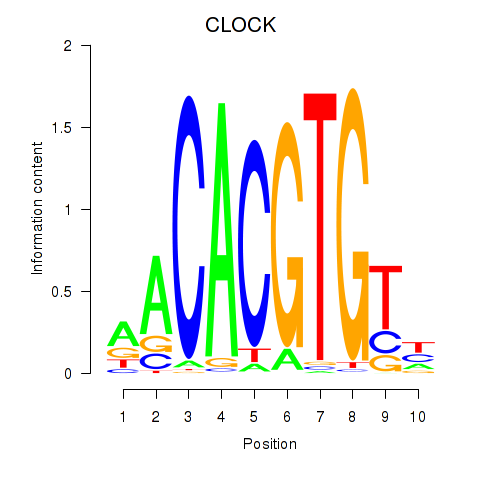
| CLOCK | 0.81 |
|
|
|
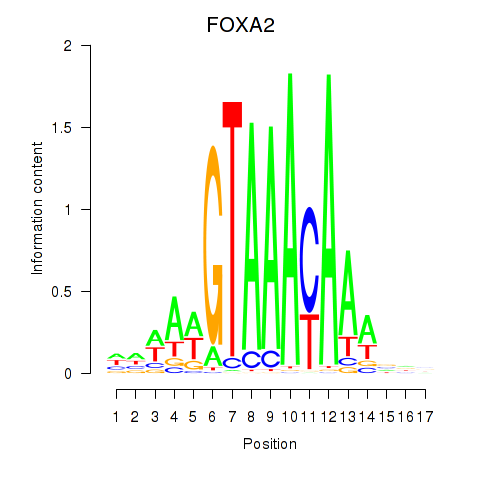
| FOXA2_FOXJ3 | 0.81 |
|

|
|
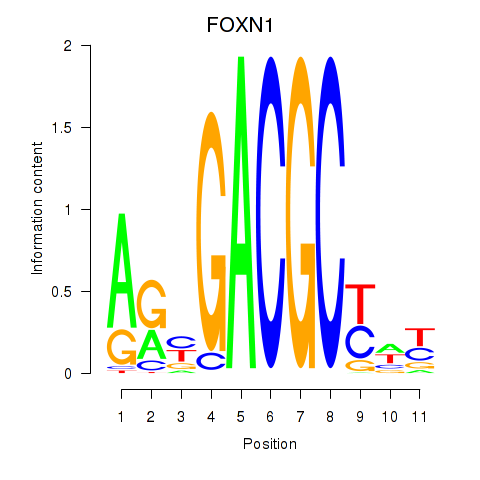
| FOXN1 | 0.81 |
|

|
|
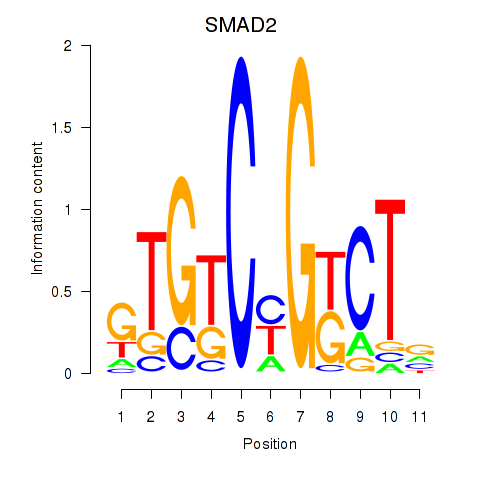
| SMAD2 | 0.80 |
|
|
|
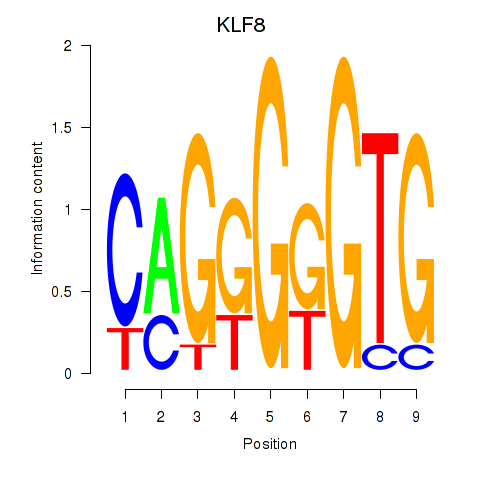
| KLF8 | 0.80 |
|
|
|
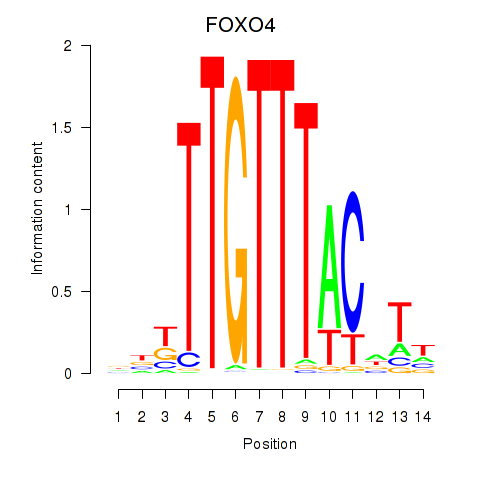
| FOXO4 | 0.80 |
|

|
|
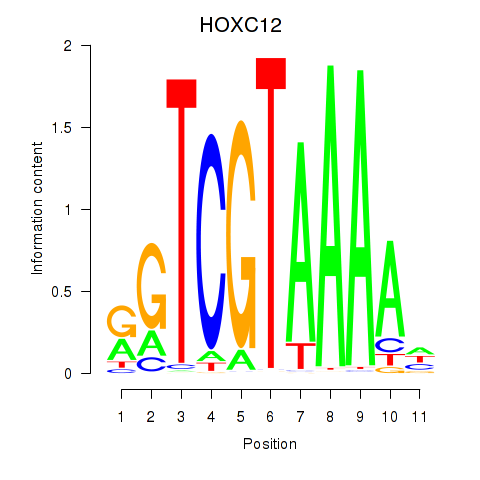
| HOXC12_HOXD12 | 0.80 |
|

|
|
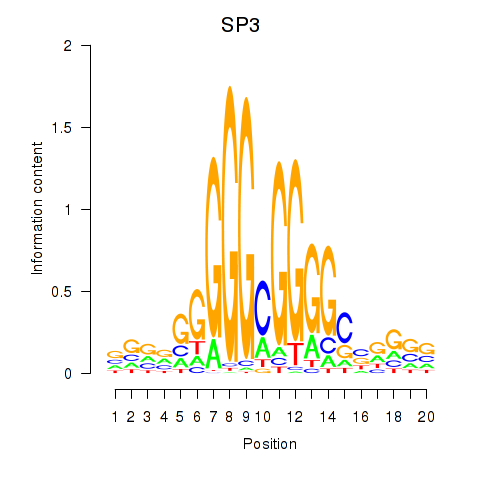
| SP3 | 0.79 |
|
|
|
| AACACUG | 0.79 |
|

|

|
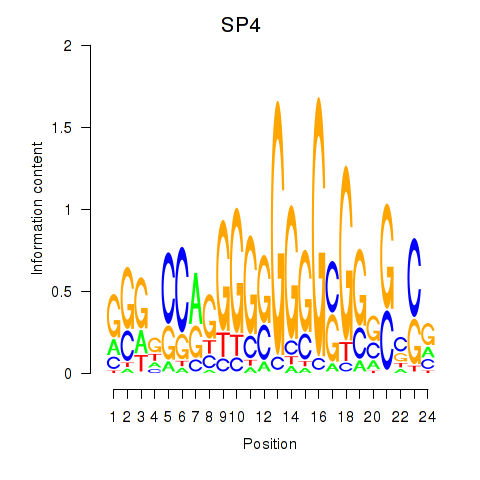
| SP4_PML | 0.79 |
|
|
|
| UGGUCCC | 0.79 |
|
|

|
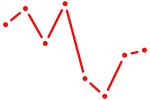
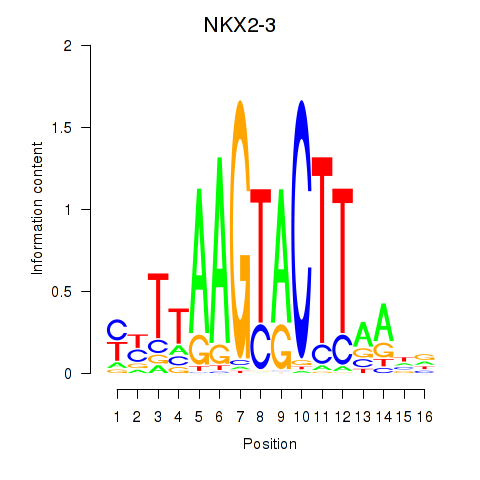
| NKX2-3 | 0.79 |
|
|
|
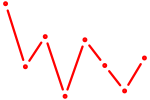
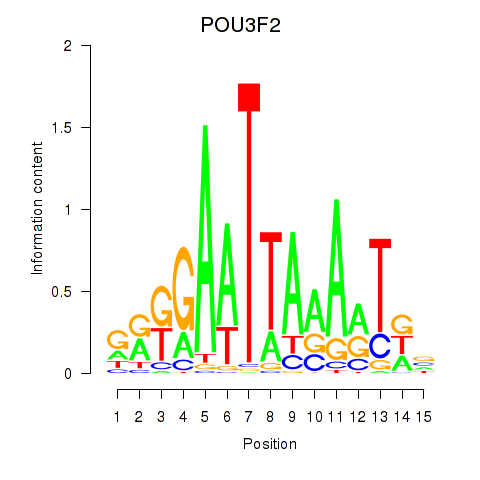
| POU3F2 | 0.79 |
|
|
|
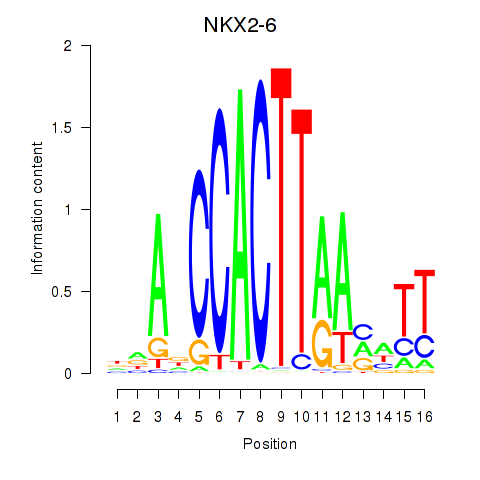
| NKX2-6 | 0.79 |
|

|
|
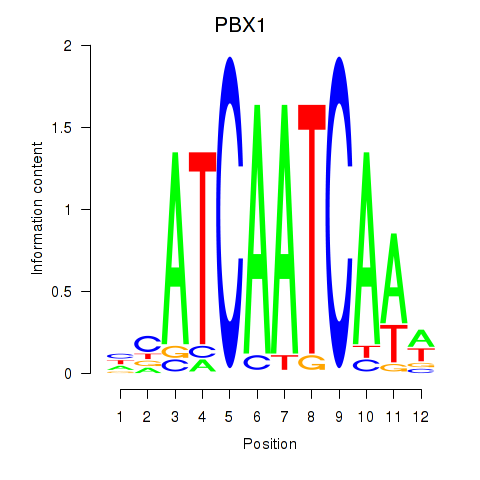
| PBX1 | 0.79 |
|

|
|
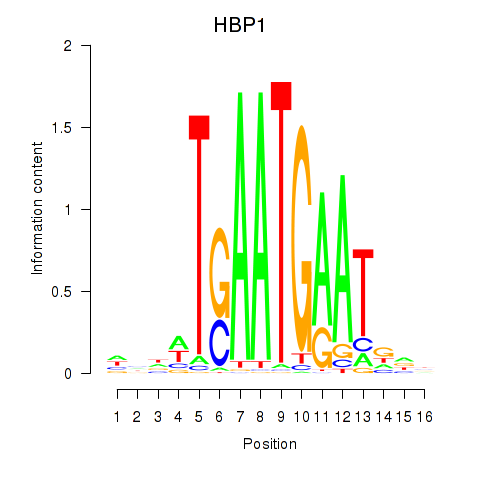
| HBP1 | 0.79 |
|
|
|
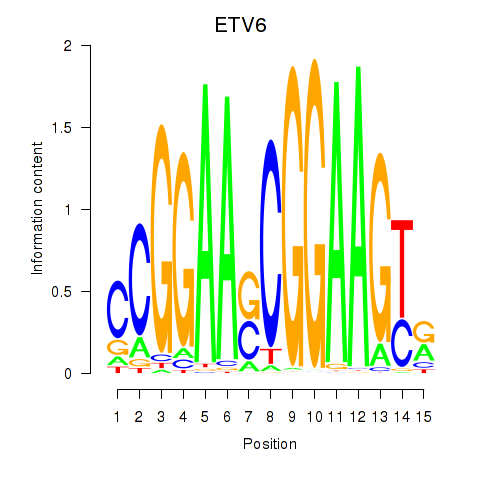
| ETV6 | 0.78 |
|

|
|
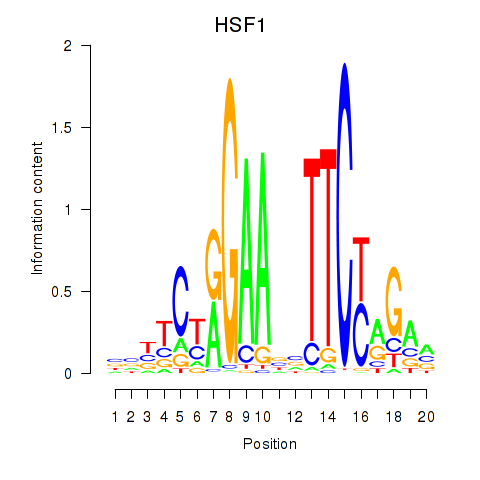
| HSF1 | 0.78 |
|

|
|
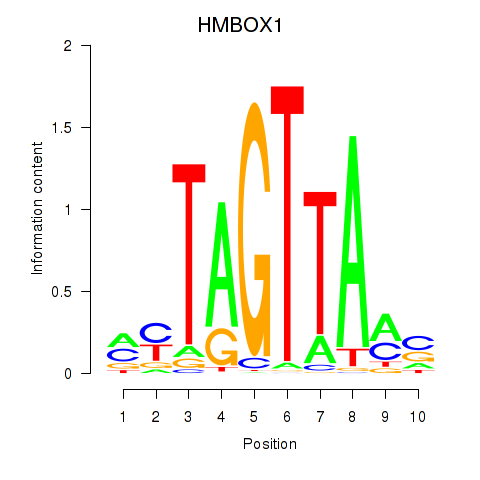
| HMBOX1 | 0.78 |
|
|
|
| RELB | 0.78 |
|

|
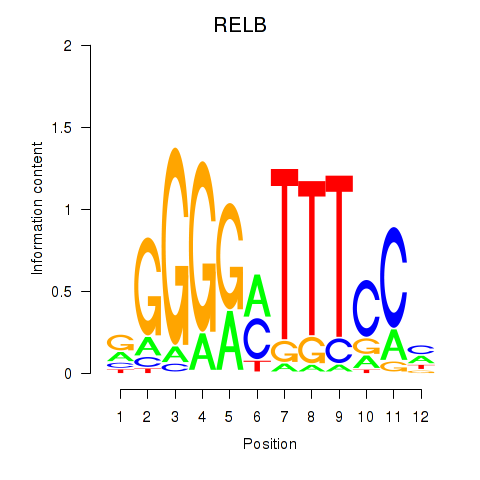
|
| DBX2_HLX | 0.78 |
|

|
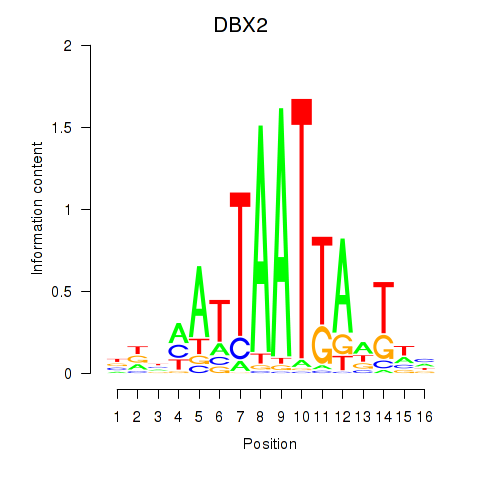
|
| IRF9 | 0.77 |
|
|
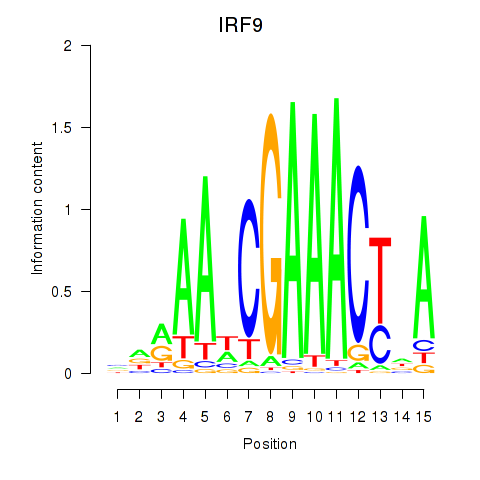
|
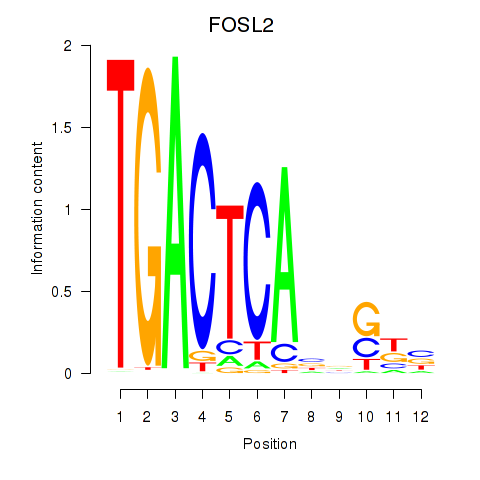
| FOSL2_SMARCC1 | 0.77 |
|

|
|
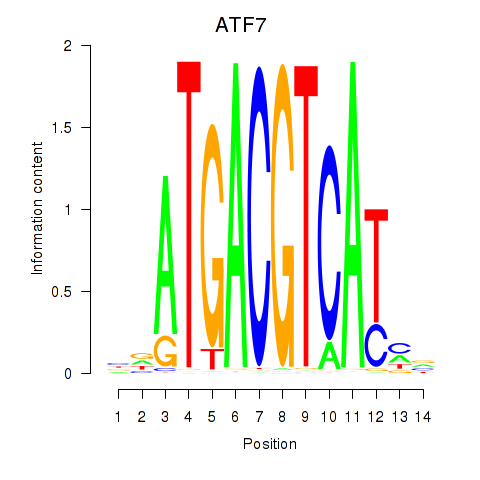
| ATF7 | 0.77 |
|

|
|
| UCAAGUA | 0.77 |
|
|

|
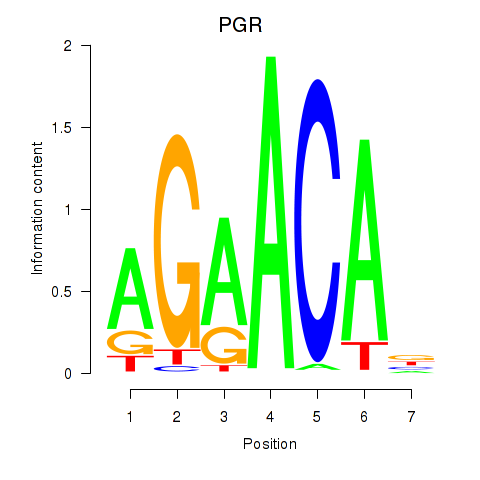
| PGR | 0.76 |
|
|
|
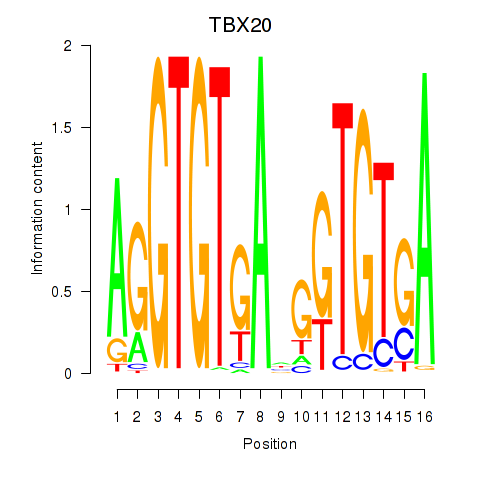
| TBX20 | 0.76 |
|

|
|
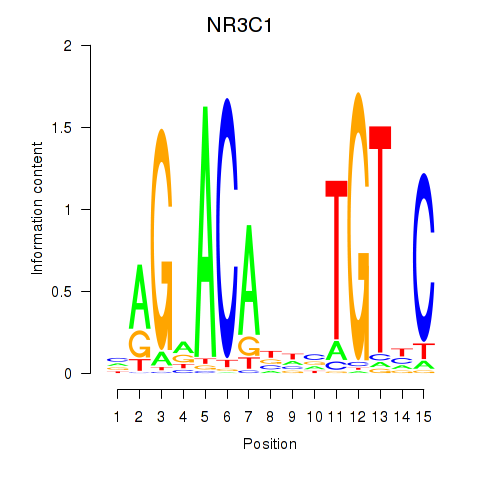
| NR3C1 | 0.75 |
|
|
|
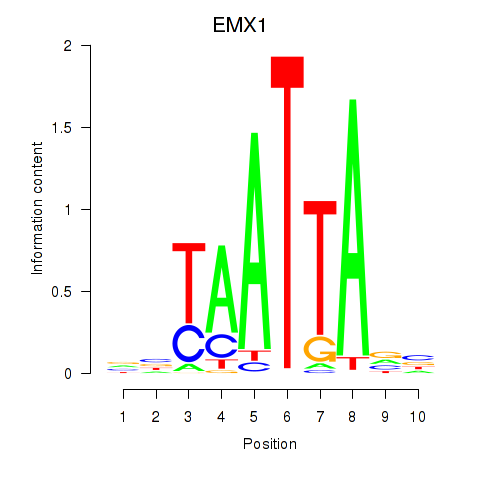
| EMX1 | 0.75 |
|
|
|
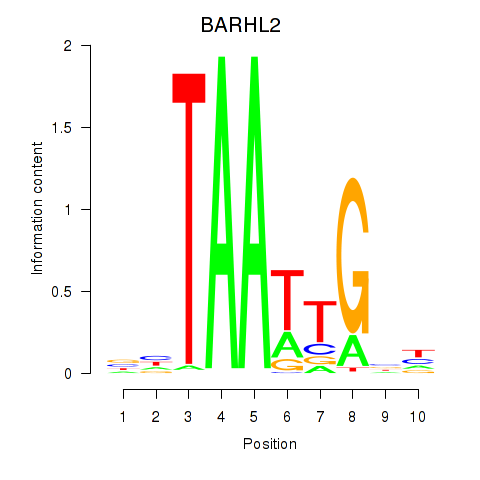
| BARHL2 | 0.75 |
|
|
|
| MEF2D_MEF2A | 0.75 |
|

|
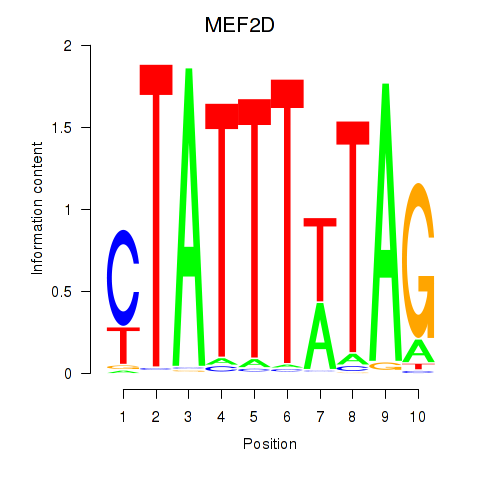
|
| ELF3_EHF | 0.75 |
|
|
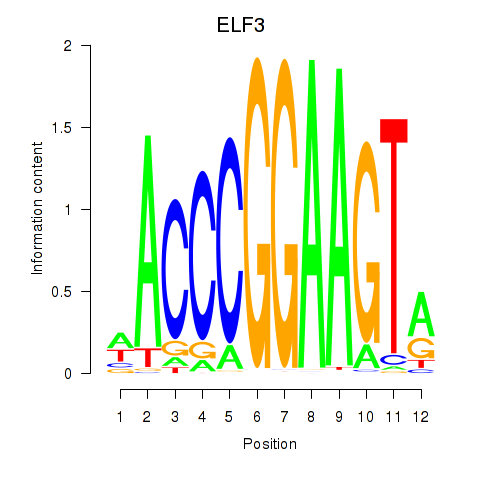
|
| CREB3L2 | 0.75 |
|
|

|
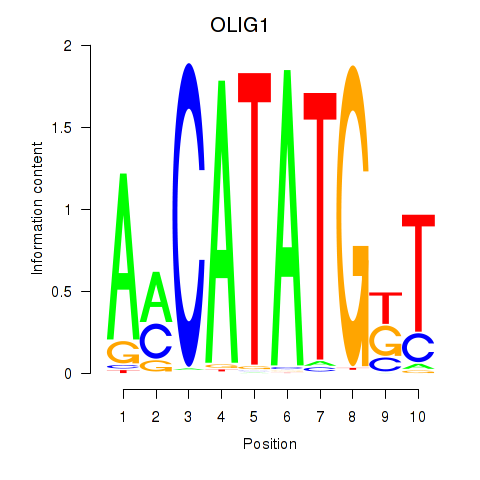
| OLIG1 | 0.75 |
|
|
|
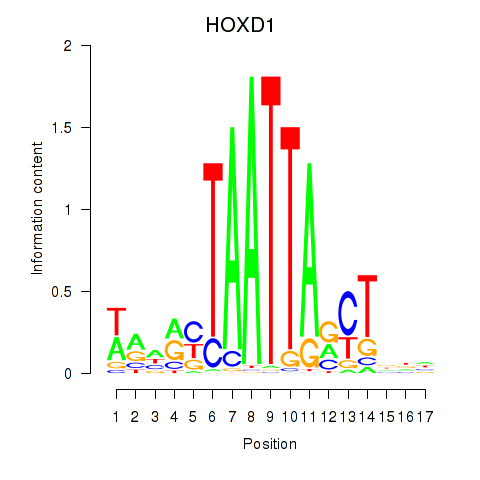
| HOXD1 | 0.75 |
|

|
|
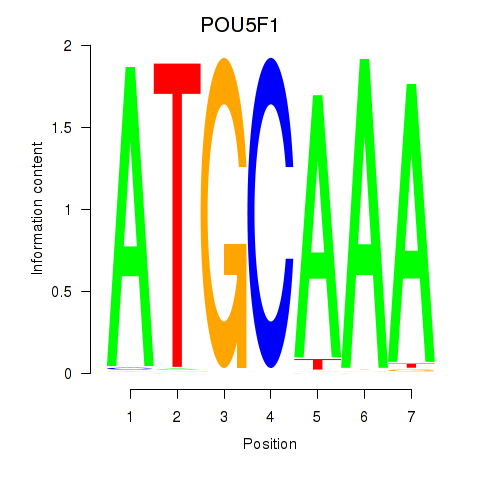
| POU5F1_POU2F3 | 0.75 |
|
|
|
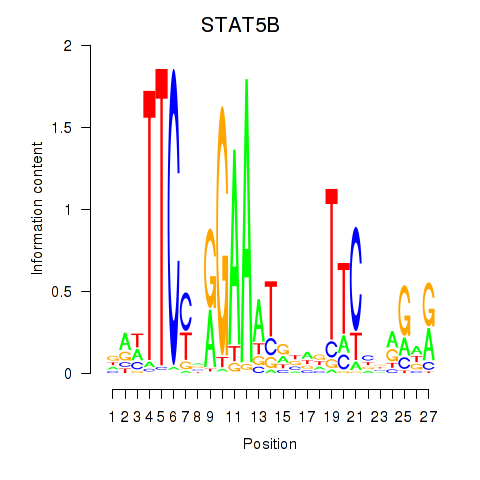
| STAT5B | 0.75 |
|

|
|
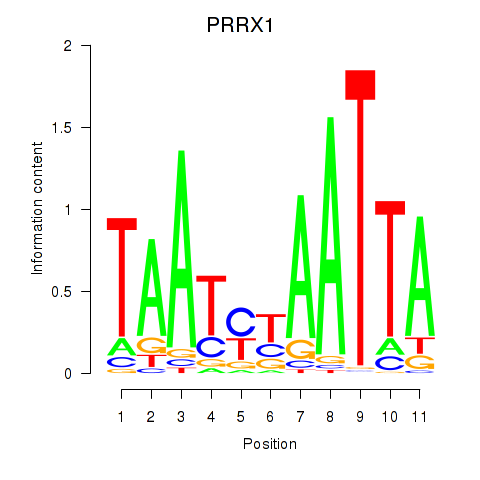
| PRRX1_ALX4_PHOX2A | 0.75 |
|

|
|
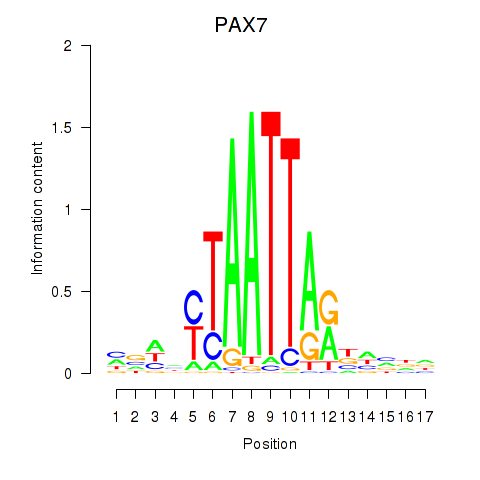
| PAX7_NOBOX | 0.74 |
|
|
|
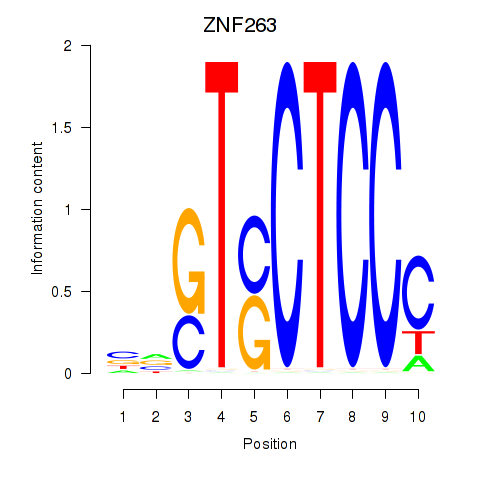
| ZNF263 | 0.74 |
|
|
|
| HMX1 | 0.74 |
|
|
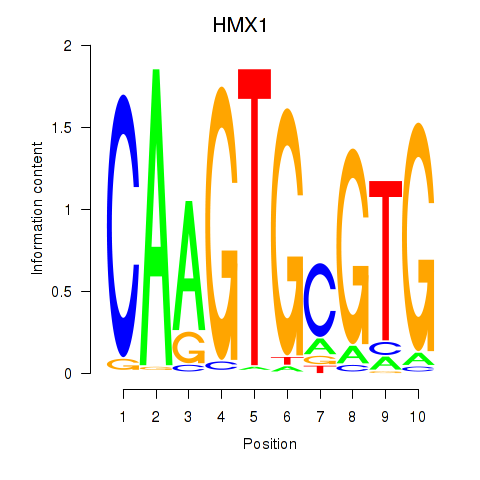
|
| SIX1_SIX3_SIX2 | 0.74 |
|

|
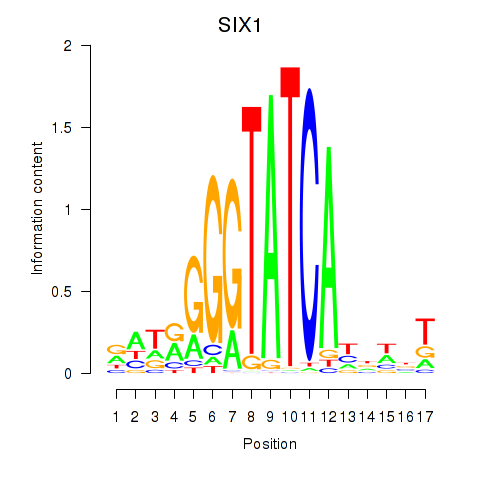
|
| OTX2_CRX | 0.74 |
|
|
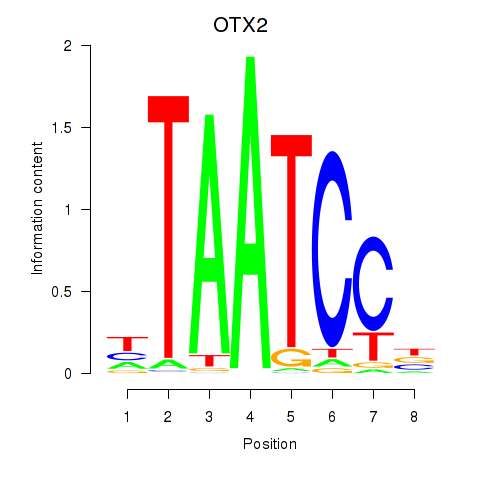
|
| AGUGCAA | 0.74 |
|

|

|
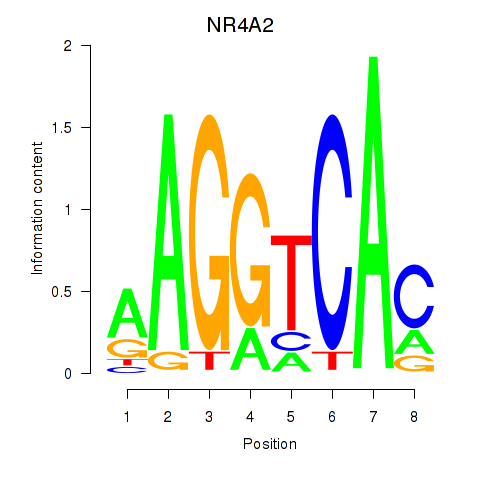
| NR4A2 | 0.74 |
|
|
|
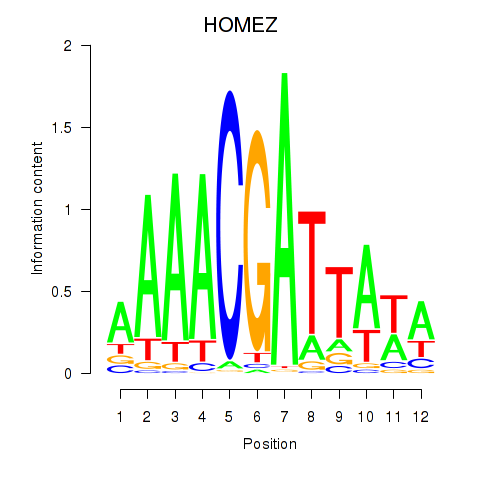
| HOMEZ | 0.73 |
|

|
|
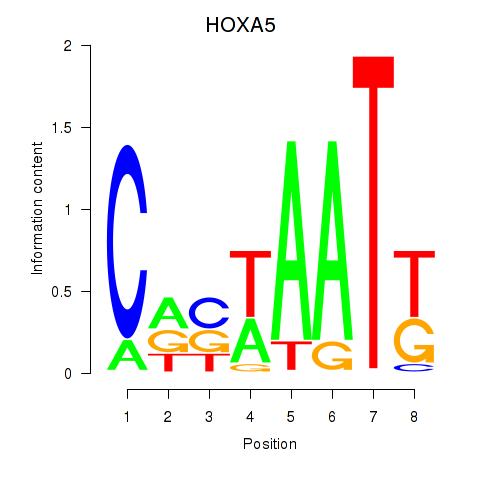
| HOXA5 | 0.73 |
|
|
|
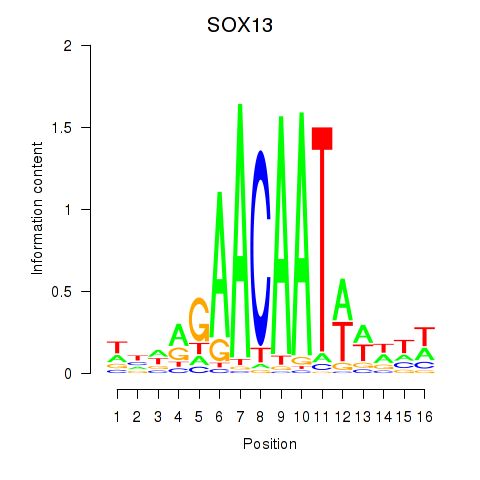
| SOX13_SOX12 | 0.73 |
|

|
|
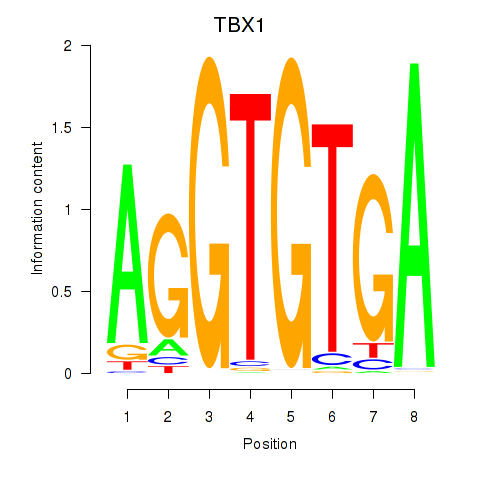
| TBX1 | 0.73 |
|

|
|
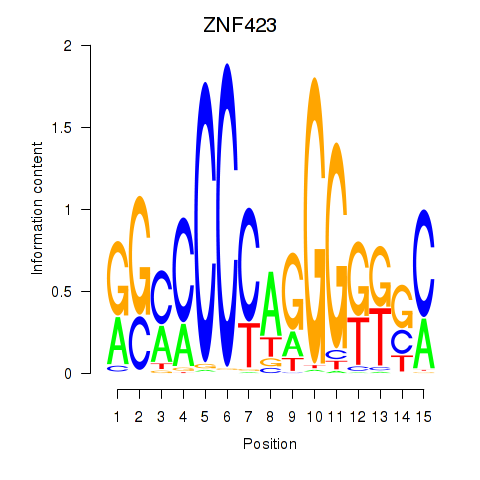
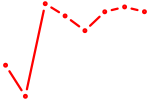
| ZNF423 | 0.73 |
|
|
|
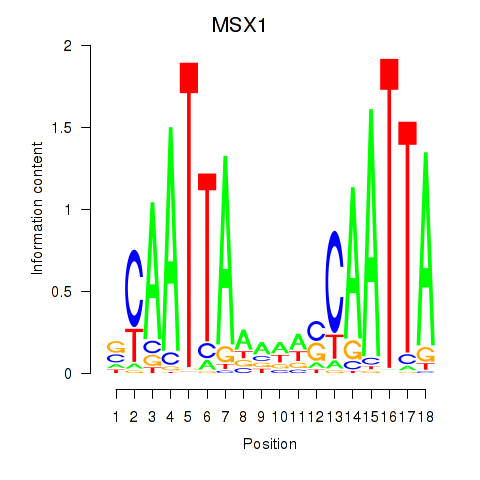
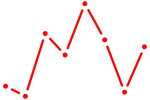
| MSX1 | 0.72 |
|
|
|
| MECOM | 0.72 |
|
|
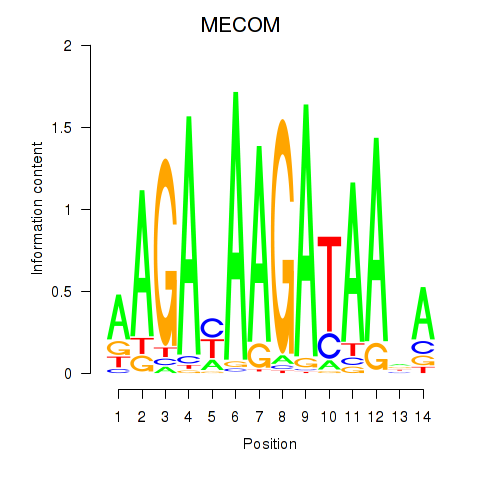
|
| E4F1 | 0.72 |
|

|
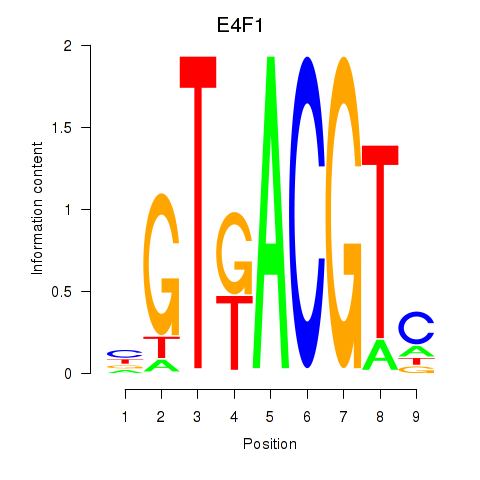
|
| EOMES | 0.72 |
|

|
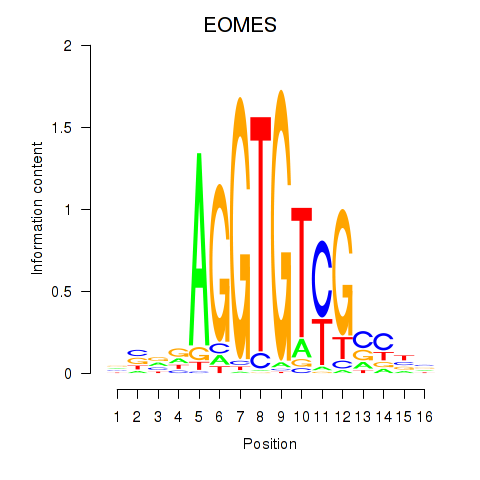
|
| CTCF_CTCFL | 0.71 |
|

|
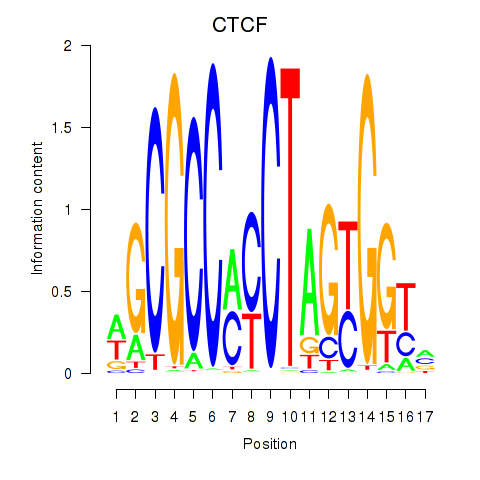
|
| ACAUUCA | 0.71 |
|
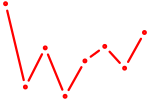
|

|
| MAFK | 0.71 |
|
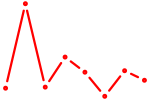
|
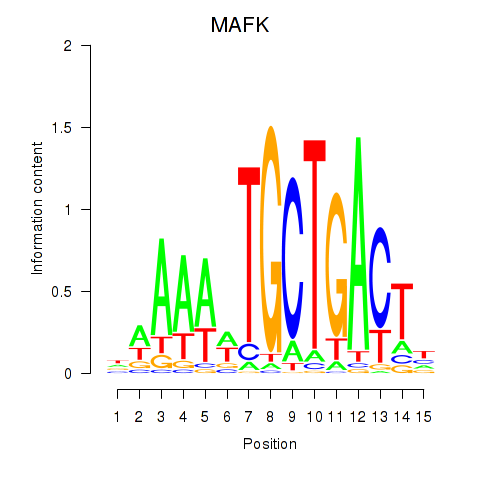
|
| HOXA4 | 0.71 |
|

|
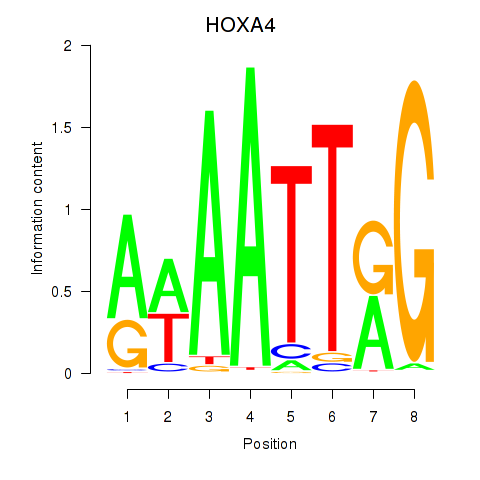
|
| HOXB4_LHX9 | 0.71 |
|

|
|
| ETV3 | 0.71 |
|

|

|
| POU6F2 | 0.71 |
|
|
|
| CREB5_CREM_JUNB | 0.71 |
|

|
|
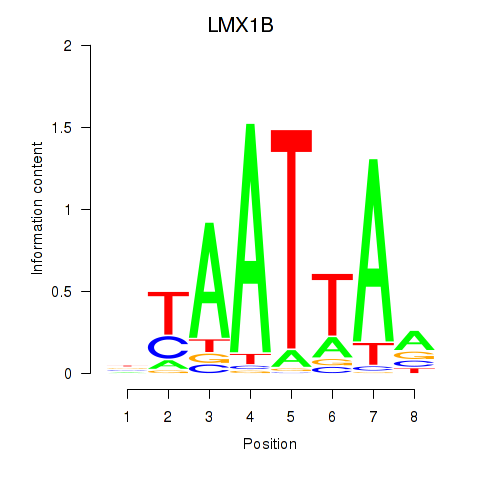
| LMX1B_MNX1_RAX2 | 0.71 |
|
|
|
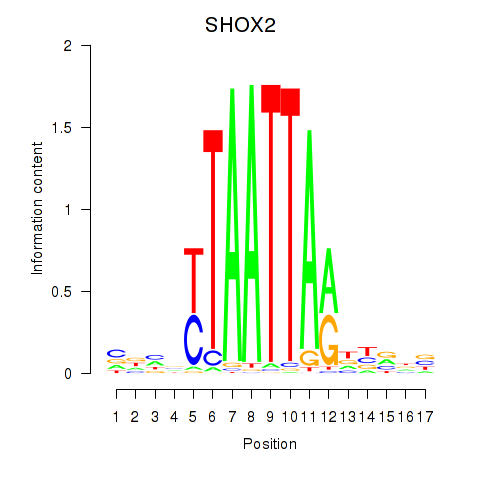
| SHOX2_HOXC5 | 0.71 |
|
|
|
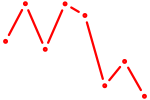
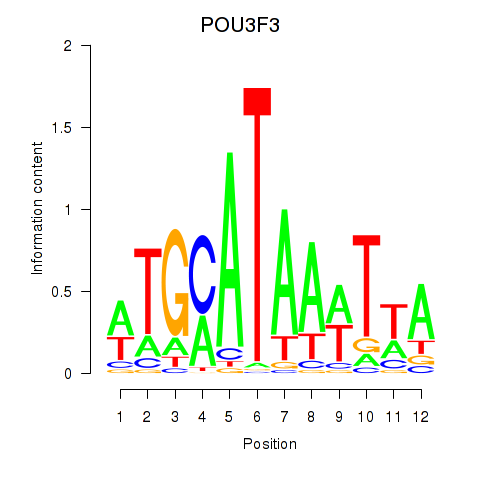
| POU3F3_POU3F4 | 0.71 |
|
|
|
| AAAGUGC | 0.70 |
|

|

|
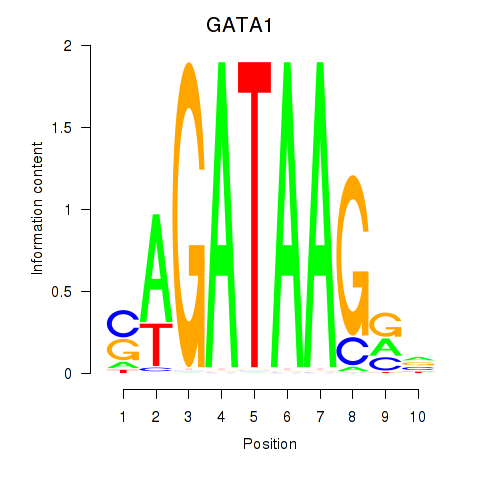
| GATA1_GATA4 | 0.70 |
|

|
|
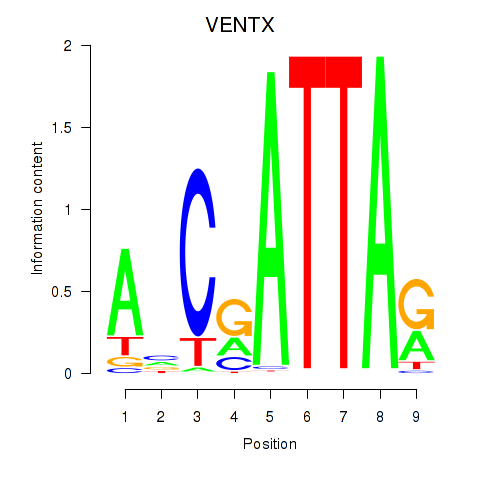
| VENTX | 0.70 |
|

|
|
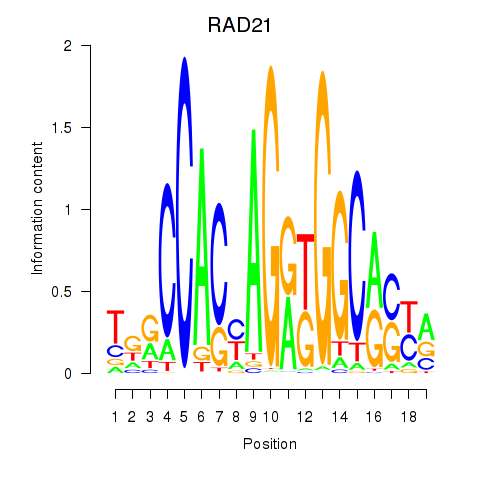
| RAD21_SMC3 | 0.69 |
|
|
|
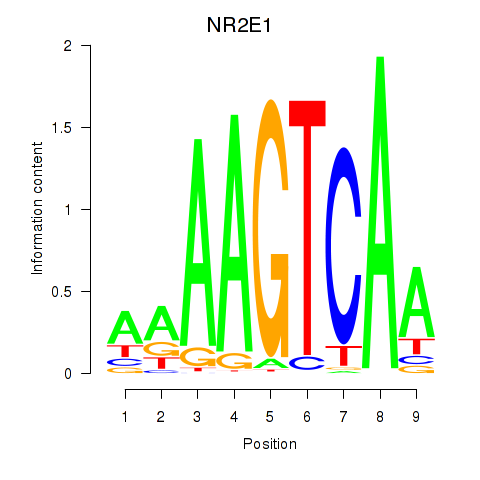
| NR2E1 | 0.69 |
|

|
|
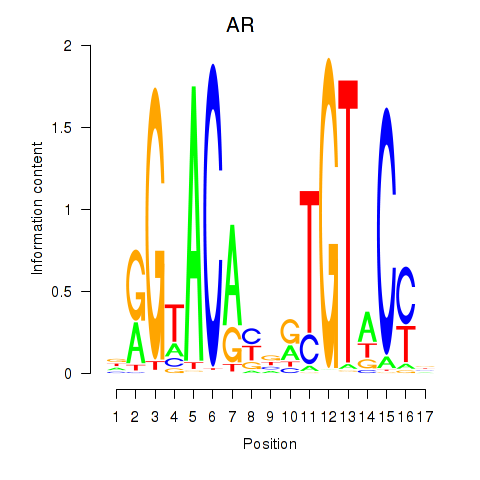
| AR_NR3C2 | 0.68 |
|

|
|
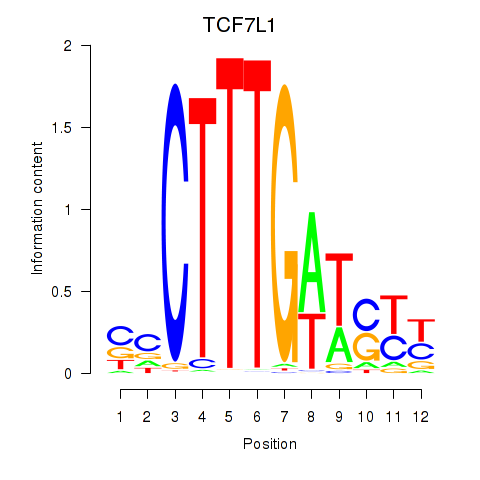
| TCF7L1 | 0.68 |
|

|
|
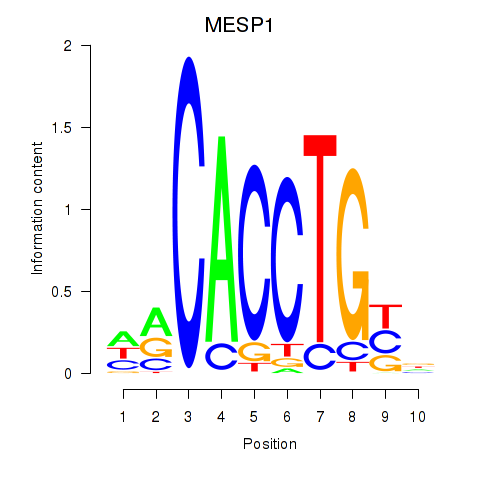
| MESP1 | 0.68 |
|
|
|
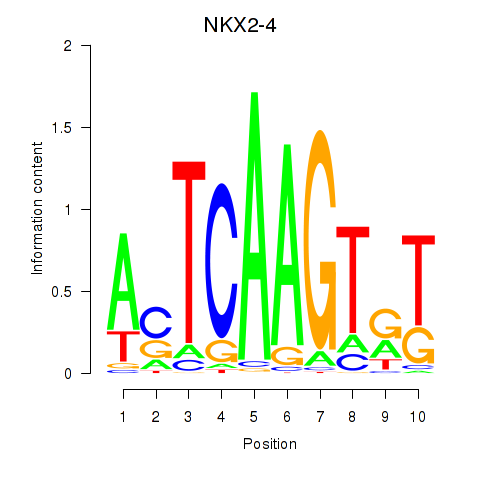
| NKX2-4 | 0.68 |
|
|
|
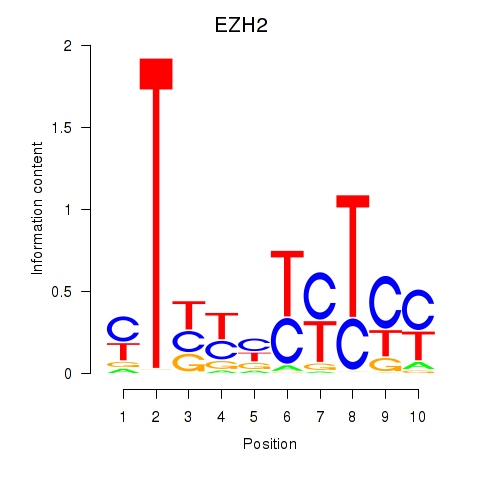
| EZH2 | 0.68 |
|
|
|
| HOXB7 | 0.67 |
|
|
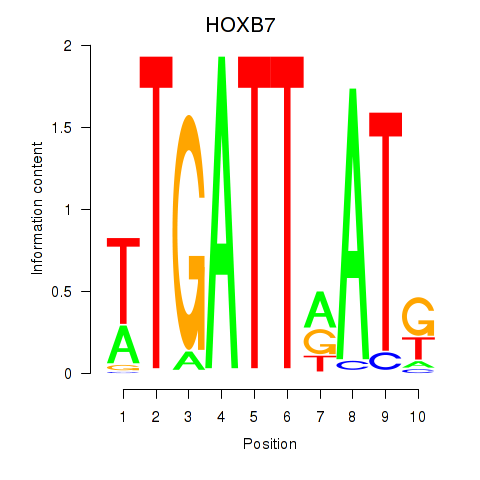
|
| AAGGCAC | 0.67 |
|

|
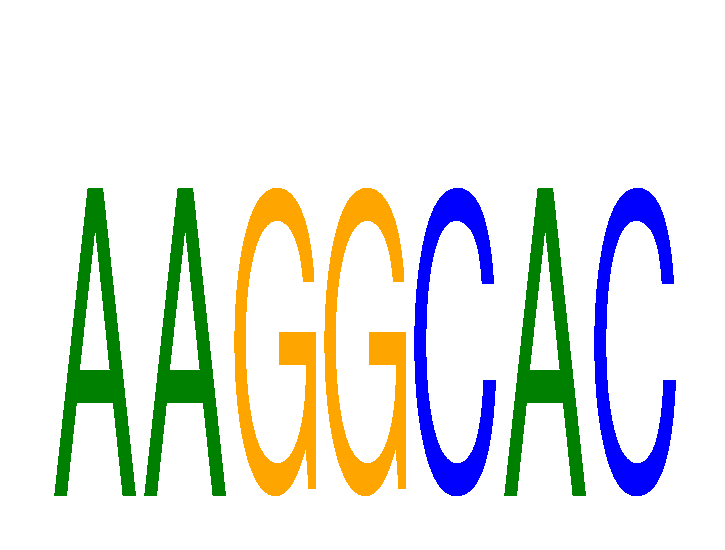
|
| DLX1_HOXA3_BARX2 | 0.67 |
|
|
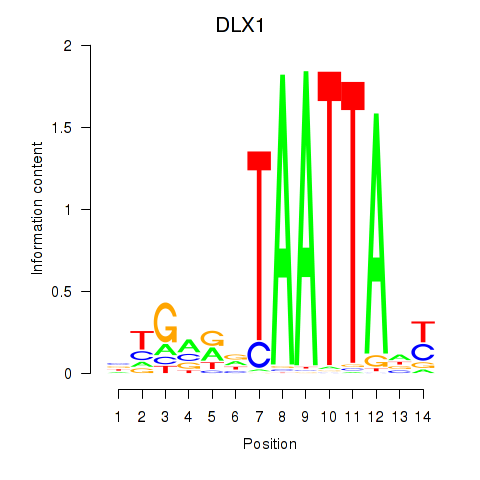
|
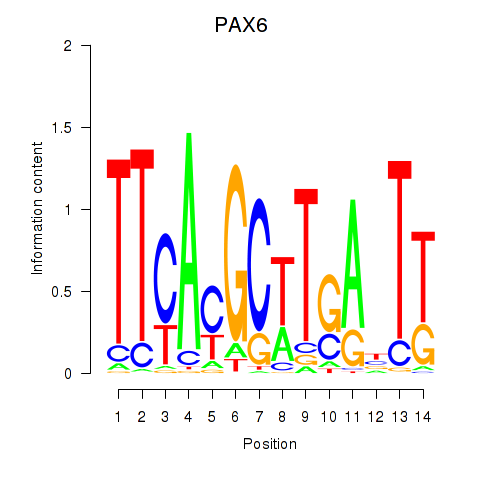
| PAX6 | 0.67 |
|

|
|
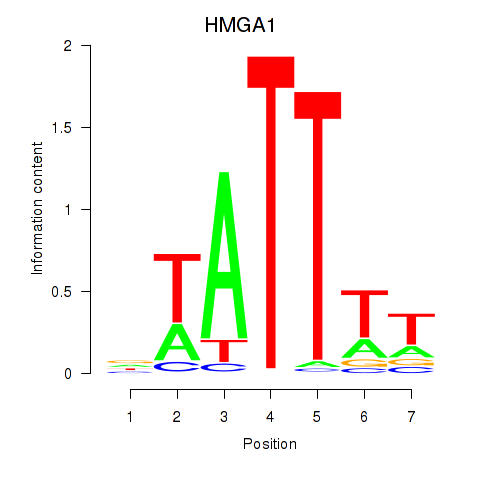
| HMGA1 | 0.66 |
|
|
|
| SP100 | 0.66 |
|
|
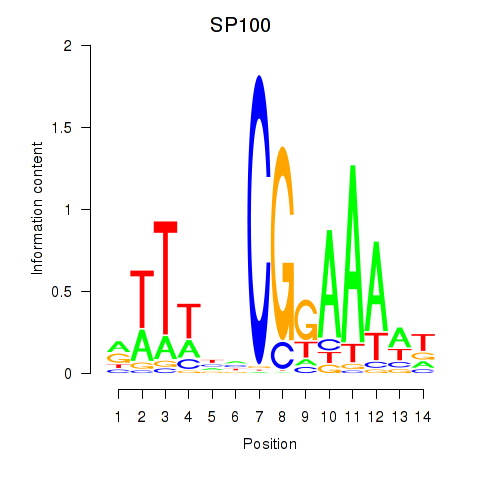
|
| UAUUGCU | 0.66 |
|
|

|
| KLF14_SP8 | 0.66 |
|
|
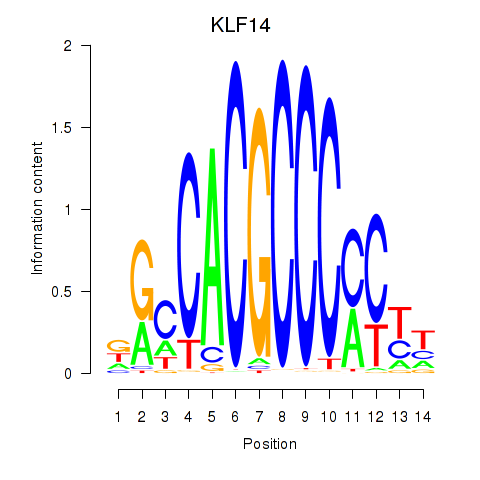
|
| AGCAGCG | 0.65 |
|

|

|
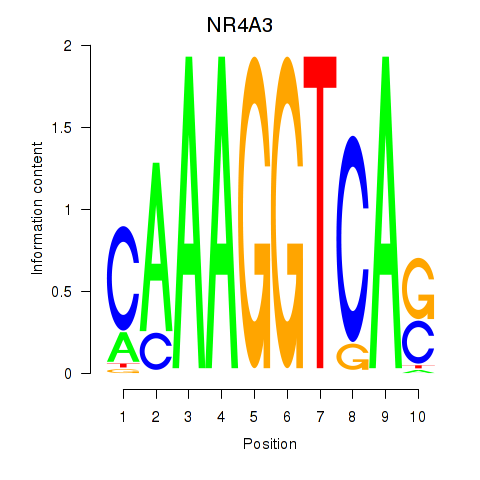
| NR4A3 | 0.65 |
|
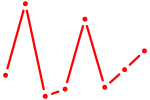
|
|
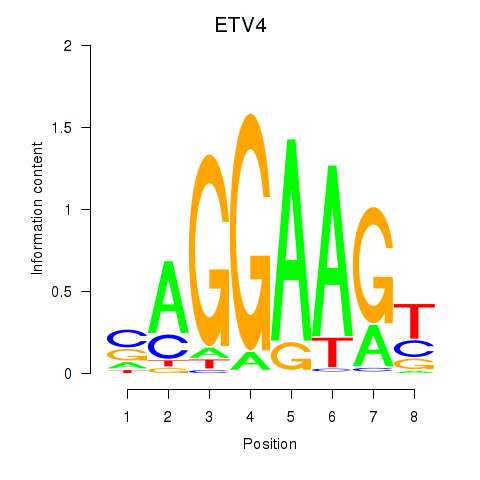
| ETV4_ETS2 | 0.65 |
|
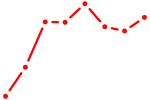
|
|
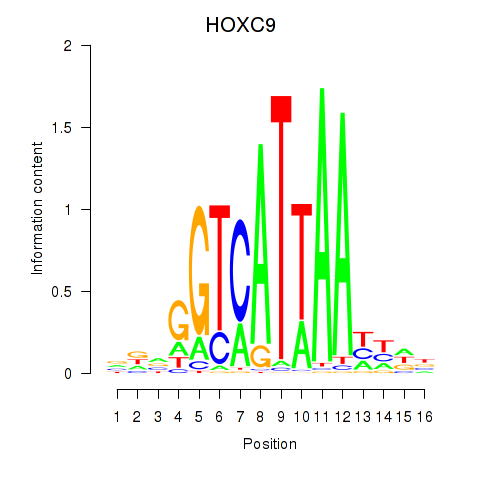
| HOXC9 | 0.65 |
|

|
|
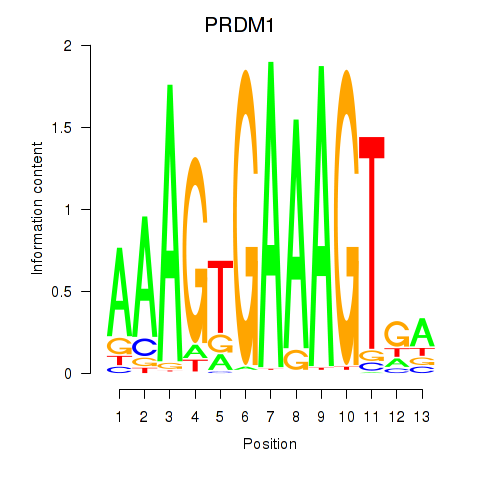
| PRDM1 | 0.65 |
|
|
|
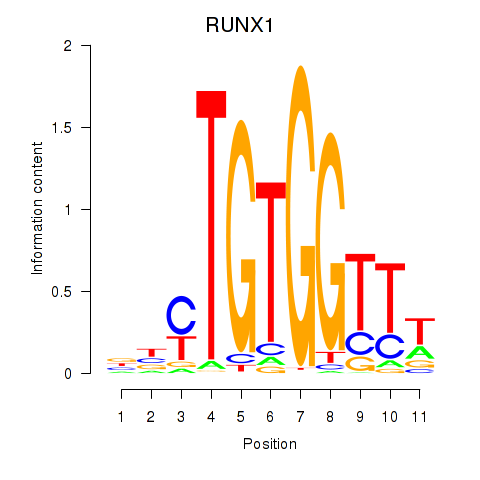
| RUNX1_RUNX2 | 0.65 |
|
|
|
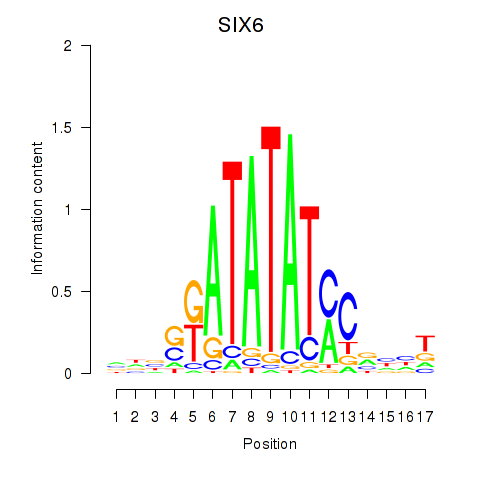
| SIX6 | 0.65 |
|
|
|
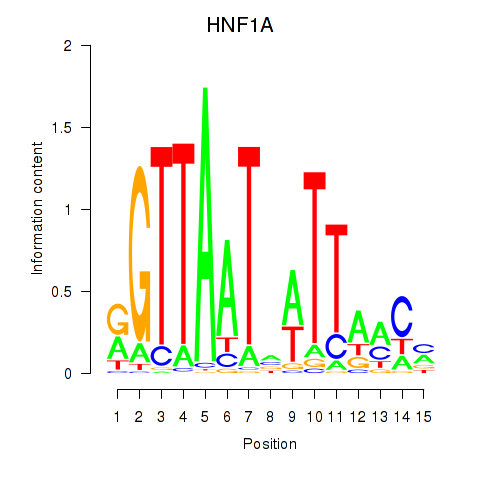
| HNF1A_HNF1B | 0.65 |
|
|
|
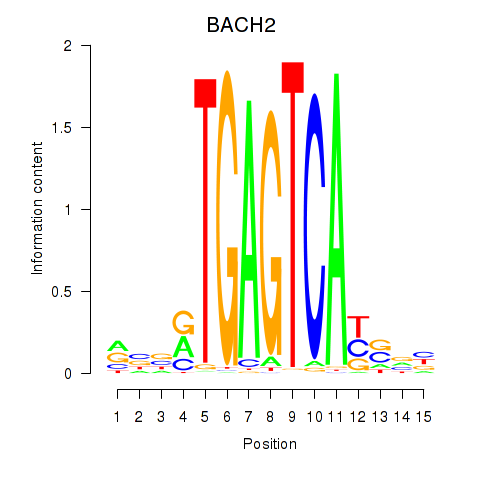
| BACH2 | 0.64 |
|
|
|
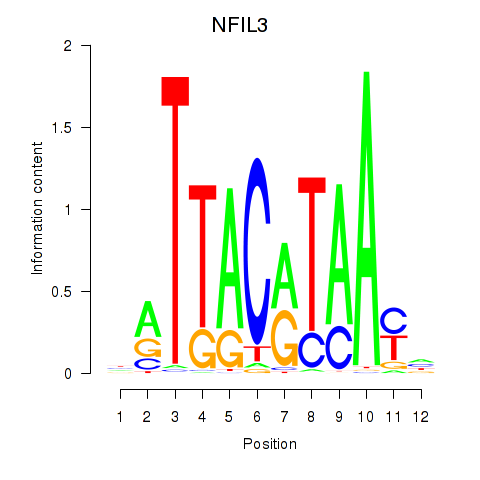
| NFIL3 | 0.64 |
|
|
|
| AGUGCUU | 0.64 |
|
|
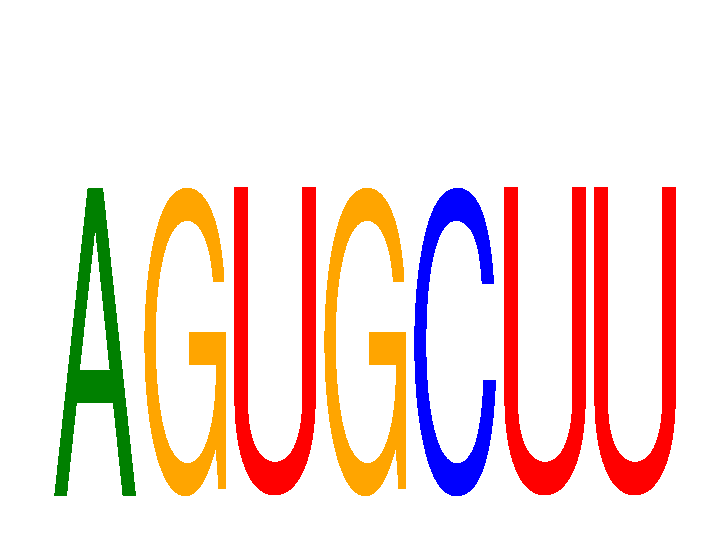
|
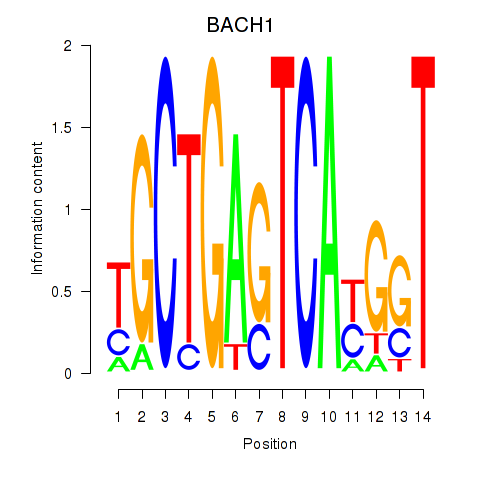
| BACH1_NFE2_NFE2L2 | 0.64 |
|

|
|
| TCF21 | 0.64 |
|

|
|
| HOXD4 | 0.63 |
|

|
|
| GUAAACA | 0.63 |
|

|

|
| RBPJ | 0.63 |
|
|
|
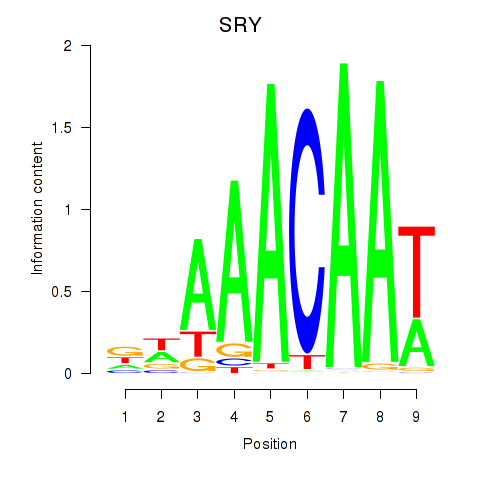
| SRY | 0.62 |
|
|
|
| CUUUGGU | 0.62 |
|

|

|
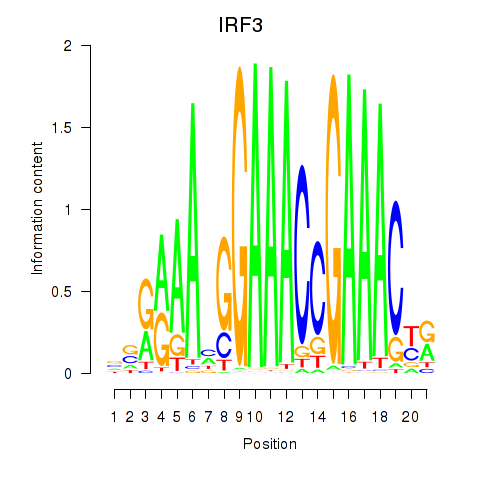
| IRF3 | 0.62 |
|
|
|
| INSM1 | 0.62 |
|

|
|
| PRDM14 | 0.62 |
|
|
|
| SOX8 | 0.61 |
|

|
|
| TBX21_TBR1 | 0.61 |
|
|
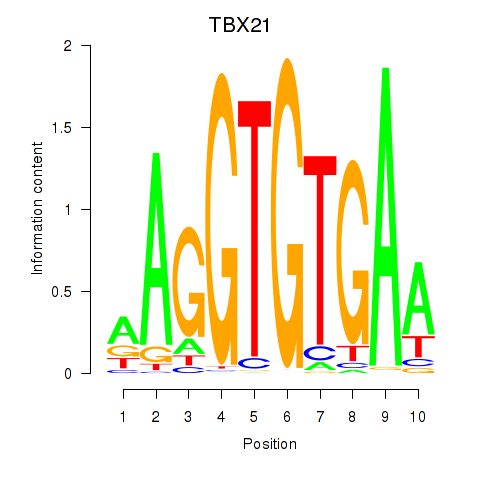
|
| SOX5 | 0.61 |
|
|
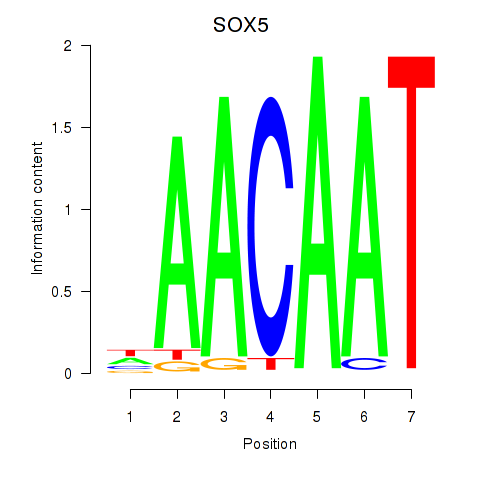
|
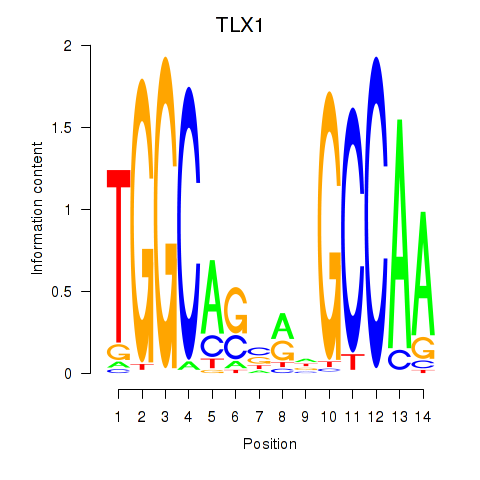
| TLX1_NFIC | 0.61 |
|
|
|
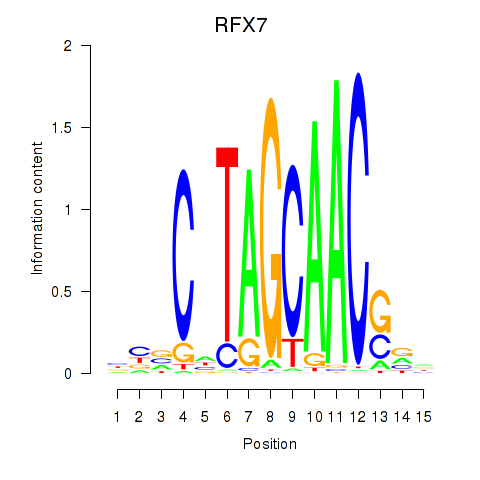
| RFX7_RFX4_RFX1 | 0.61 |
|

|
|
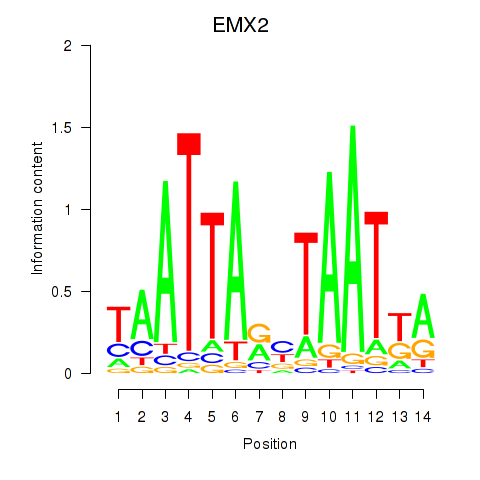
| EMX2 | 0.61 |
|
|
|
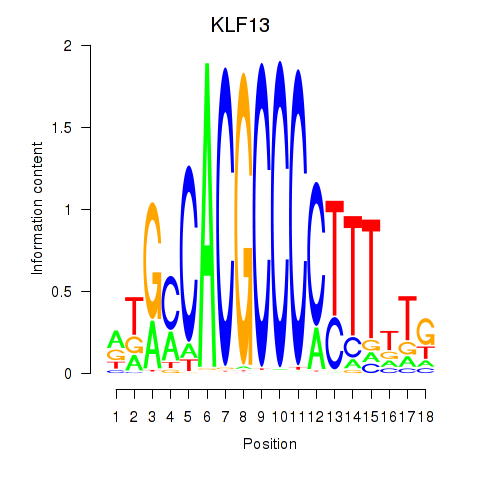
| KLF13 | 0.60 |
|
|
|
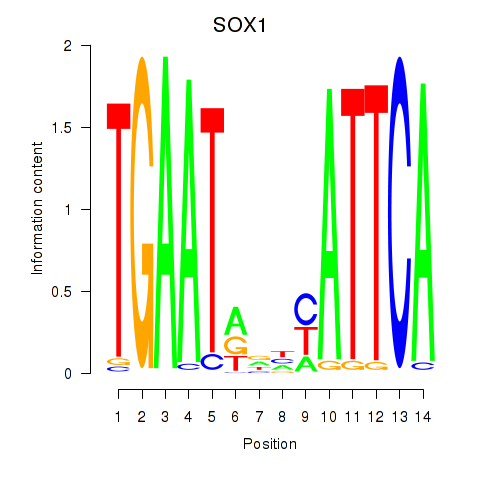
| SOX1 | 0.60 |
|
|
|
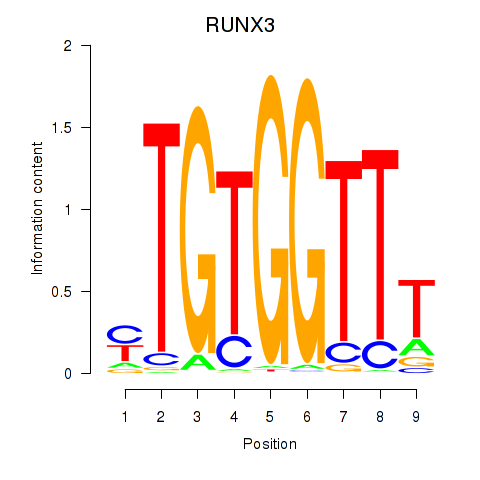
| RUNX3_BCL11A | 0.60 |
|
|
|
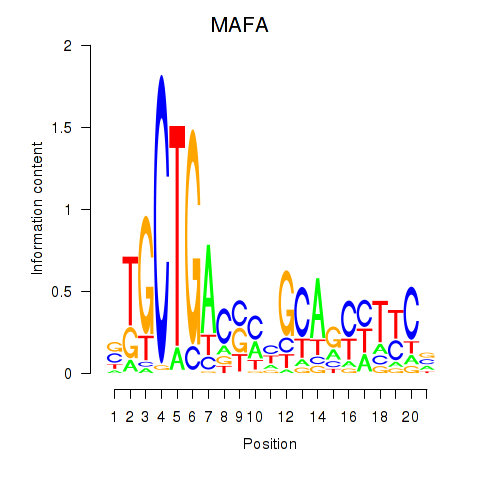
| MAFA | 0.60 |
|

|
|
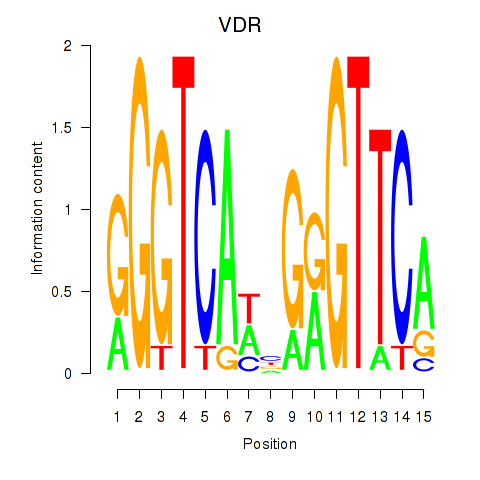
| VDR | 0.60 |
|
|
|
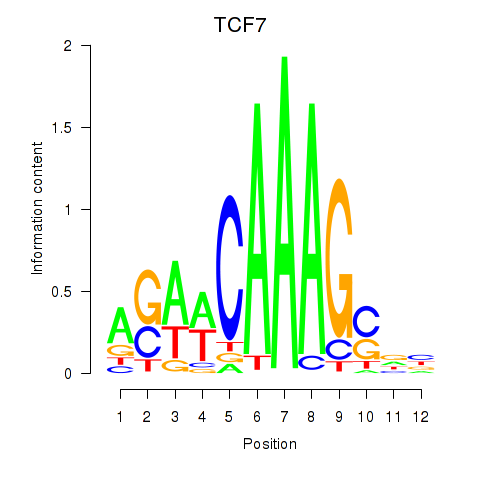
| TCF7 | 0.60 |
|
|
|
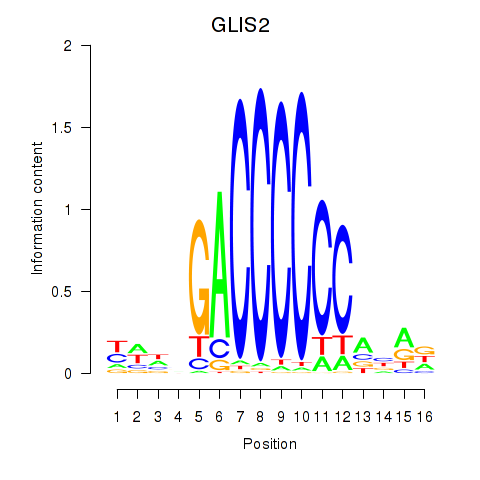
| GLIS2 | 0.59 |
|
|
|
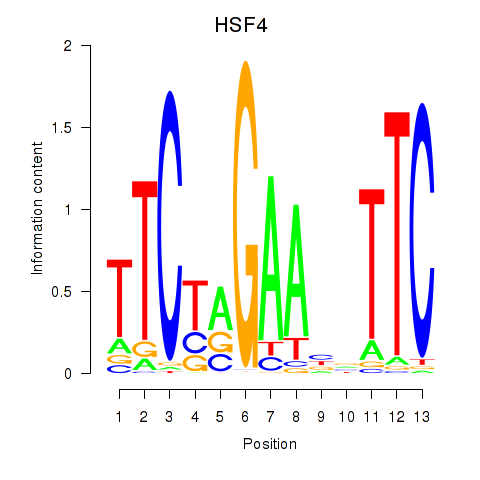
| HSF4 | 0.59 |
|
|
|
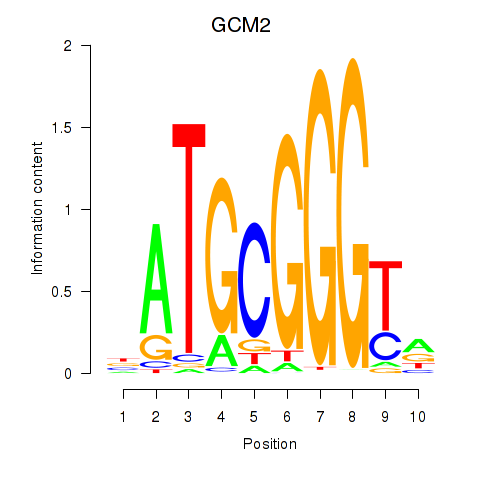
| GCM2 | 0.58 |
|
|
|
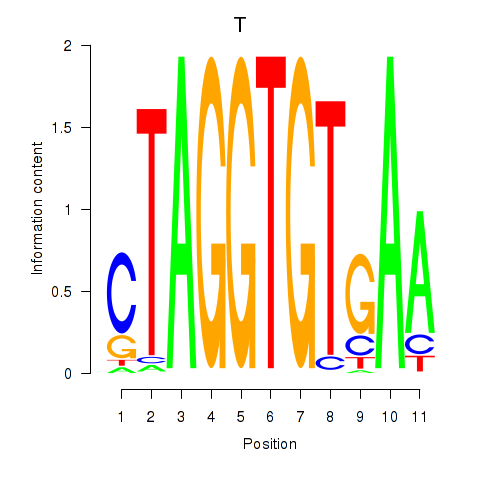
| T | 0.58 |
|

|
|
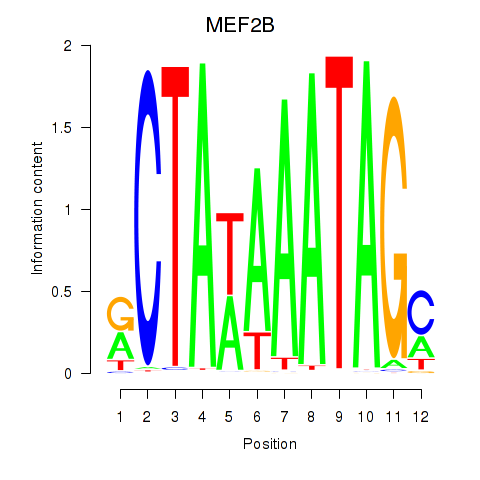
| MEF2B | 0.58 |
|
|
|
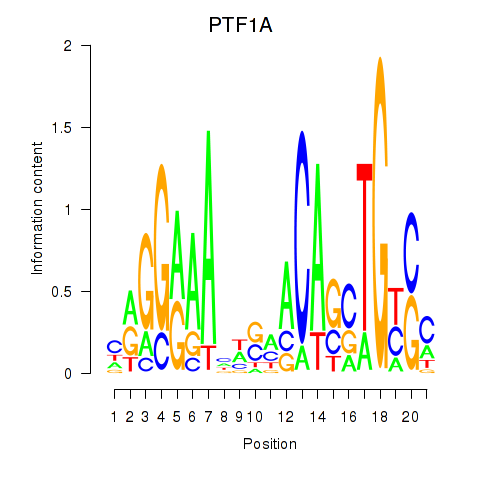
| PTF1A | 0.58 |
|
|
|
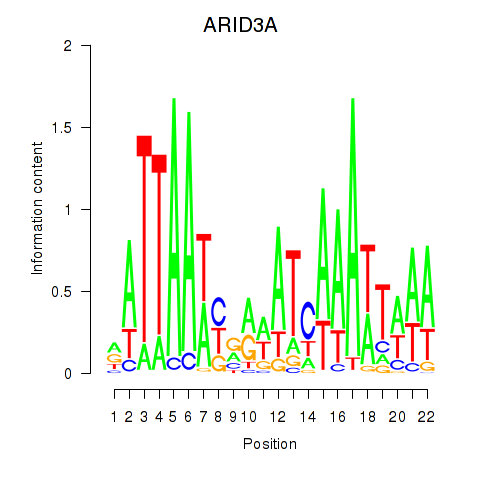
| ARID3A | 0.57 |
|

|
|
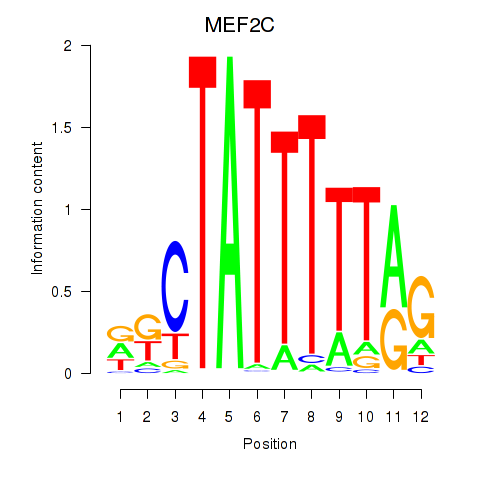
| MEF2C | 0.57 |
|
|
|
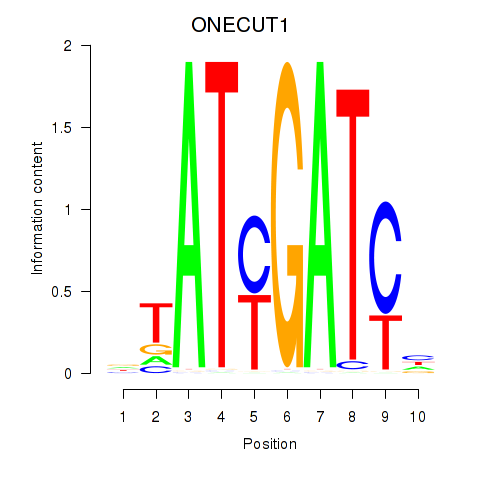
| ONECUT1 | 0.57 |
|
|
|
| GCM1 | 0.57 |
|
|
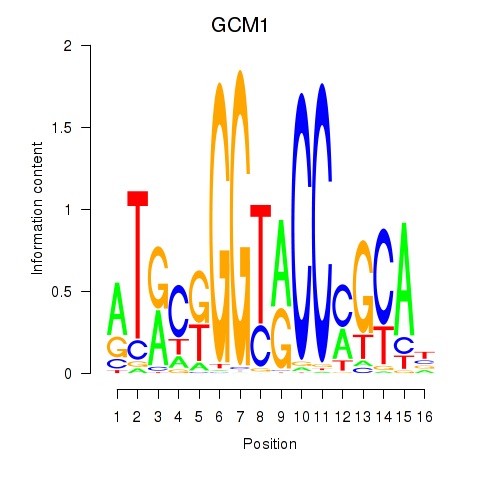
|
| DPRX | 0.57 |
|
|
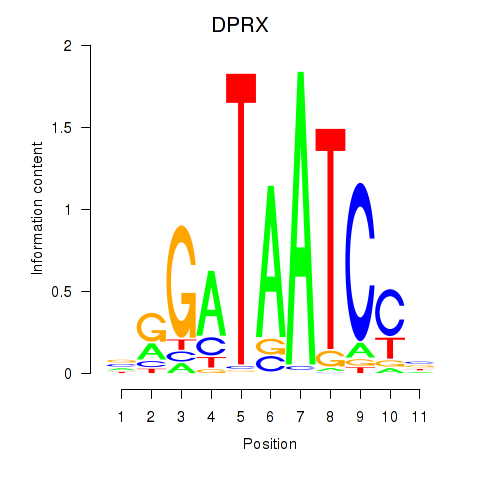
|
| HOXD11_HOXA11 | 0.57 |
|
|
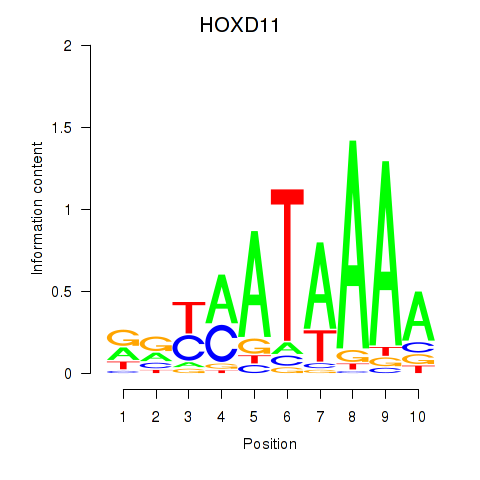
|
| UBP1 | 0.57 |
|
|
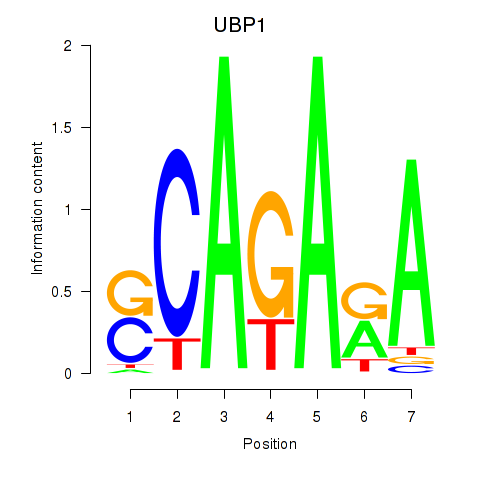
|
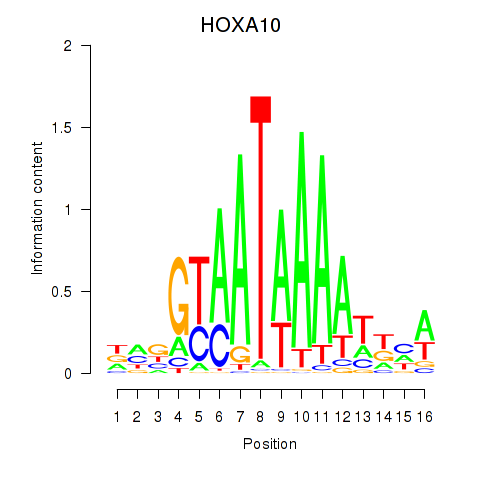
| HOXA10_HOXB9 | 0.57 |
|

|
|
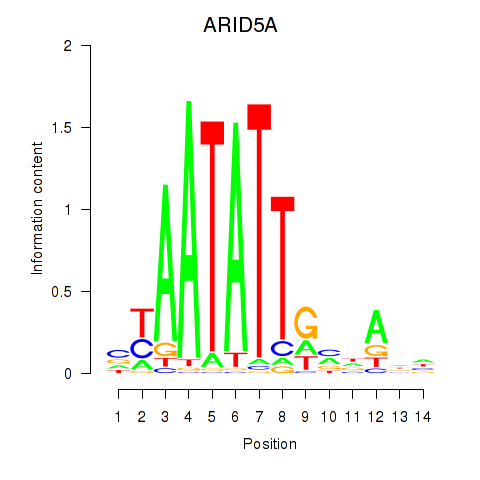
| ARID5A | 0.57 |
|
|
|
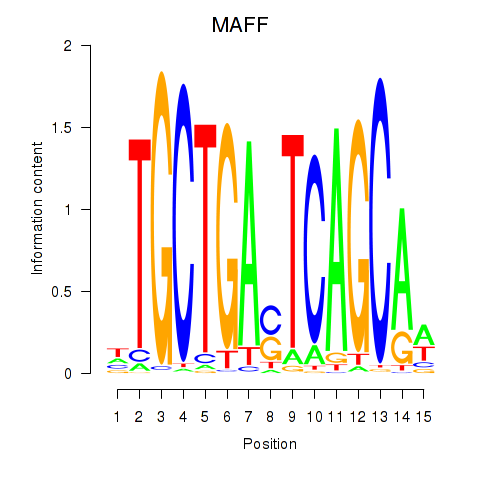
| MAFF_MAFG | 0.57 |
|
|
|
| UCCAGUU | 0.57 |
|

|

|
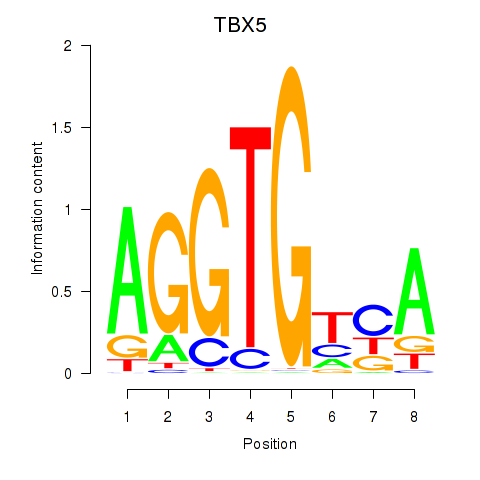
| TBX5 | 0.57 |
|
|
|
| UAAGGCA | 0.57 |
|
|

|
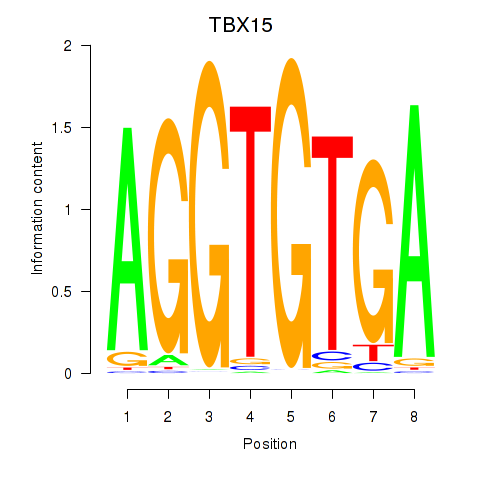
| TBX15_MGA | 0.56 |
|

|
|
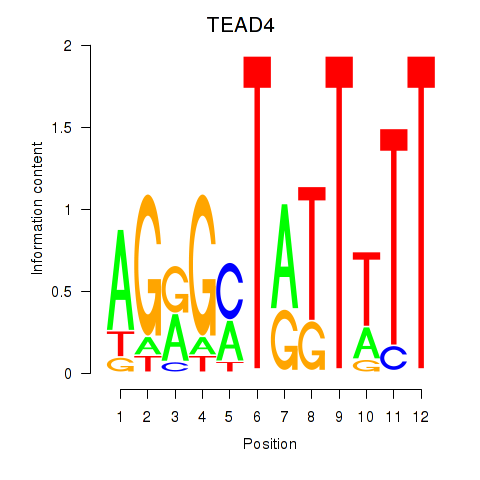
| TEAD4 | 0.56 |
|
|
|
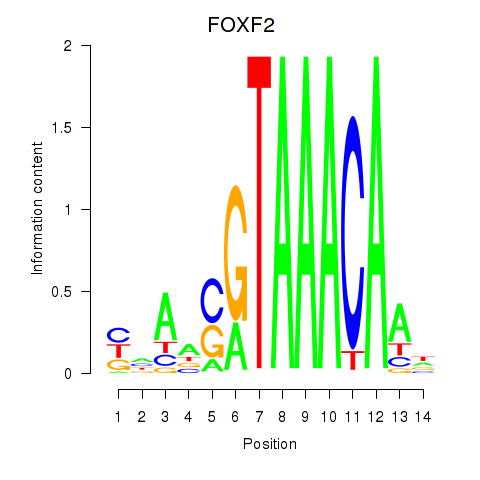
| FOXF2_FOXJ1 | 0.56 |
|

|
|
| GCUACAU | 0.56 |
|

|

|
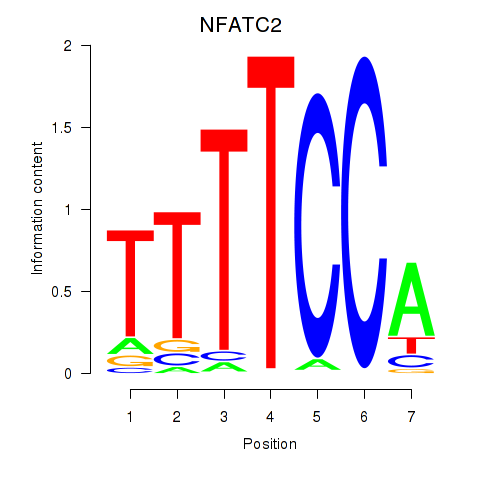
| NFATC2_NFATC3 | 0.56 |
|
|
|
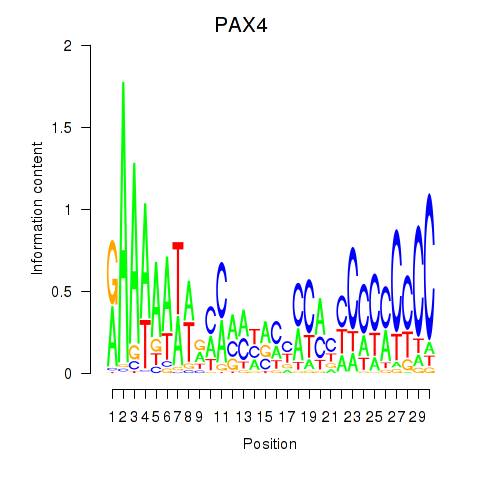
| PAX4 | 0.56 |
|

|
|
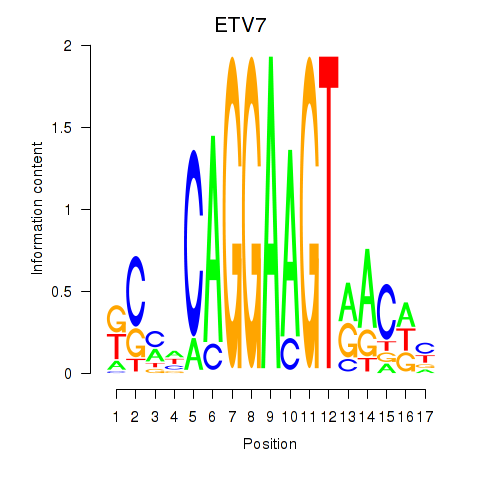
| ETV7 | 0.56 |
|
|
|
| CAGUGCA | 0.56 |
|
|

|
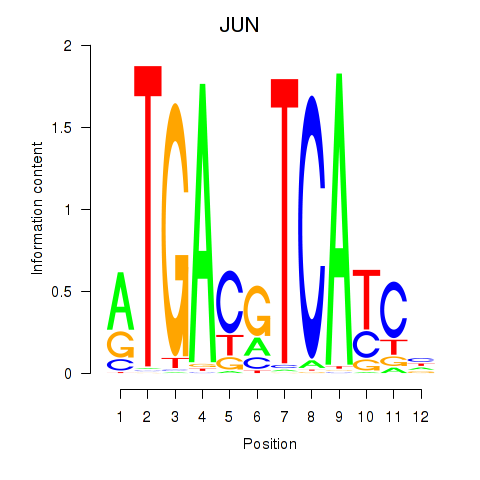
| JUN | 0.55 |
|
|
|
| UGCAUAG | 0.55 |
|

|

|
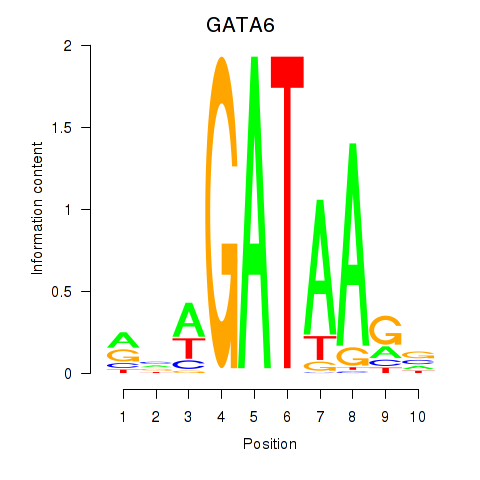
| GATA6 | 0.54 |
|

|
|
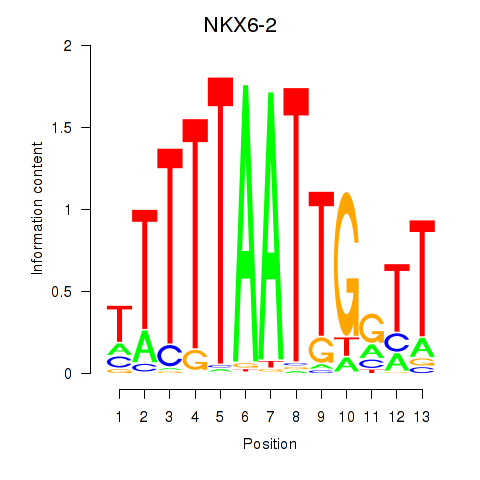
| NKX6-2 | 0.54 |
|
|
|
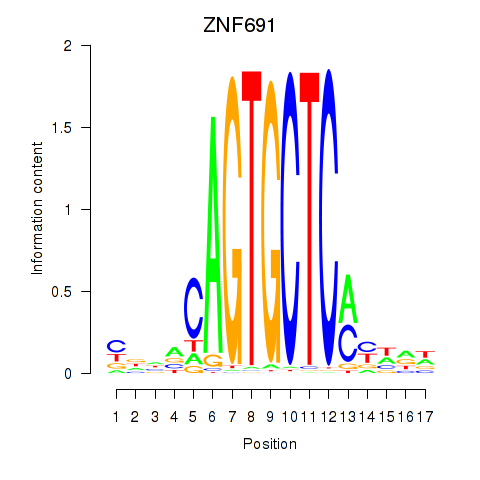
| ZNF691 | 0.54 |
|

|
|
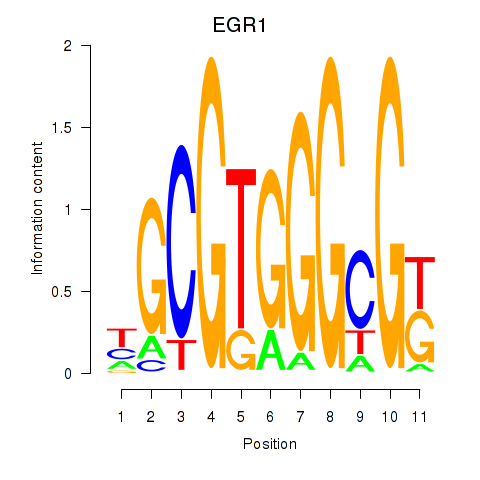
| EGR1_EGR4 | 0.54 |
|

|
|
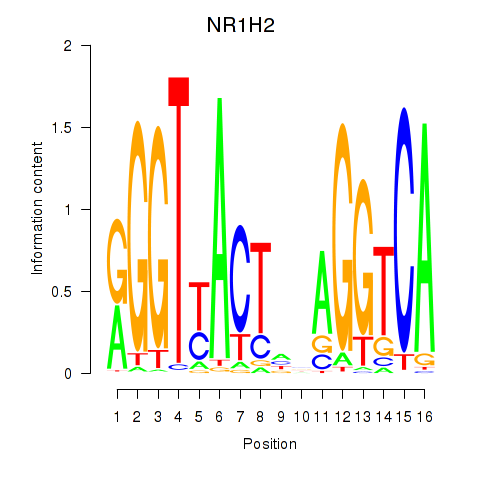
| NR1H2 | 0.54 |
|

|
|
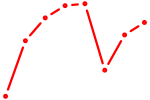
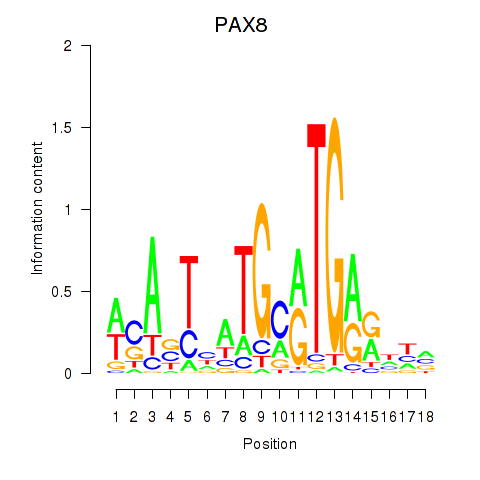
| PAX8 | 0.54 |
|
|
|
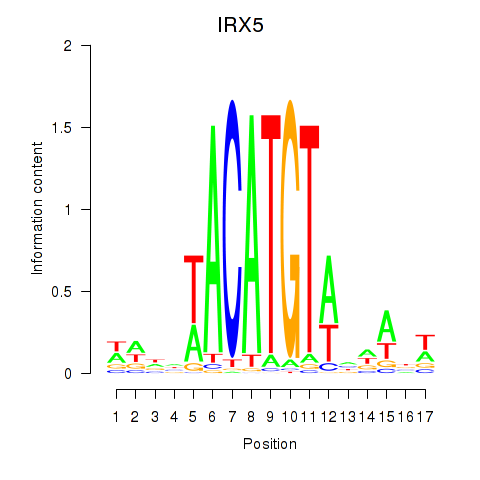
| IRX5 | 0.54 |
|
|
|
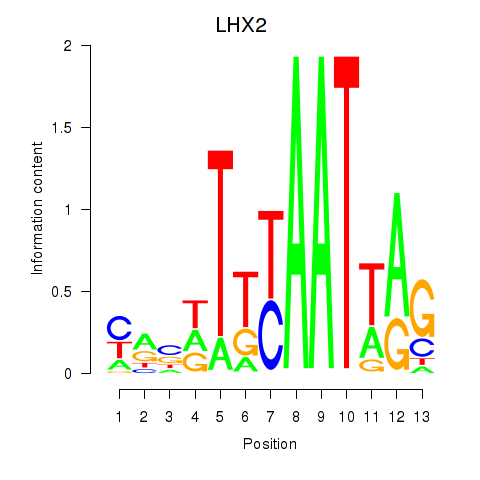
| LHX2 | 0.54 |
|
|
|
| NR6A1 | 0.54 |
|

|
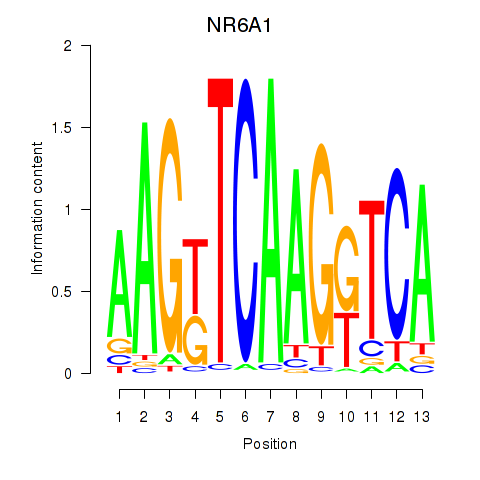
|
| VAX2_RHOXF2 | 0.54 |
|
|
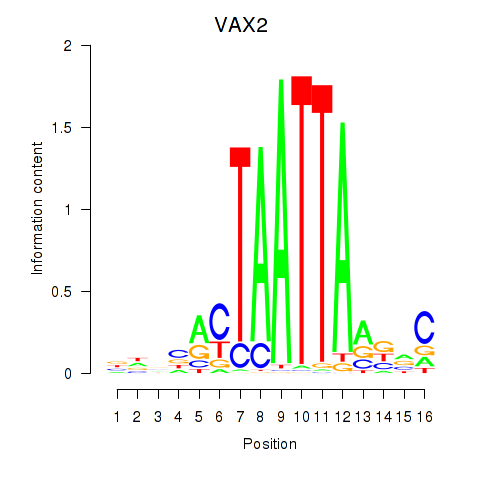
|
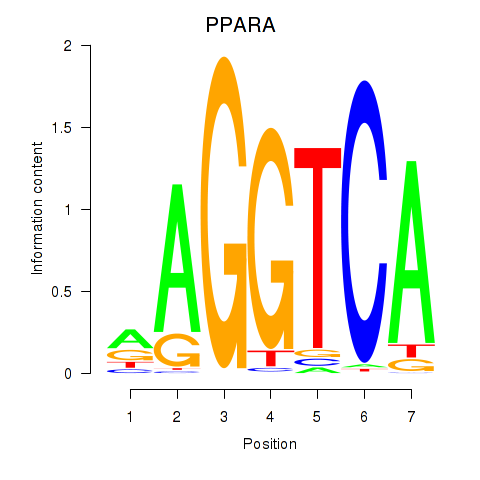
| PPARA | 0.53 |
|
|
|
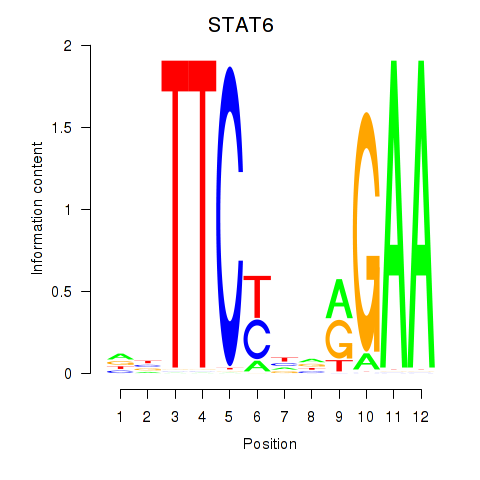
| STAT6 | 0.53 |
|
|
|
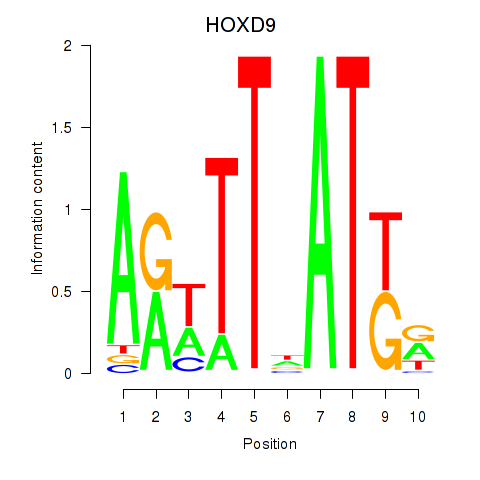
| HOXD9 | 0.52 |
|
|
|
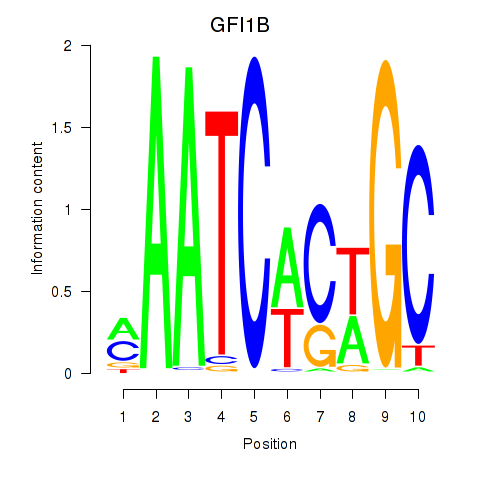
| GFI1B | 0.52 |
|
|
|
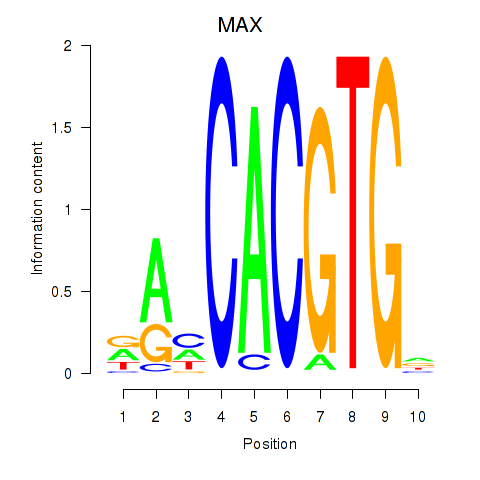
| MAX_TFEB | 0.52 |
|
|
|
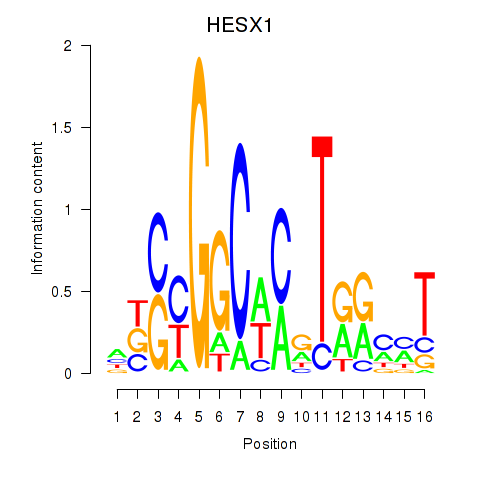
| HESX1 | 0.52 |
|

|
|
| GCUGGUG | 0.52 |
|

|

|
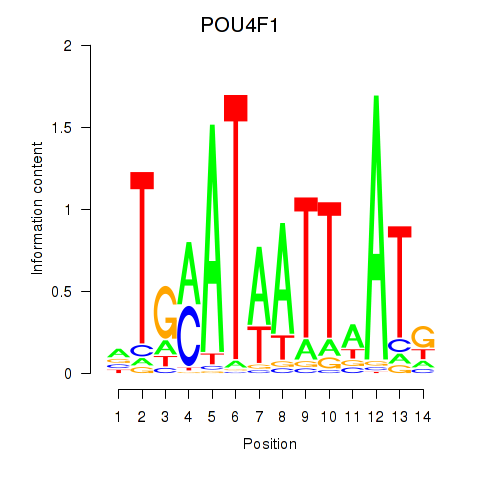
| POU4F1_POU4F3 | 0.52 |
|

|
|
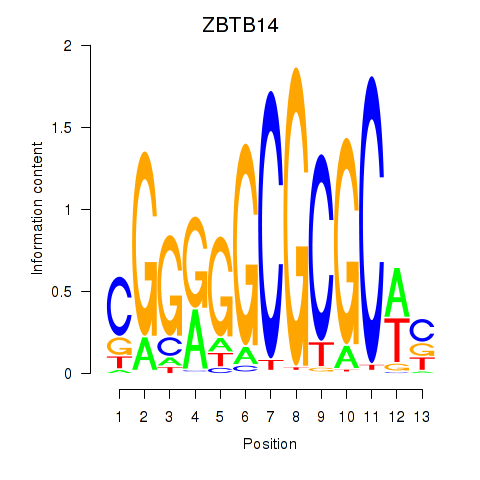
| ZBTB14 | 0.52 |
|
|
|
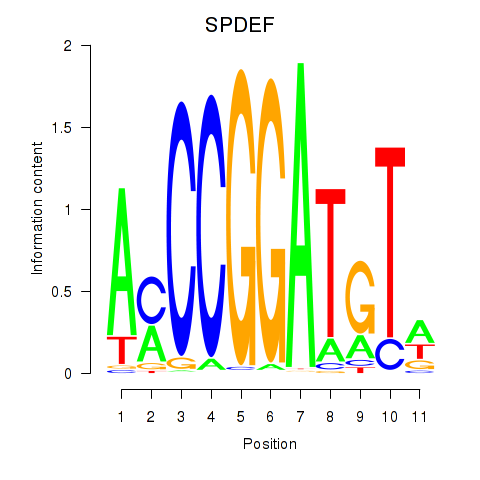
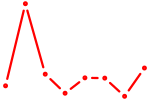
| SPDEF | 0.52 |
|

|
|
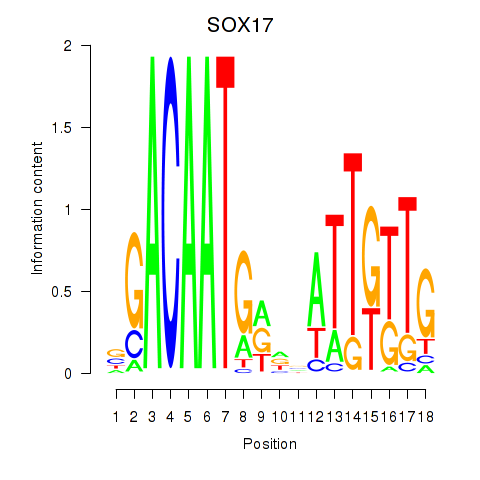
| SOX17 | 0.52 |
|
|
|
| IRX3 | 0.51 |
|
|
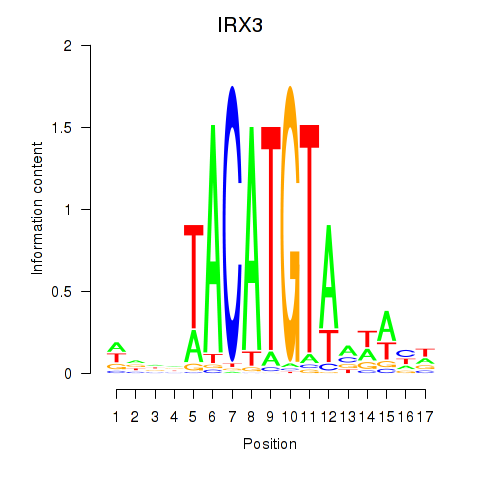
|
| ZNF784 | 0.51 |
|
|
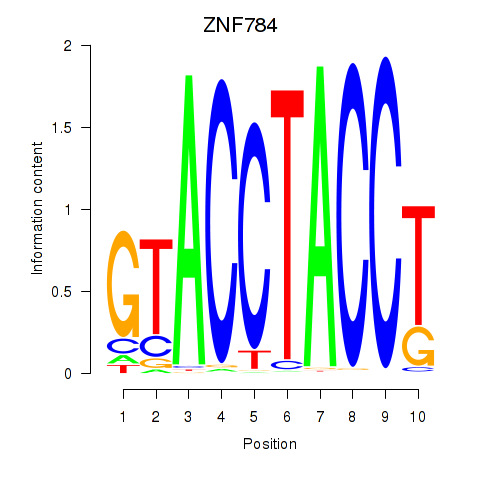
|
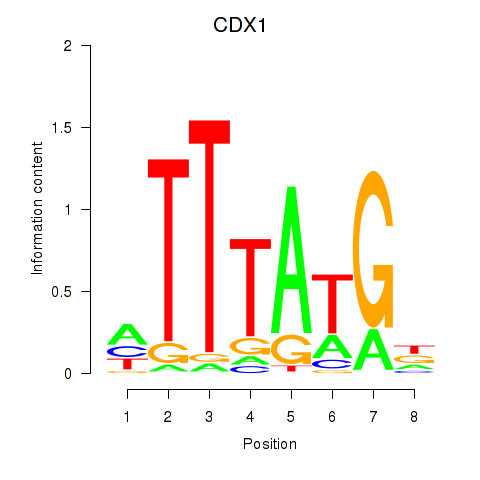
| CDX1 | 0.51 |
|
|
|
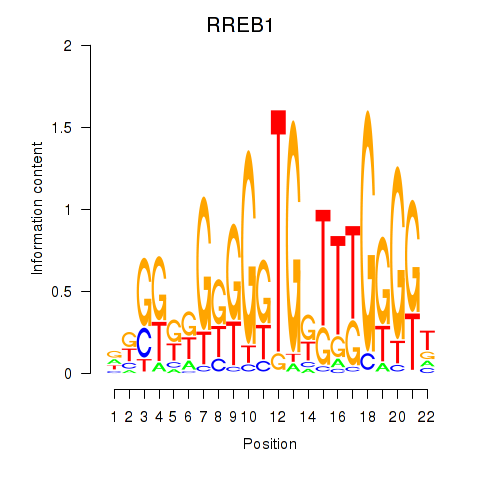
| RREB1 | 0.51 |
|
|
|
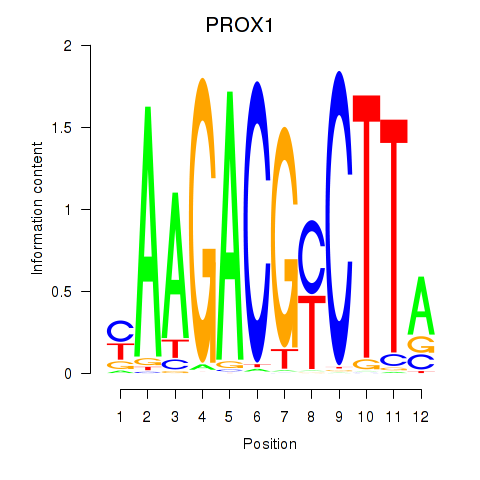
| PROX1 | 0.51 |
|

|
|
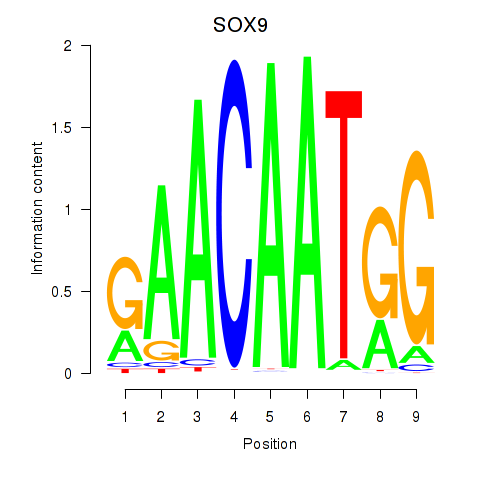
| SOX9 | 0.51 |
|

|
|
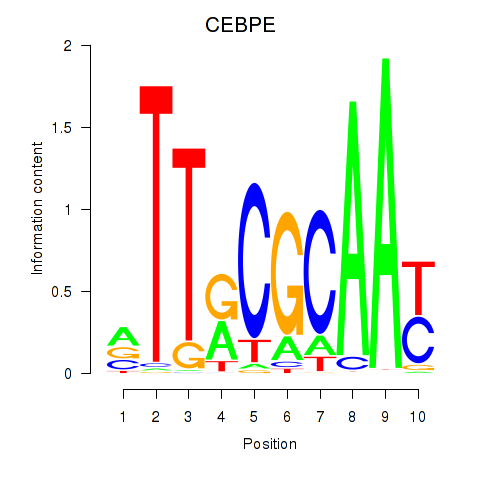
| CEBPE_CEBPD | 0.51 |
|

|
|
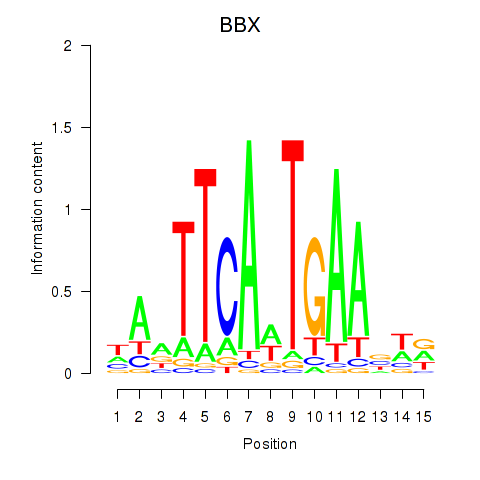
| BBX | 0.51 |
|
|
|
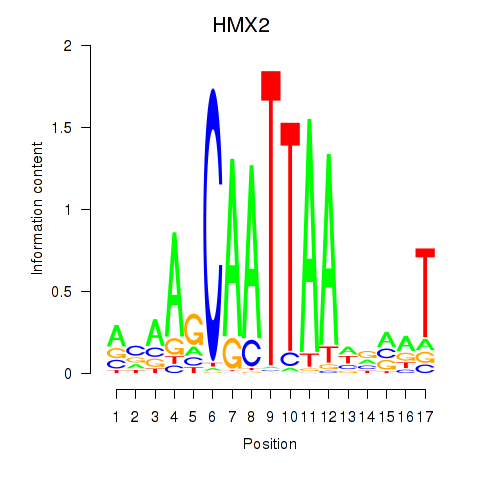
| HMX2 | 0.50 |
|
|
|
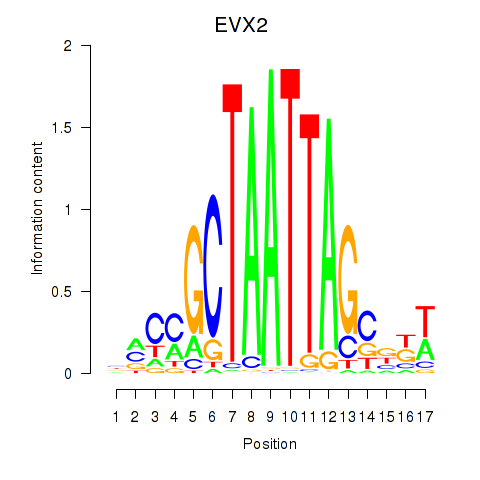
| EVX2 | 0.50 |
|
|
|
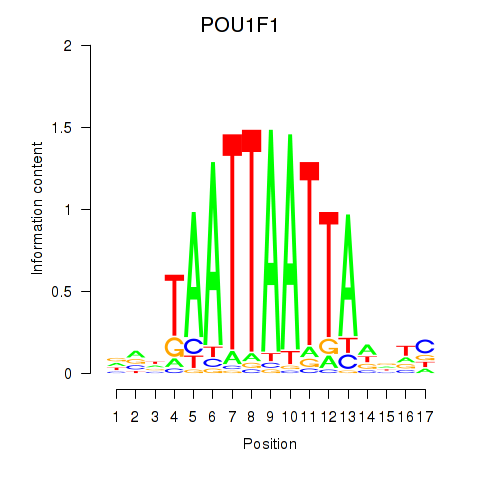
| POU1F1 | 0.50 |
|
|
|
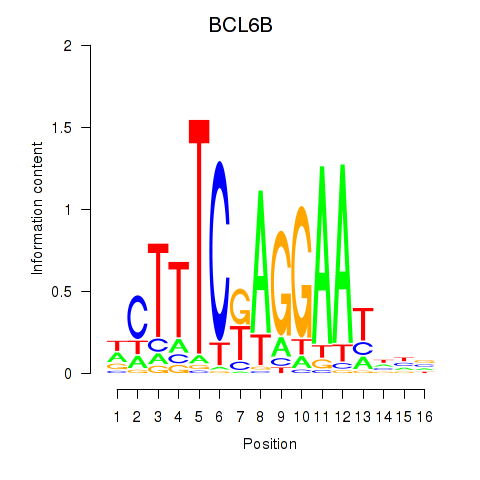
| BCL6B | 0.50 |
|

|
|
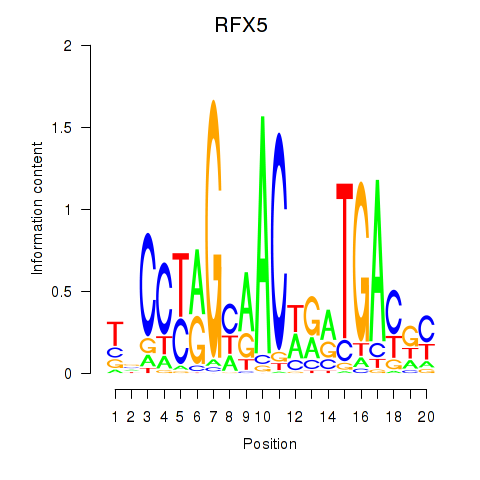
| RFX5 | 0.50 |
|

|
|
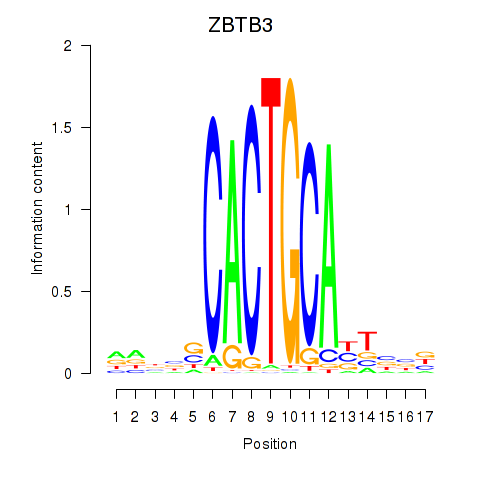
| ZBTB3 | 0.50 |
|
|
|
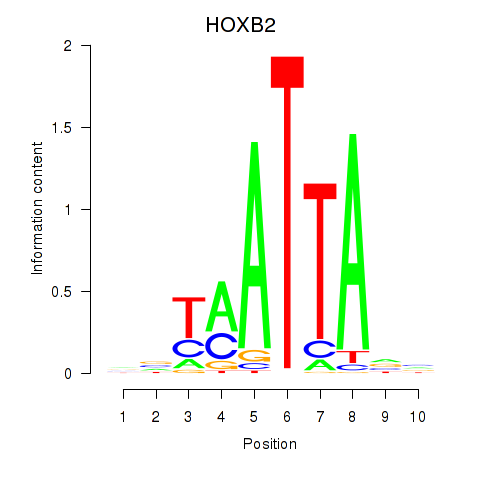
| HOXB2_UNCX_HOXD3 | 0.50 |
|
|
|
| AUGGCUU | 0.50 |
|
|
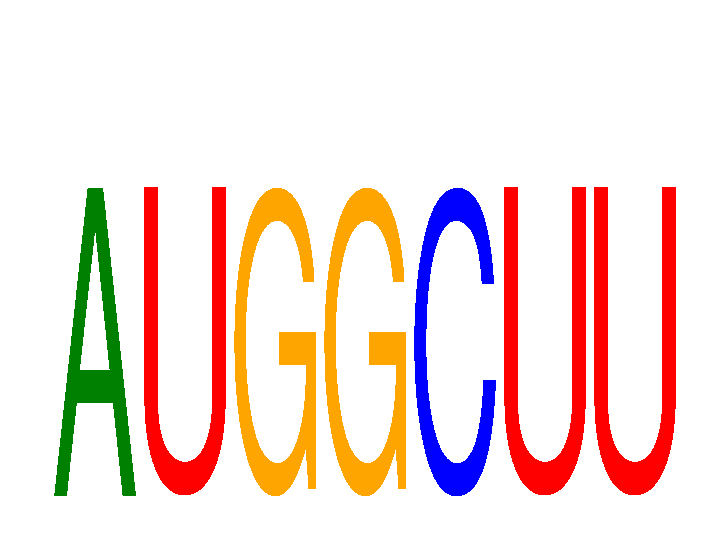
|
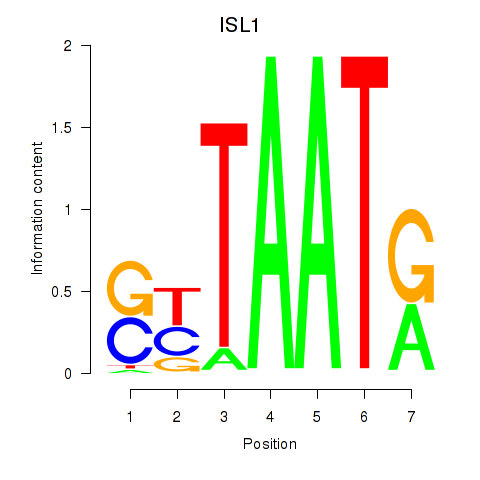
| ISL1 | 0.50 |
|
|
|
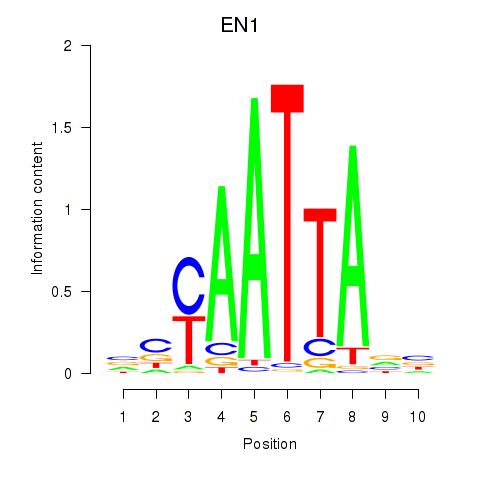
| EN1_ESX1_GBX1 | 0.49 |
|
|
|
| PKNOX2 | 0.49 |
|

|

|
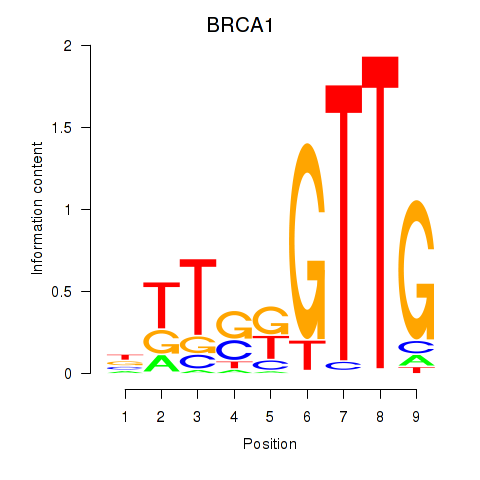
| BRCA1 | 0.49 |
|
|
|
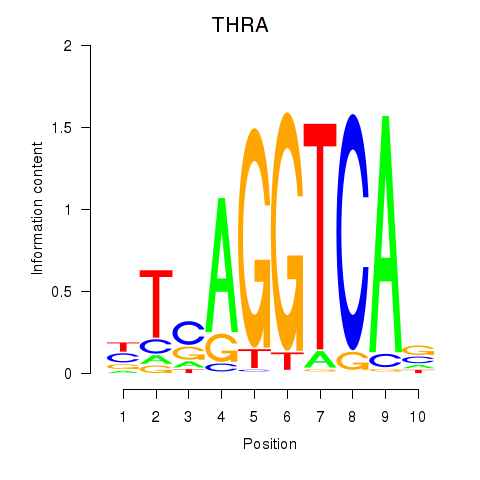
| THRA_RXRB | 0.49 |
|
|
|
| KLF15 | 0.49 |
|
|
|
| FOXJ2 | 0.49 |
|

|
|
| UCACAGU | 0.49 |
|
|

|
| ZIC2_GLI1 | 0.49 |
|

|
|
| AACAGUC | 0.48 |
|
|

|
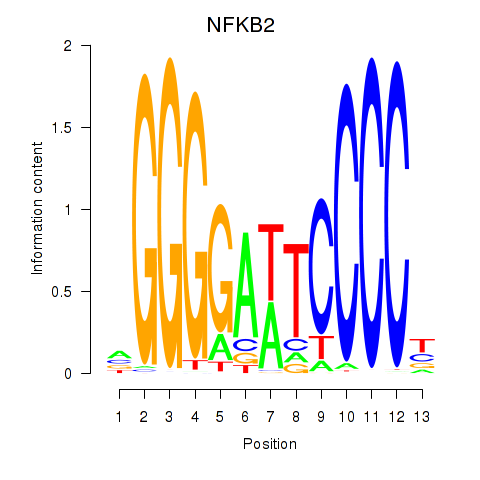
| NFKB2 | 0.48 |
|
|
|
| PKNOX1_TGIF2 | 0.48 |
|
|
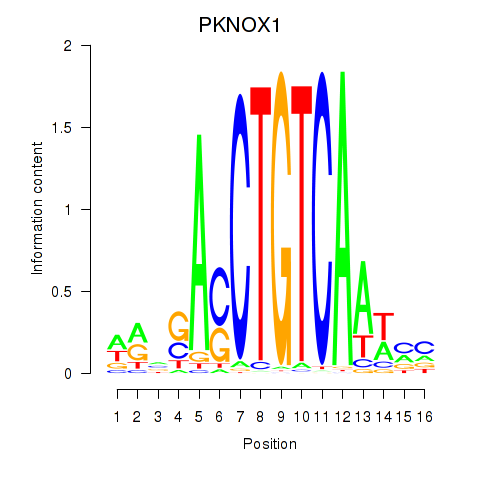
|
| GFI1 | 0.48 |
|

|
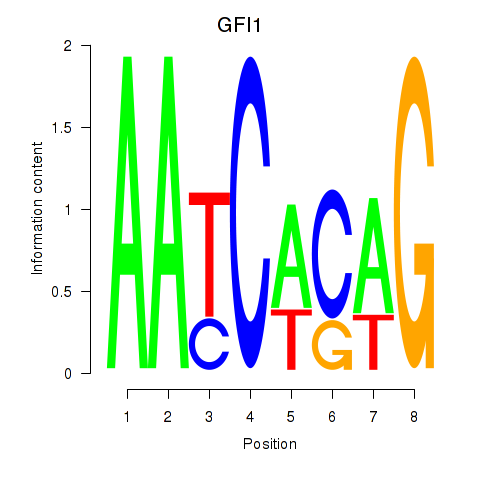
|
| MYOD1 | 0.48 |
|
|
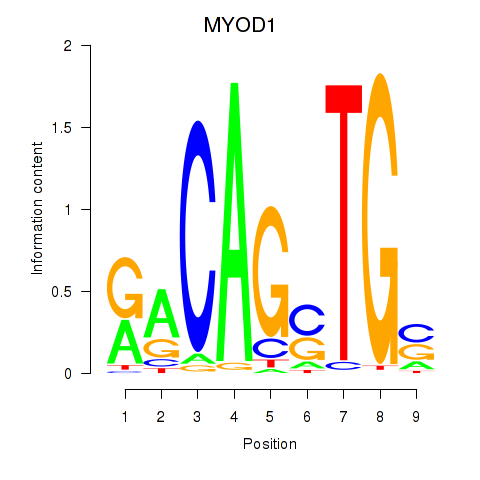
|
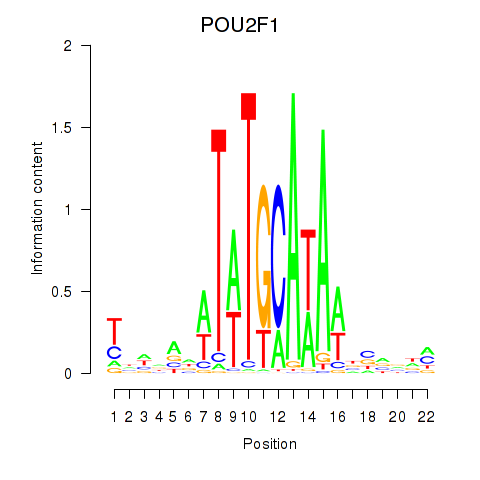
| POU2F1 | 0.48 |
|
|
|
| ACAGUAU | 0.47 |
|

|

|
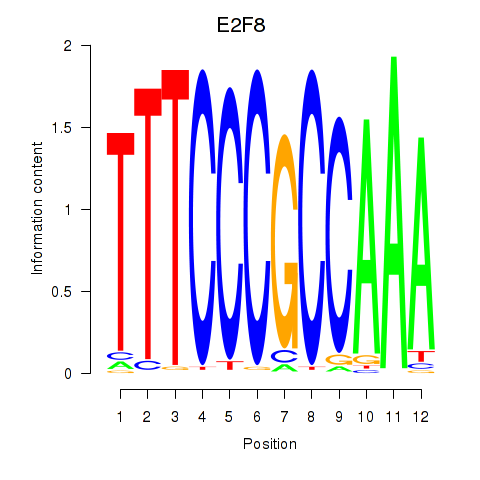
| E2F8 | 0.47 |
|
|
|
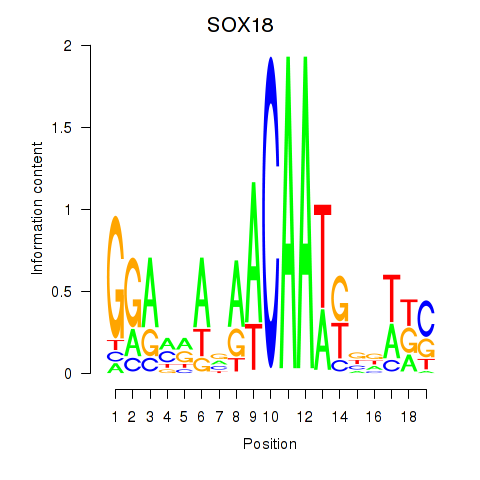
| SOX18 | 0.47 |
|
|
|
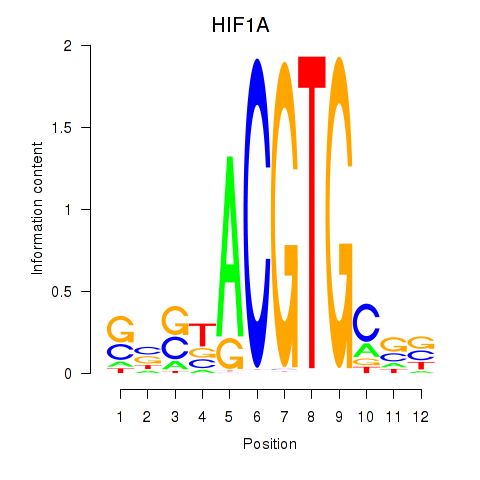
| HIF1A | 0.47 |
|
|
|
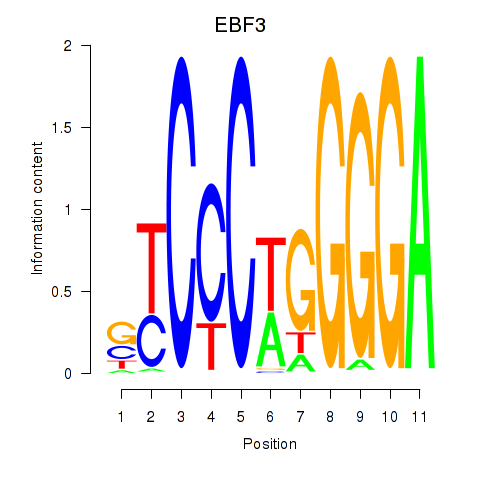
| EBF3 | 0.47 |
|

|
|
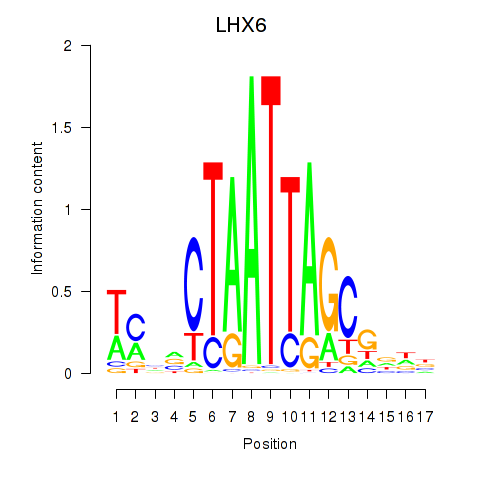
| LHX6 | 0.47 |
|
|
|
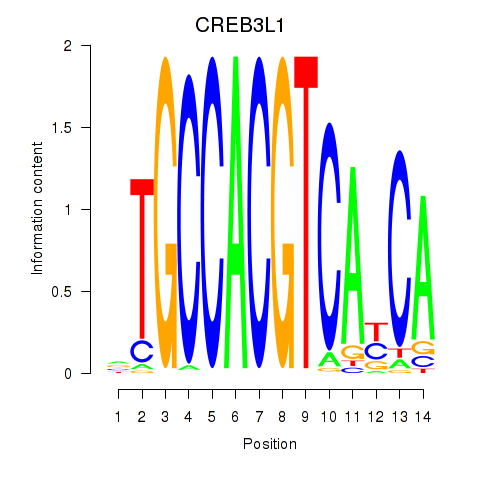
| CREB3L1_CREB3 | 0.47 |
|
|
|
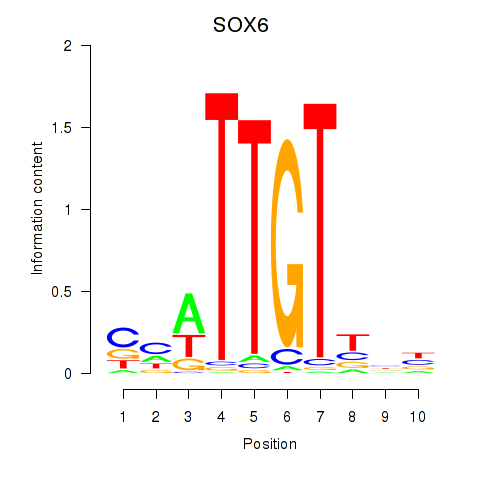
| SOX6 | 0.47 |
|

|
|
| UGGCACU | 0.46 |
|

|

|
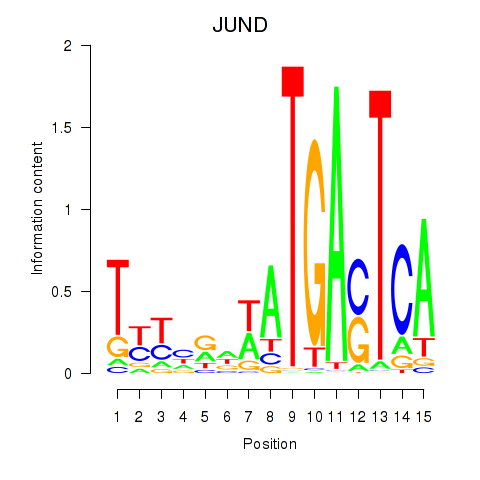
| JUND | 0.46 |
|

|
|
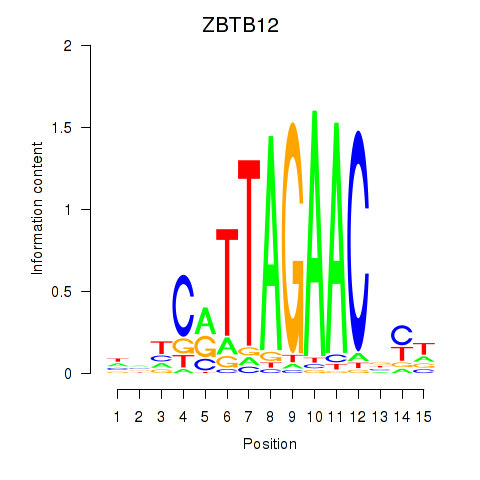
| ZBTB12 | 0.46 |
|
|
|
| GCAGCAU | 0.46 |
|

|

|
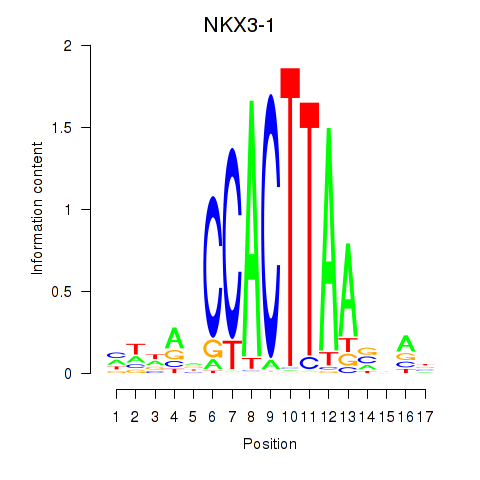
| NKX3-1 | 0.46 |
|
|
|
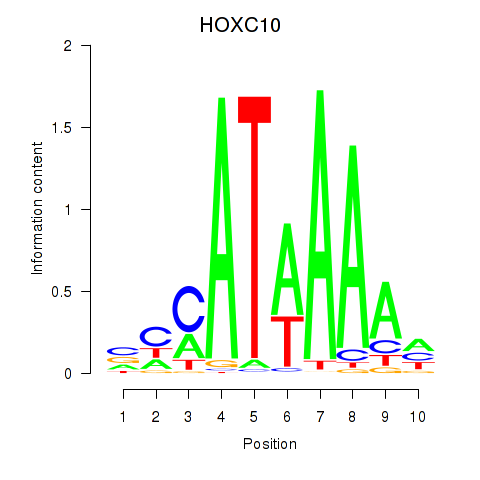
| HOXC10_HOXD13 | 0.46 |
|
|
|
| GUAGUGU | 0.45 |
|
|

|
| UCACAUU | 0.45 |
|
|

|
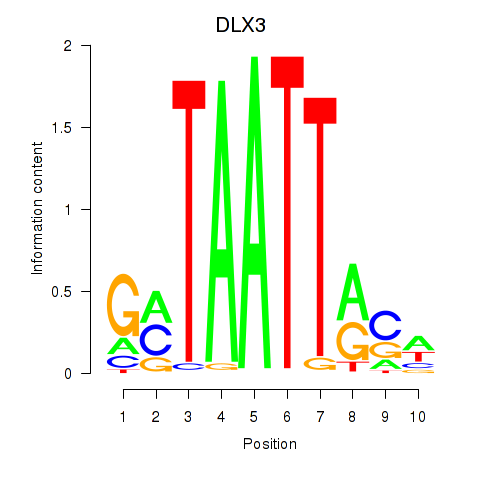
| DLX3_EVX1_MEOX1 | 0.45 |
|
|
|
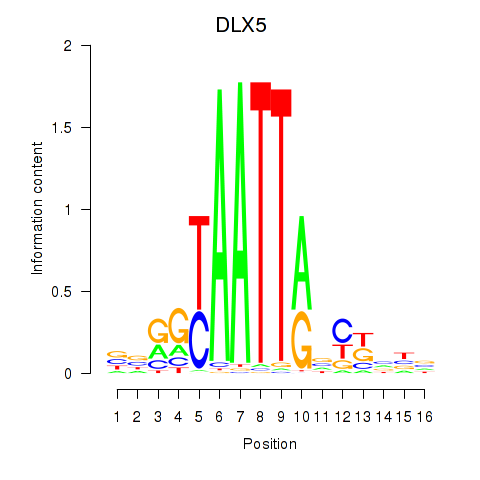
| DLX5 | 0.45 |
|
|
|
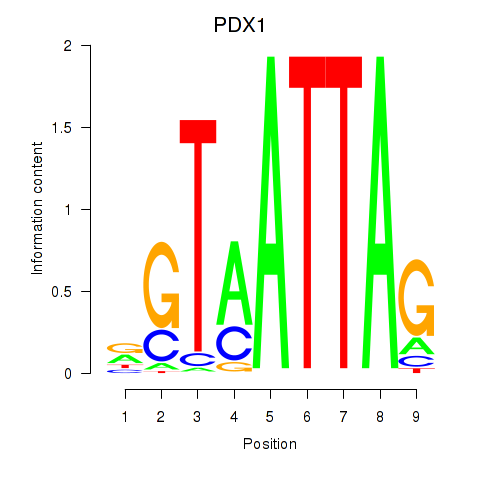
| PDX1 | 0.45 |
|
|
|
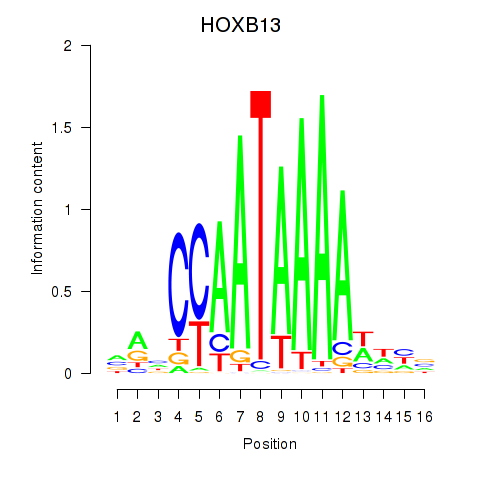
| HOXB13 | 0.45 |
|

|
|
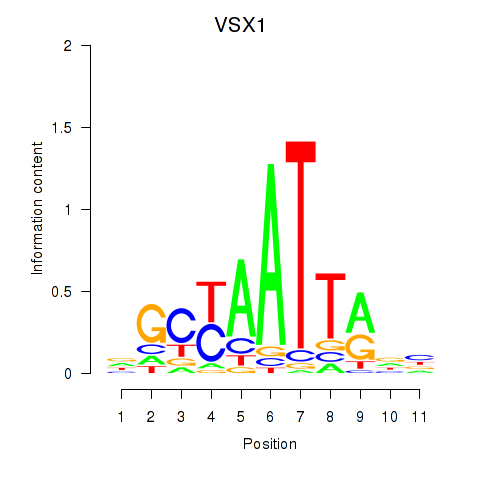
| VSX1 | 0.45 |
|

|
|
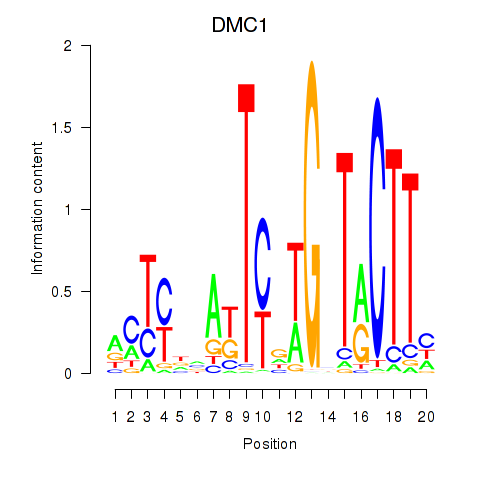
| DMC1 | 0.44 |
|
|
|
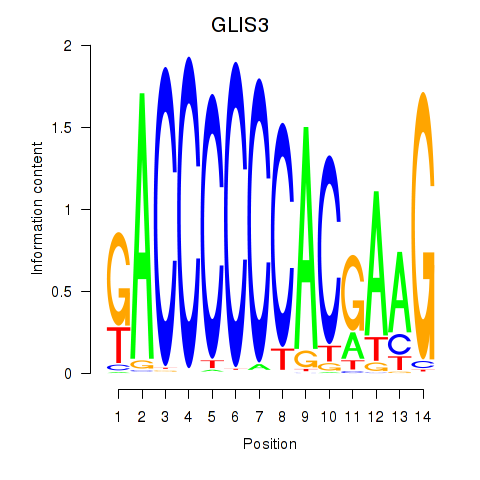
| GLIS3 | 0.44 |
|
|
|
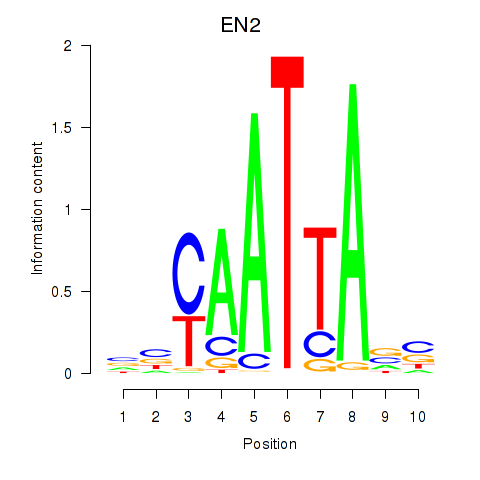
| EN2_GBX2_LBX2 | 0.44 |
|

|
|
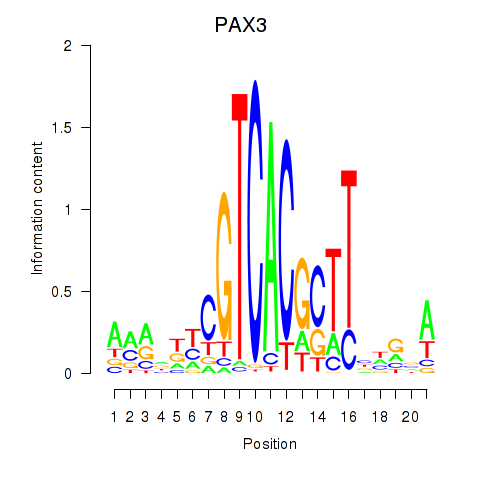
| PAX3 | 0.44 |
|
|
|
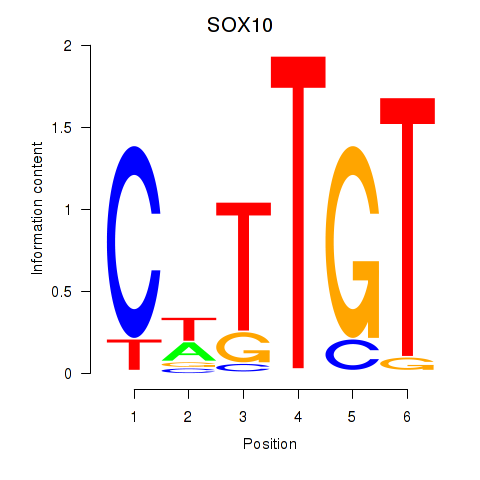
| SOX10_SOX15 | 0.43 |
|

|
|
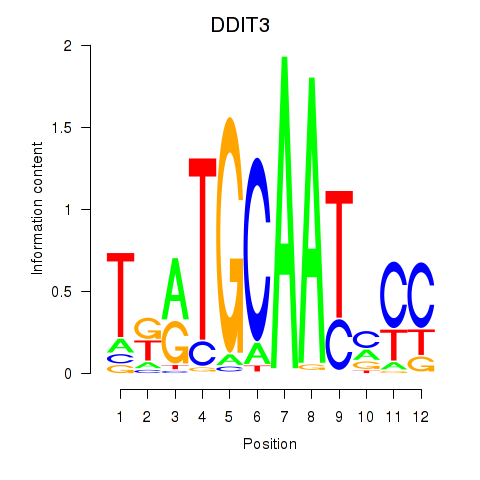
| DDIT3 | 0.43 |
|
|
|
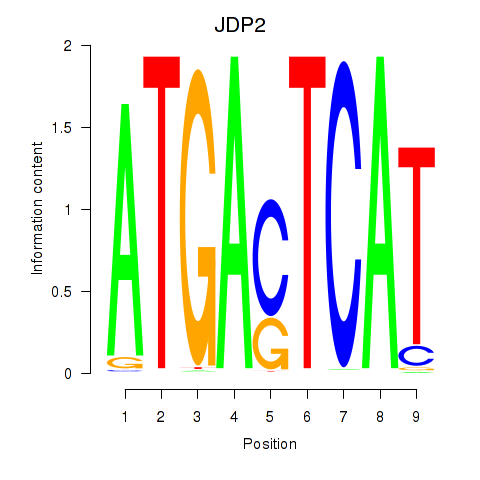
| JDP2 | 0.43 |
|
|
|
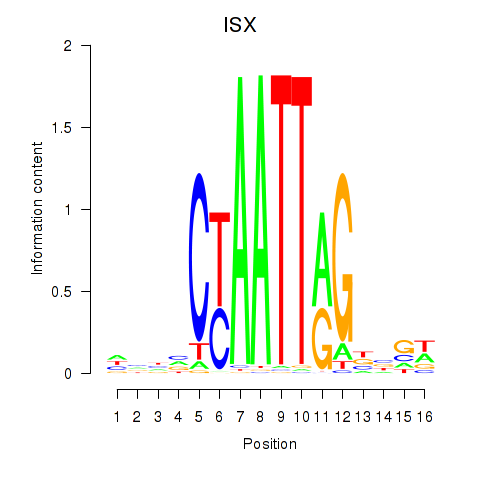
| ISX | 0.43 |
|

|
|
| GUCAGUU | 0.43 |
|
|

|
| ZNF333 | 0.43 |
|
|
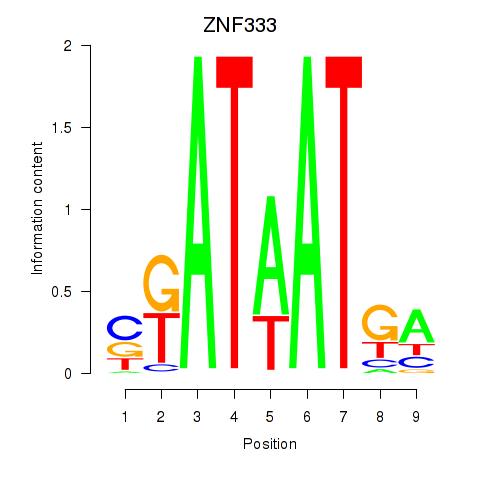
|
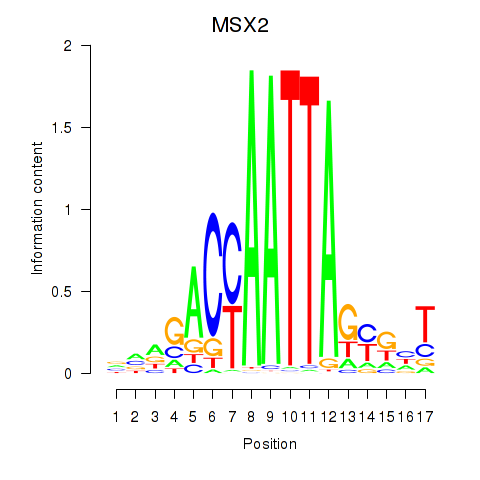
| MSX2 | 0.42 |
|
|
|
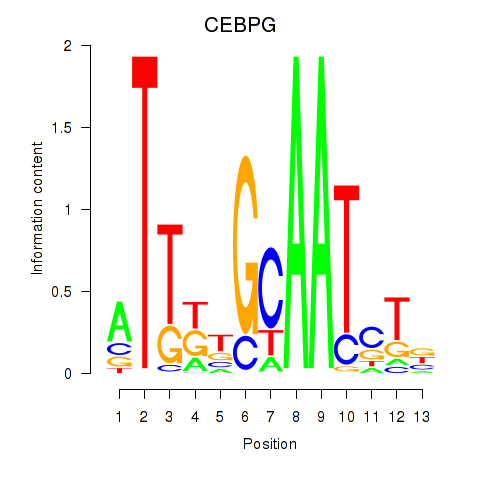
| CEBPG | 0.42 |
|
|
|
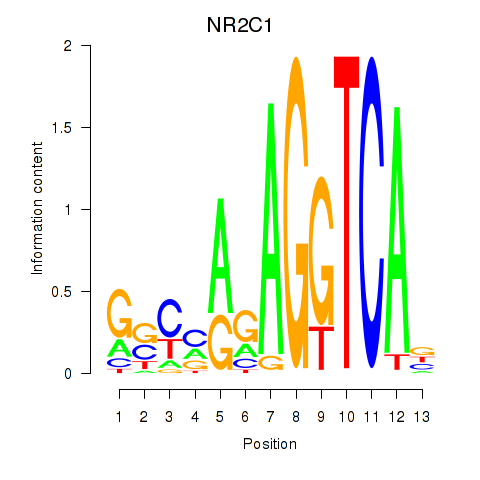
| NR2C1 | 0.42 |
|

|
|
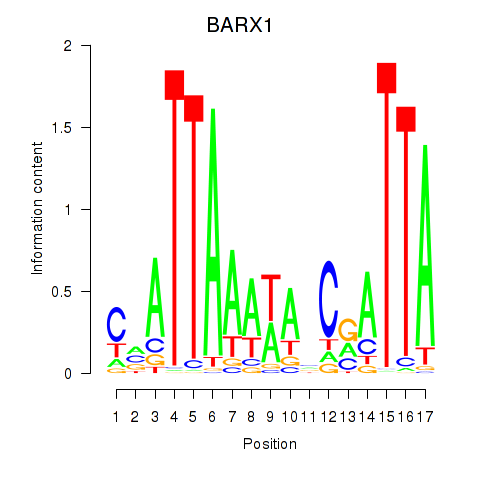
| BARX1 | 0.42 |
|
|
|
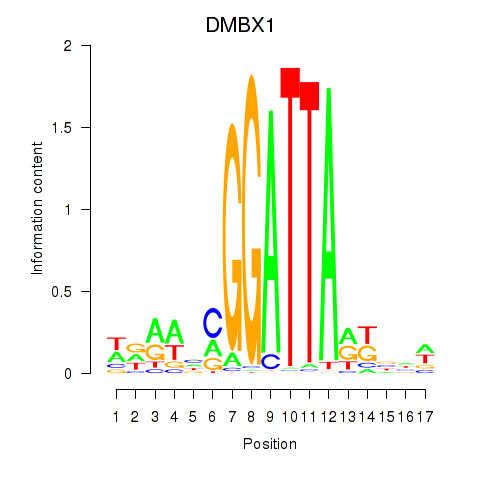
| DMBX1 | 0.42 |
|

|
|
| UGAAAUG | 0.42 |
|

|

|
| UUGGCAC | 0.42 |
|
|

|
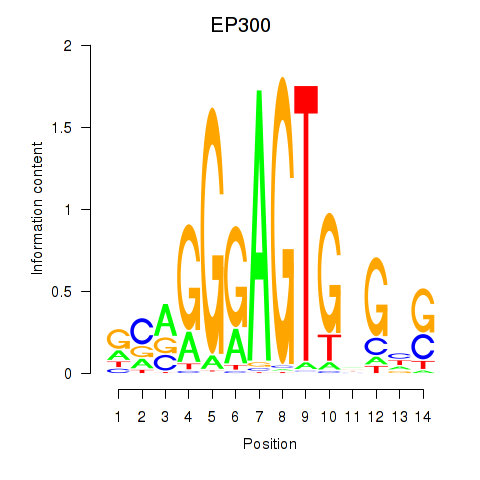
| EP300 | 0.41 |
|
|
|
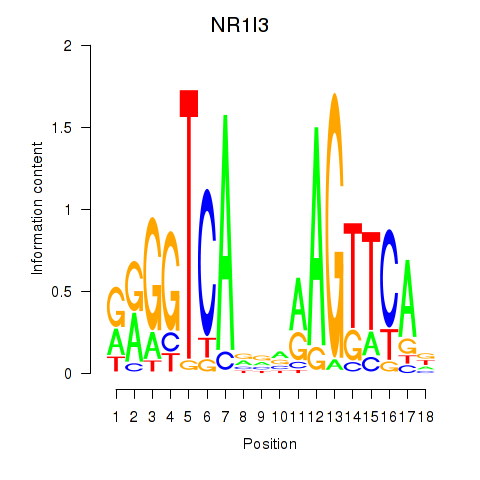
| NR1I3 | 0.41 |
|
|
|
| AUAAAGU | 0.41 |
|
|

|
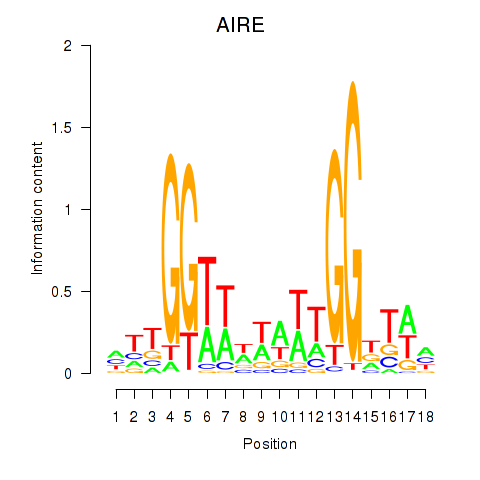
| AIRE | 0.41 |
|
|
|
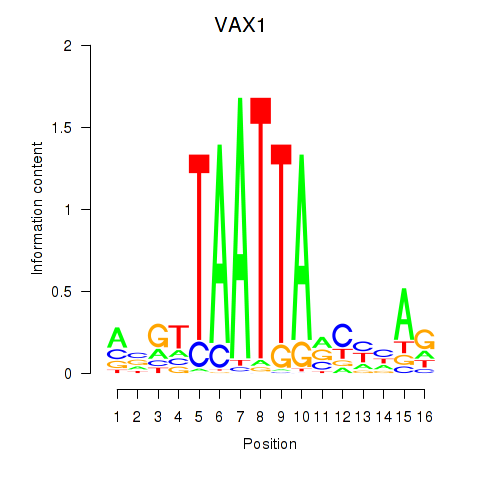
| VAX1_GSX2 | 0.41 |
|

|
|
| NANOG | 0.40 |
|
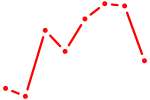
|
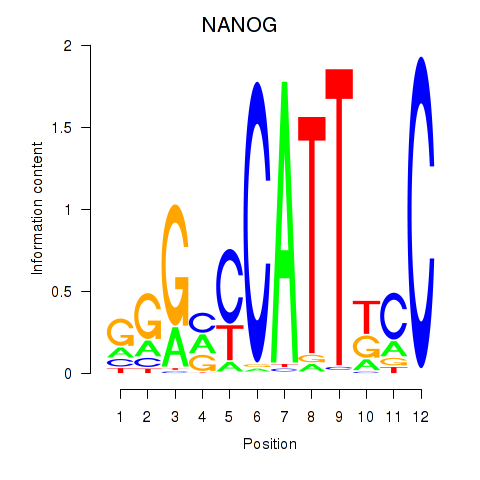
|
| TCF12_ASCL2 | 0.40 |
|
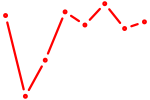
|
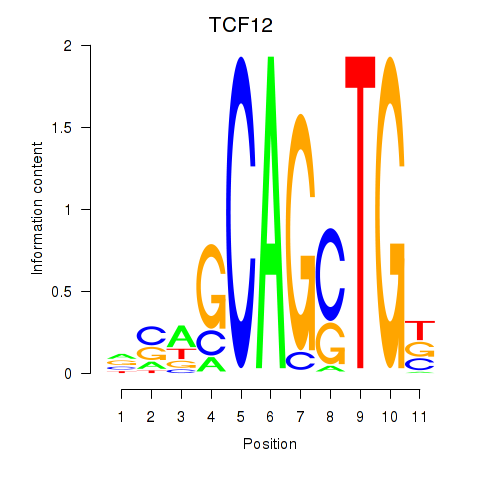
|
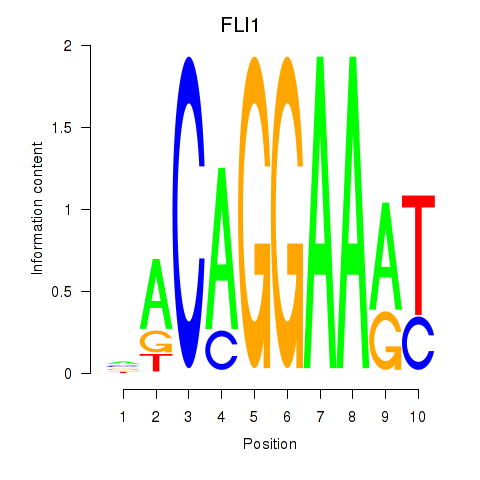
| FLI1 | 0.40 |
|
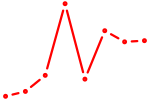
|
|
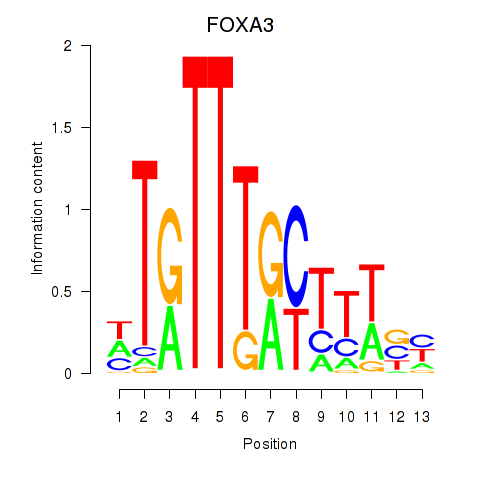
| FOXA3_FOXC2 | 0.40 |
|
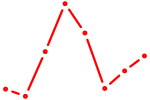
|
|
| AGCUGCC | 0.40 |
|
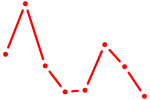
|

|
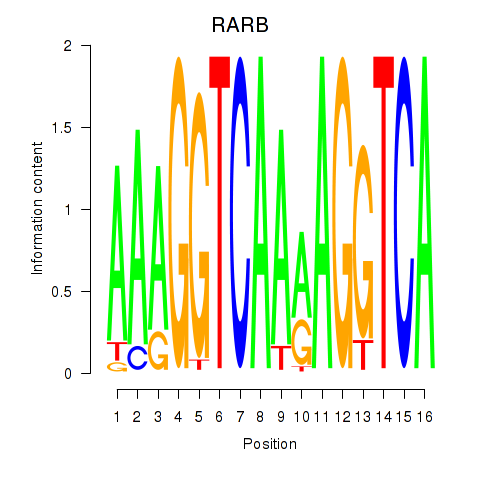
| RARB | 0.40 |
|

|
|
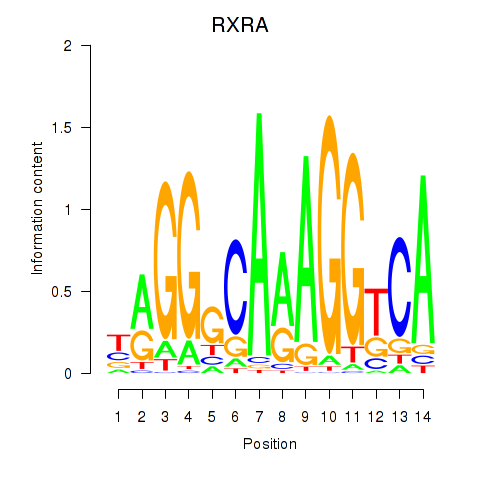
| RXRA_NR2F6_NR2C2 | 0.40 |
|
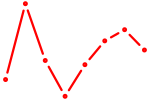
|
|
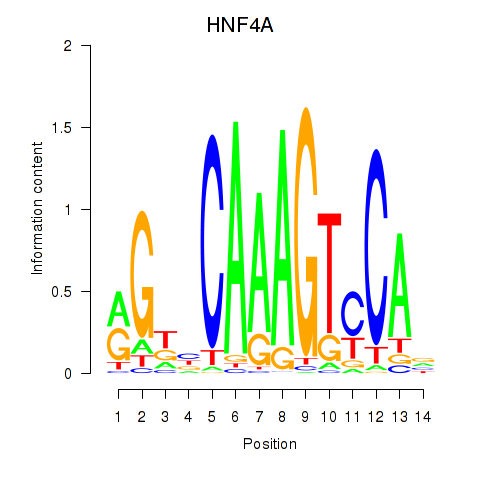
| HNF4A | 0.40 |
|
|
|
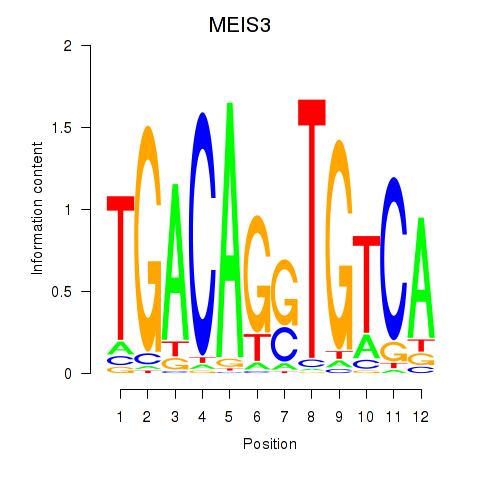
| MEIS3_TGIF2LX | 0.40 |
|

|
|
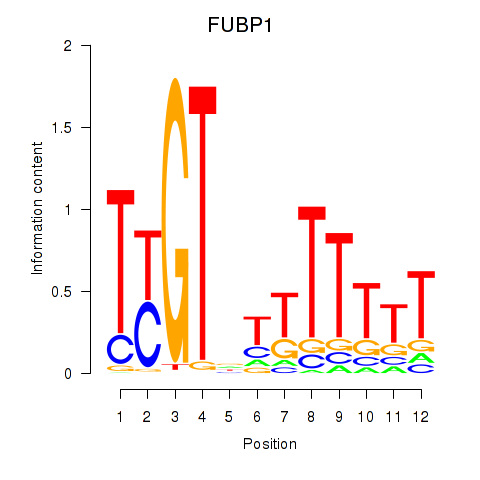
| FUBP1 | 0.40 |
|
|
|
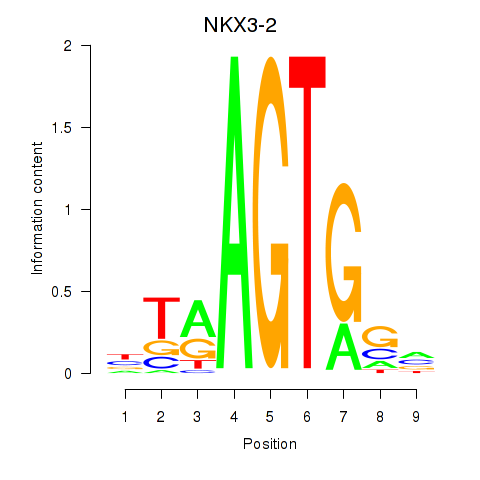
| NKX3-2 | 0.40 |
|
|
|
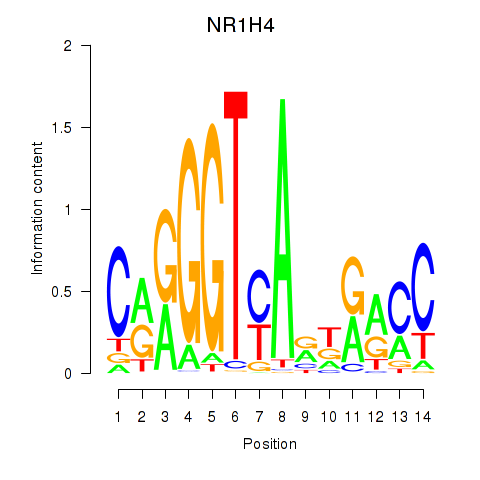
| NR1H4 | 0.39 |
|
|
|
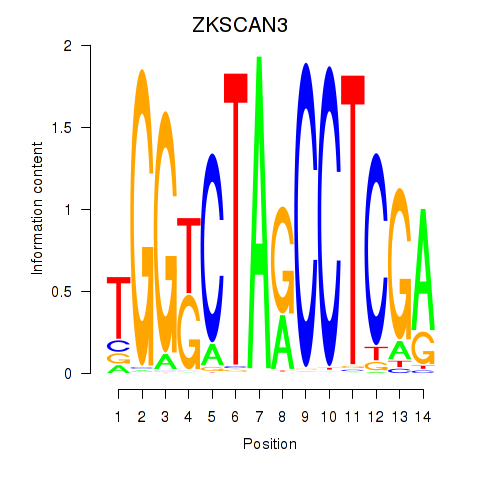
| ZKSCAN3 | 0.39 |
|
|
|
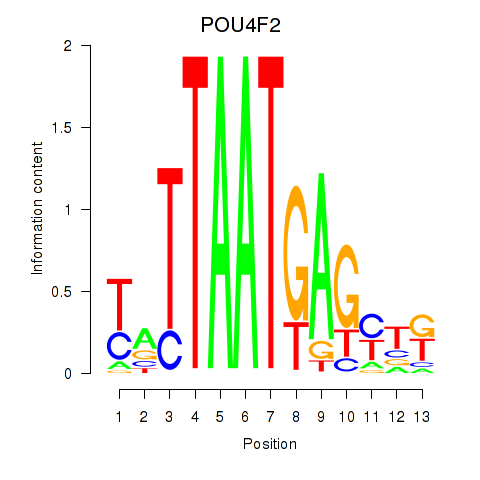
| POU4F2 | 0.39 |
|

|
|
| GUGCAAA | 0.38 |
|
|

|
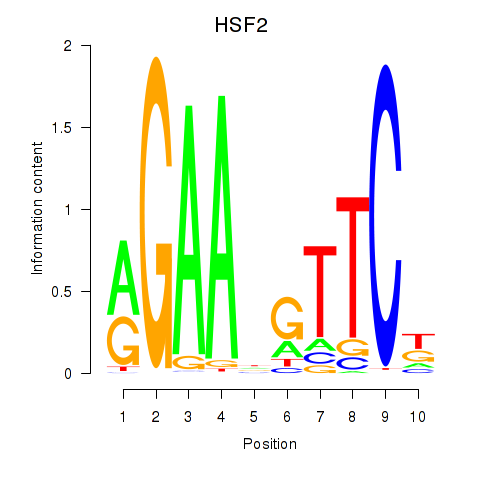
| HSF2 | 0.38 |
|
|
|
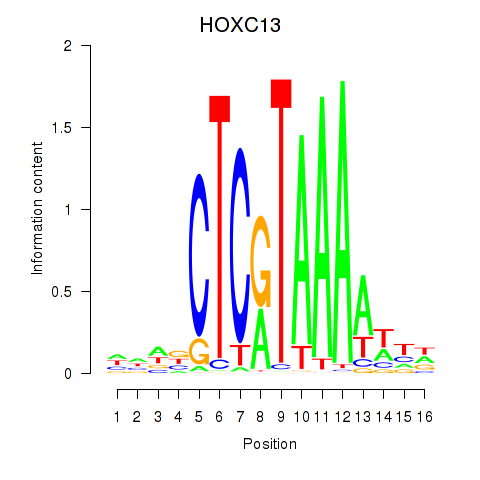
| HOXC13 | 0.38 |
|
|
|
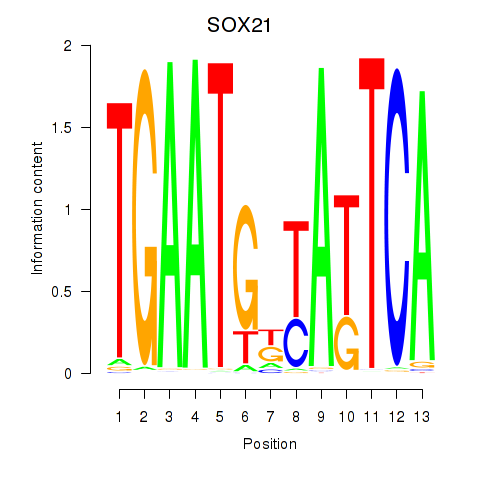
| SOX21 | 0.38 |
|

|
|
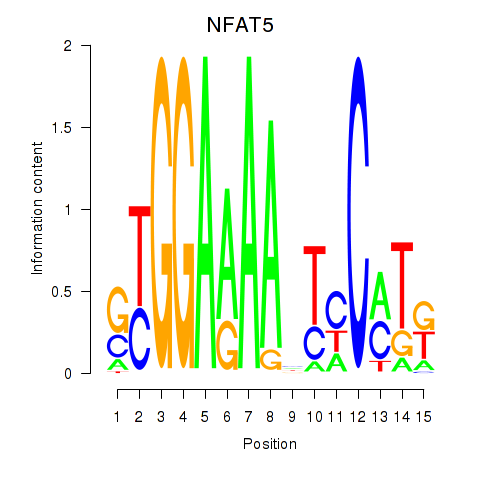
| NFAT5 | 0.38 |
|
|
|
| GAAAUGU | 0.38 |
|
|

|
| UGUGCUU | 0.38 |
|

|

|
| AGGUAGU | 0.38 |
|

|
|
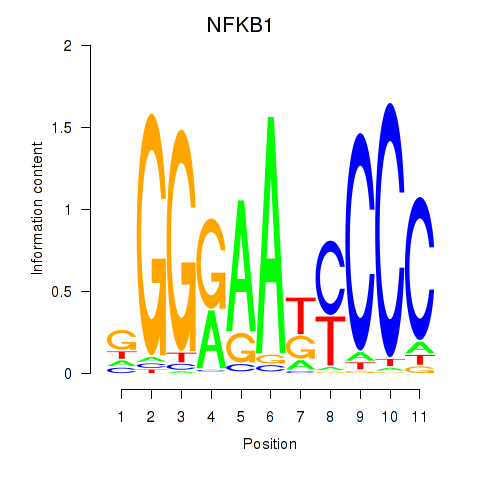
| NFKB1 | 0.38 |
|
|
|
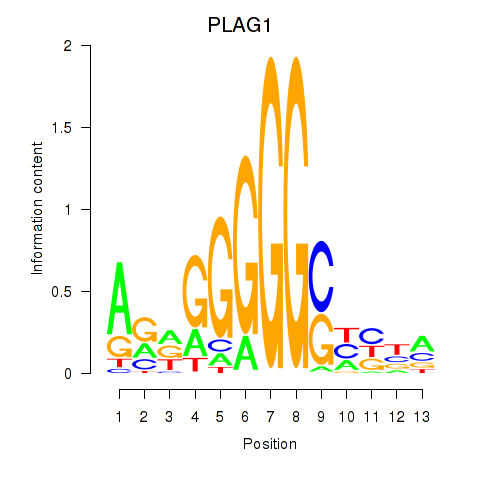
| PLAG1 | 0.37 |
|
|
|
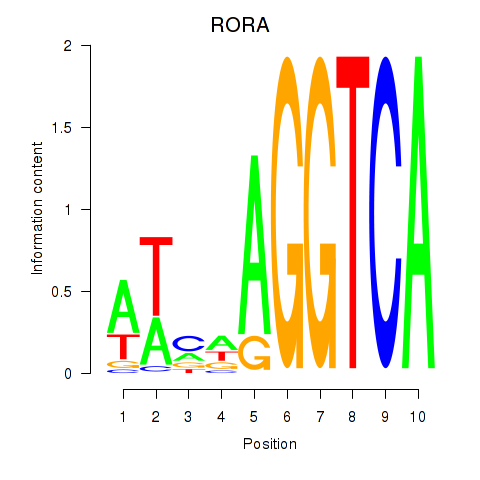
| RORA | 0.37 |
|
|
|
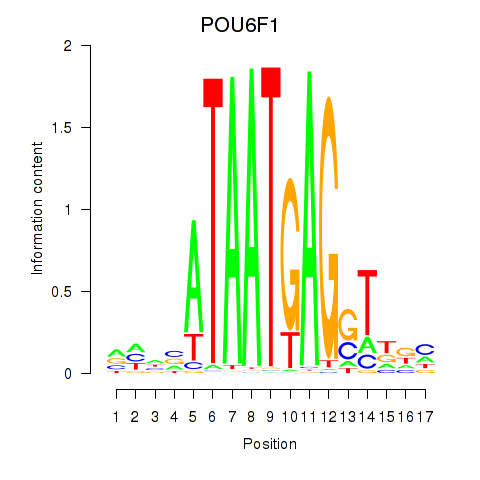
| POU6F1 | 0.37 |
|
|
|
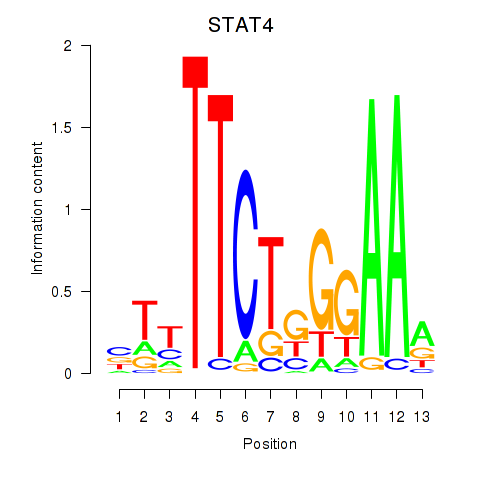
| STAT4 | 0.37 |
|
|
|
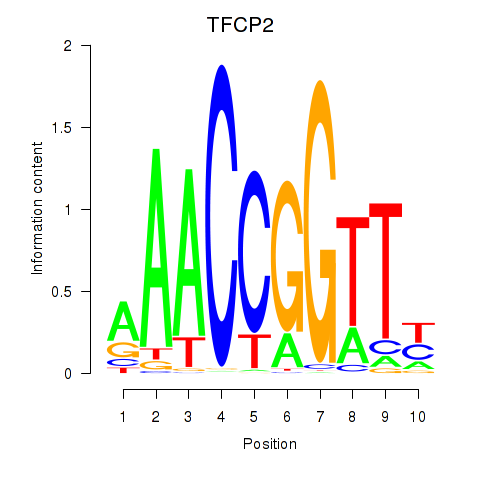
| TFCP2 | 0.37 |
|
|
|
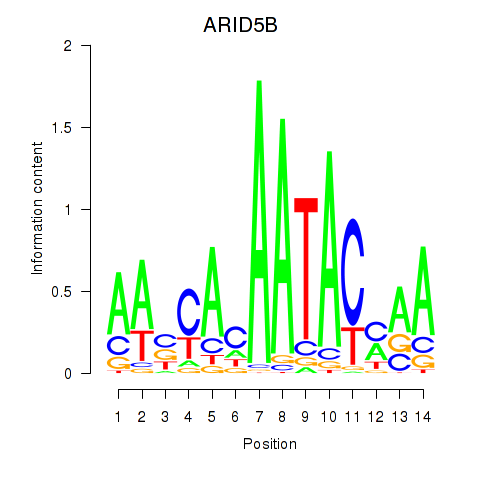
| ARID5B | 0.37 |
|
|
|
| GGAAGAC | 0.37 |
|

|

|
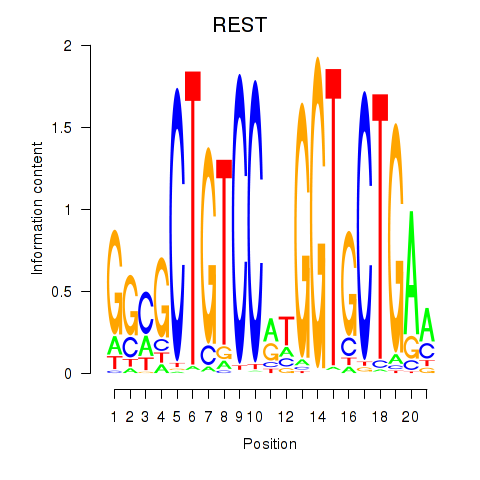
| REST | 0.37 |
|
|
|
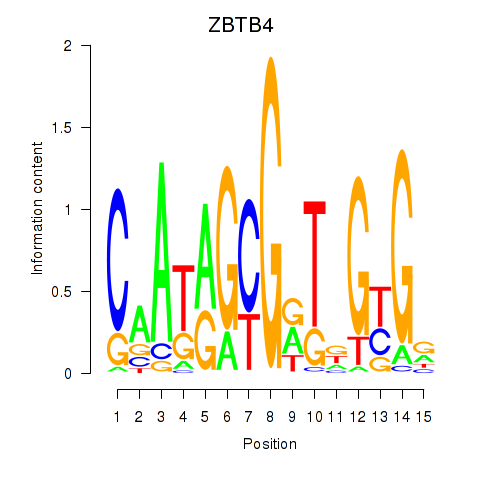
| ZBTB4 | 0.37 |
|

|
|
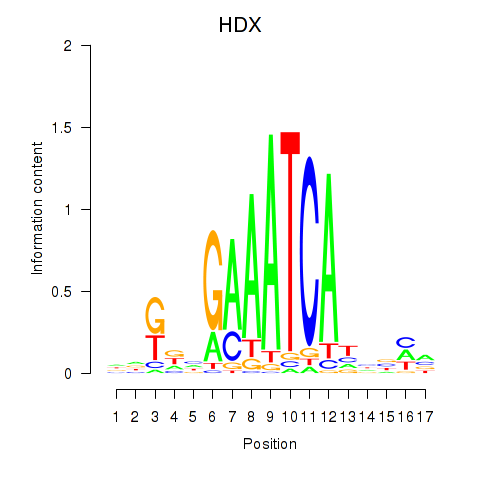
| HDX | 0.36 |
|
|
|
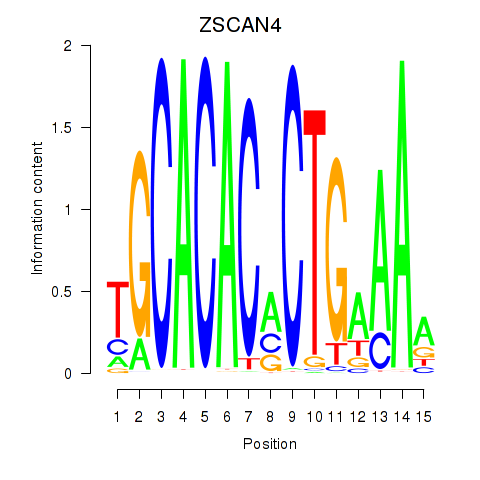
| ZSCAN4 | 0.36 |
|
|
|
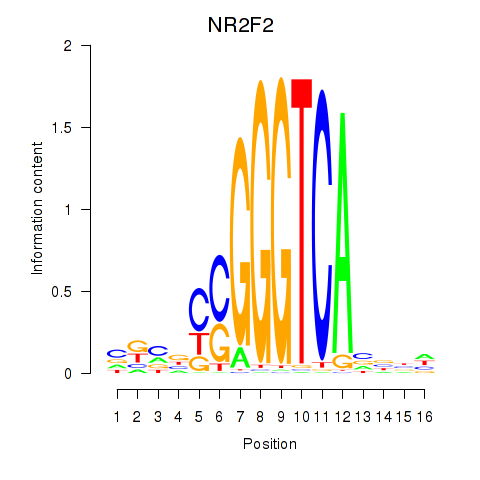
| NR2F2 | 0.36 |
|

|
|
| CAGUAAC | 0.36 |
|
|

|
| SOX4 | 0.36 |
|
|
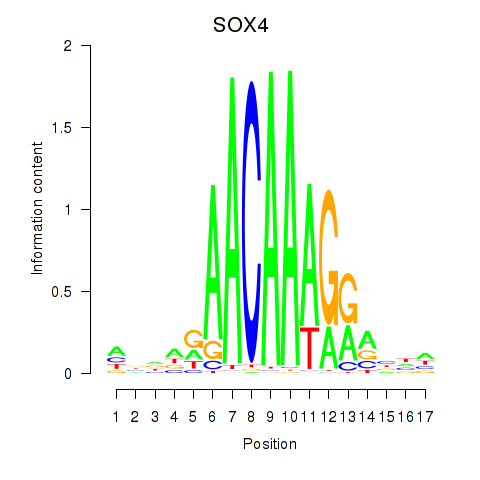
|
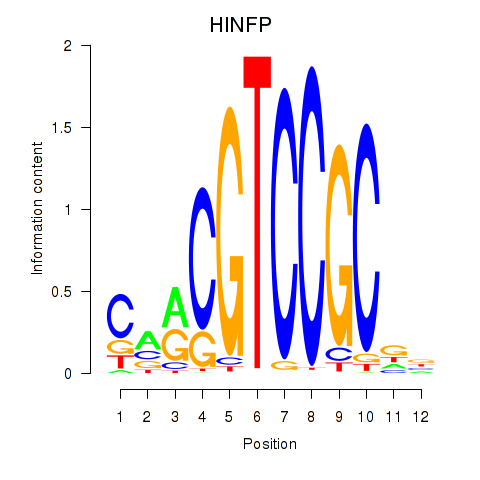
| HINFP | 0.36 |
|
|
|
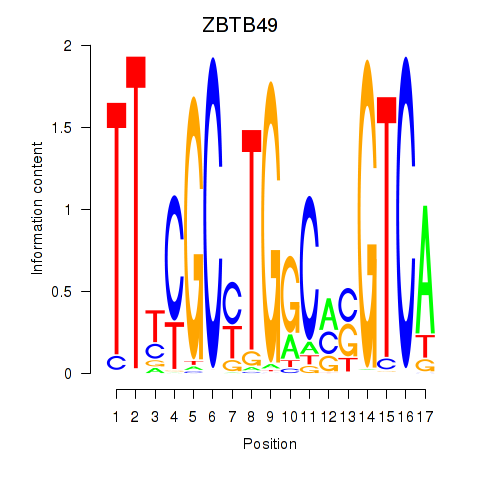
| ZBTB49 | 0.36 |
|
|
|
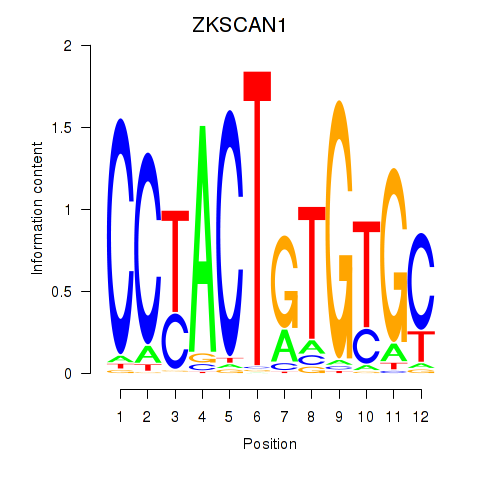
| ZKSCAN1 | 0.35 |
|
|
|
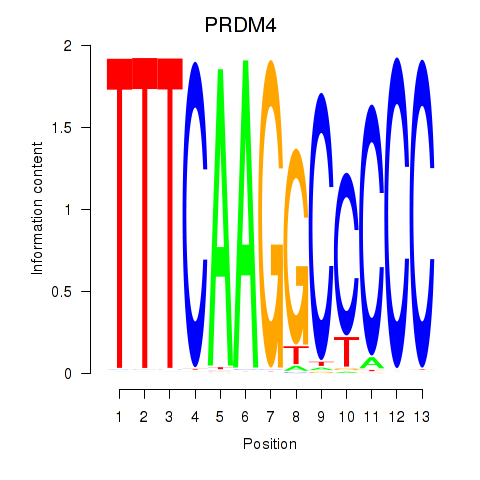
| PRDM4 | 0.35 |
|
|
|
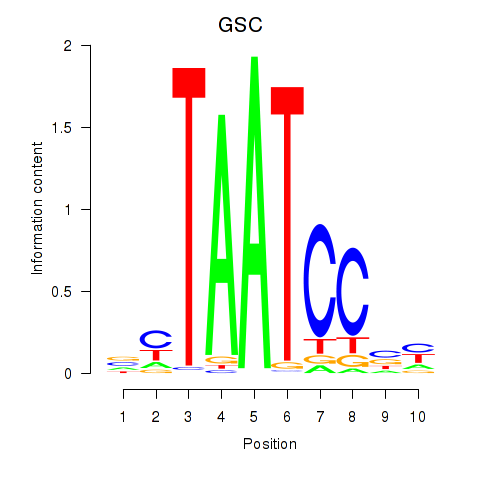
| GSC_GSC2 | 0.35 |
|

|
|
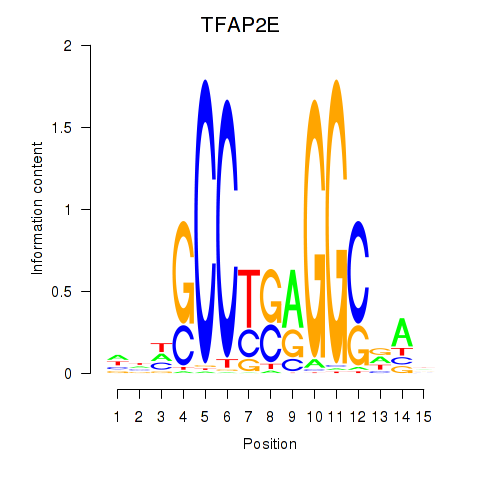
| TFAP2E | 0.35 |
|
|
|
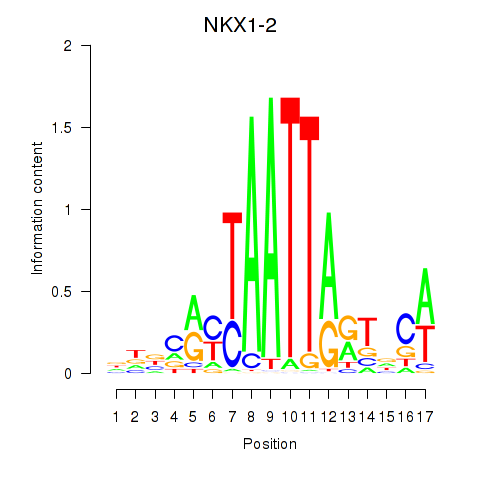
| NKX1-2_RAX | 0.35 |
|

|
|
| UACAGUA | 0.35 |
|
|

|
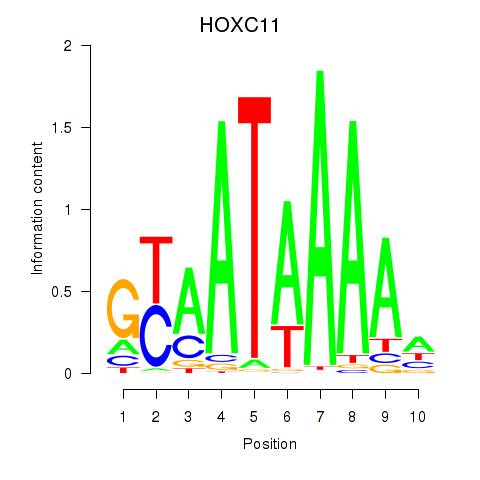
| HOXC11 | 0.35 |
|
|
|
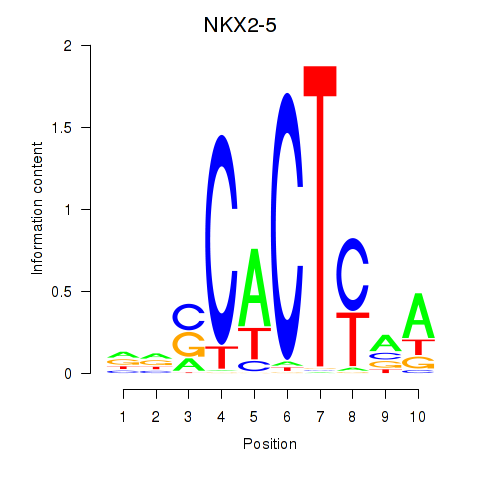
| NKX2-5 | 0.35 |
|
|
|
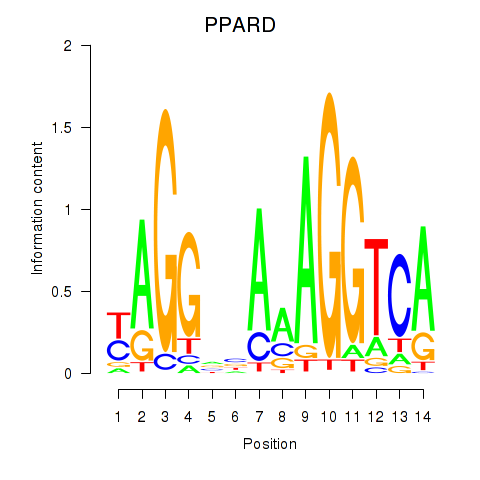
| PPARD | 0.35 |
|

|
|
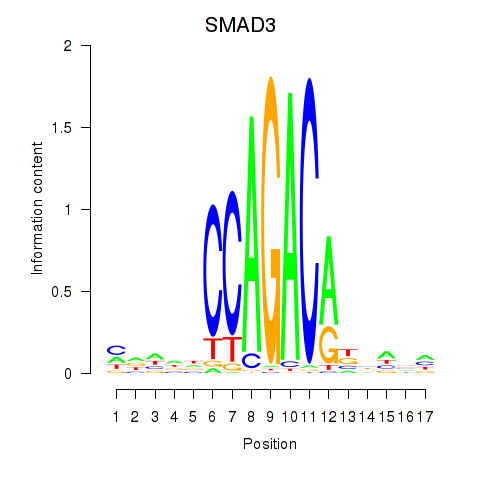
| SMAD3 | 0.34 |
|
|
|
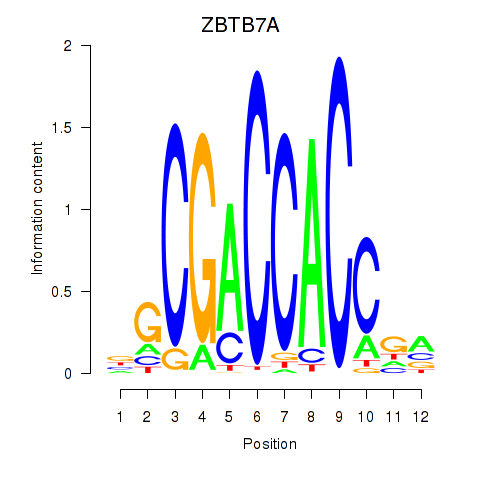
| ZBTB7A_ZBTB7C | 0.34 |
|
|
|
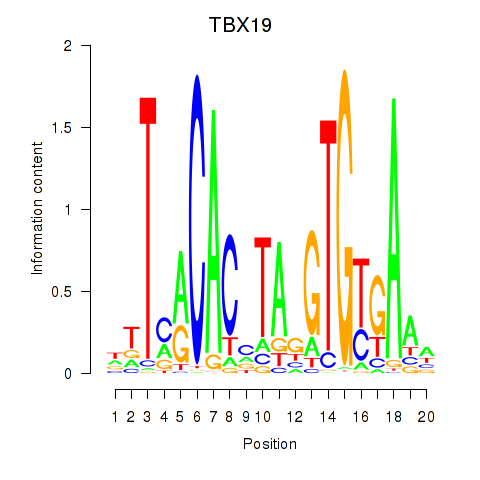
| TBX19 | 0.34 |
|
|
|
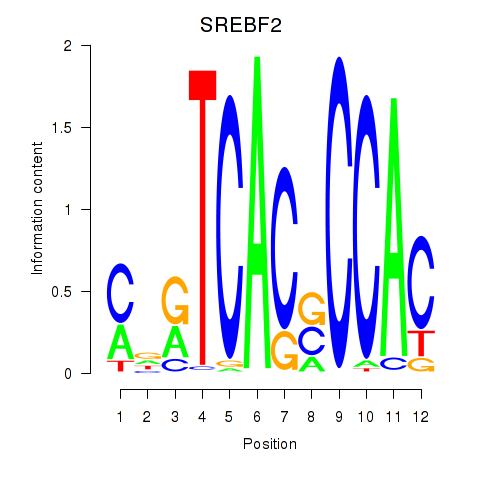
| SREBF2 | 0.34 |
|
|
|
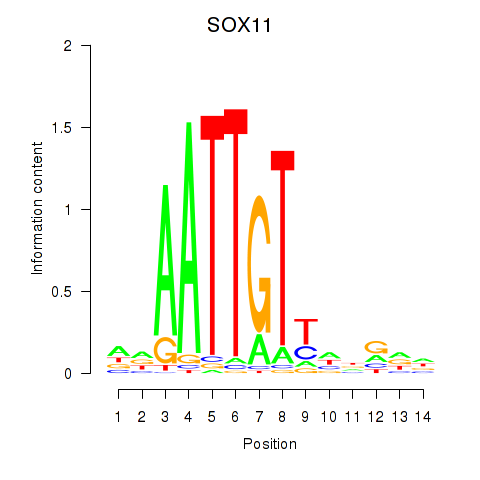
| SOX11 | 0.34 |
|
|
|
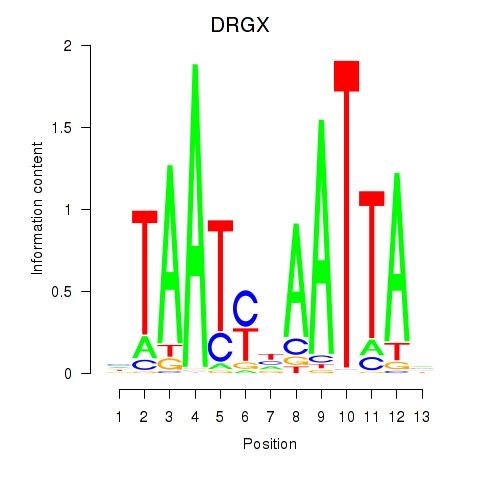
| DRGX_PROP1 | 0.34 |
|
|
|
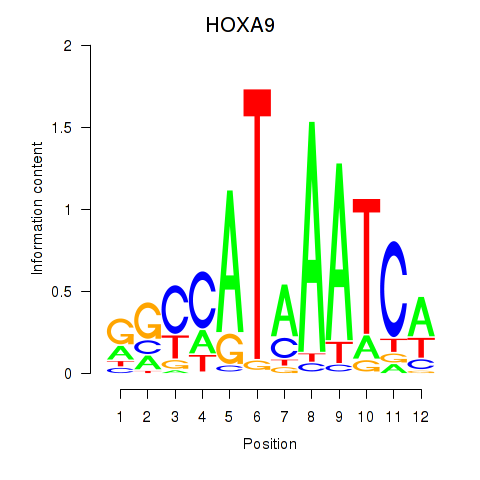
| HOXA9 | 0.34 |
|
|
|
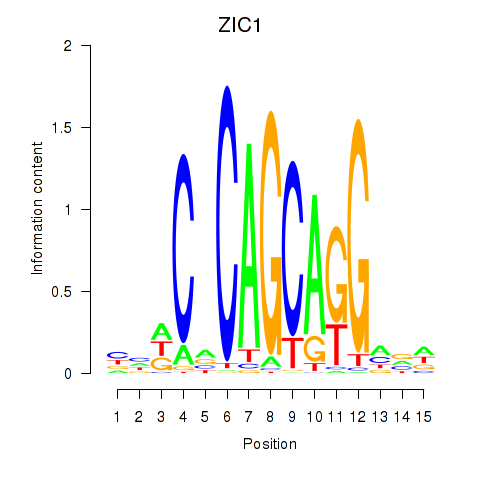
| ZIC1 | 0.34 |
|
|
|
| CCAGUGU | 0.33 |
|

|

|
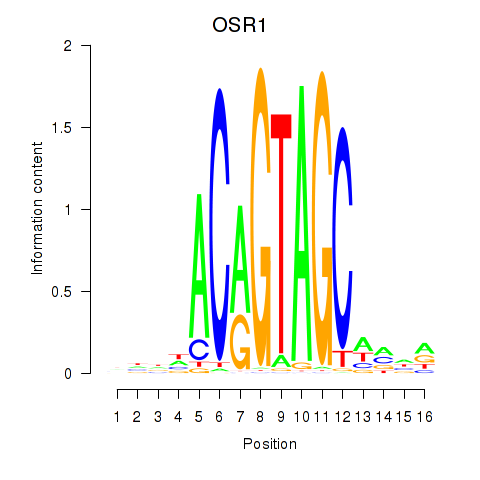
| OSR1_OSR2 | 0.33 |
|

|
|
| UAGUGUU | 0.33 |
|

|

|
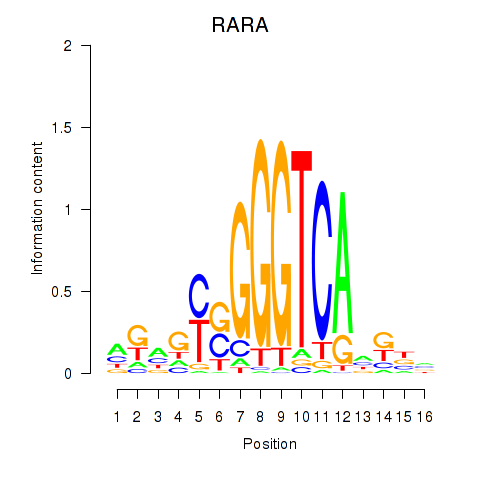
| RARA | 0.33 |
|

|
|
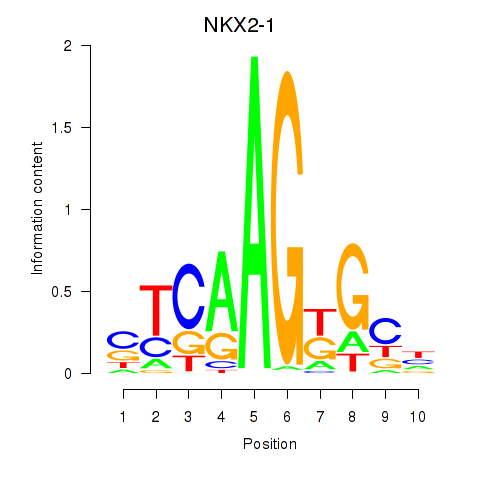
| NKX2-1 | 0.32 |
|

|
|
| ACCCUGU | 0.32 |
|

|

|
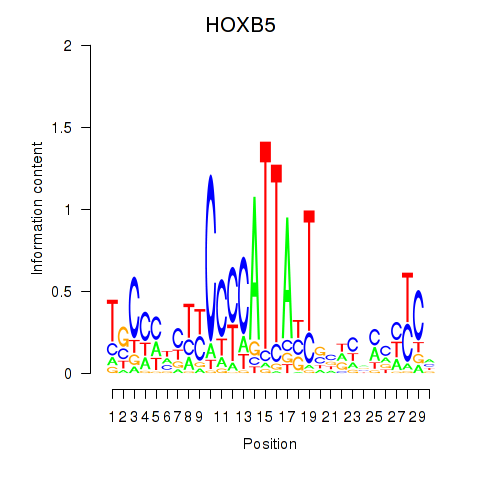
| HOXB5 | 0.32 |
|
|
|
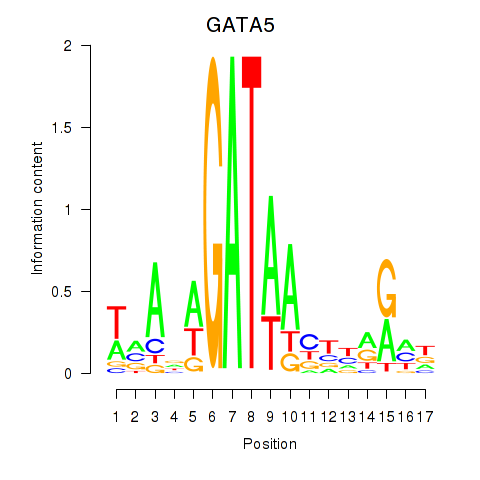
| GATA5 | 0.32 |
|
|
|
| GGCUCAG | 0.32 |
|
|

|
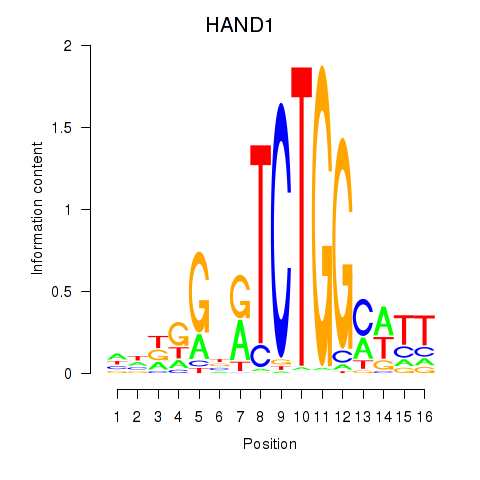
| HAND1 | 0.32 |
|
|
|
| ZNF350 | 0.32 |
|
|
|
| LHX8 | 0.32 |
|
|
|
| ZNF8 | 0.32 |
|

|
|
| AGCCCUU | 0.31 |
|
|
|
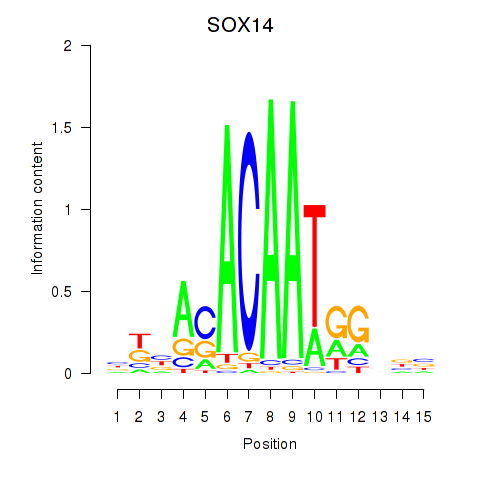
| SOX14 | 0.31 |
|
|
|
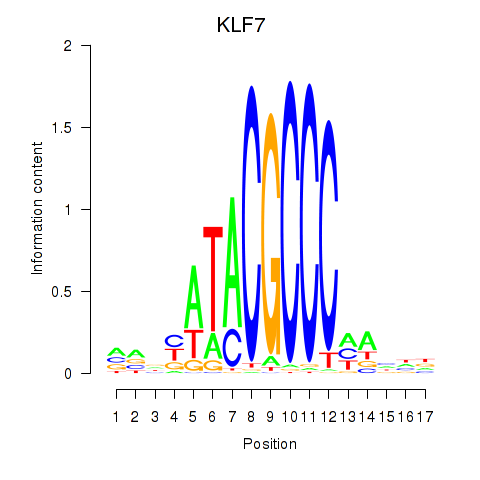
| KLF7 | 0.31 |
|
|
|
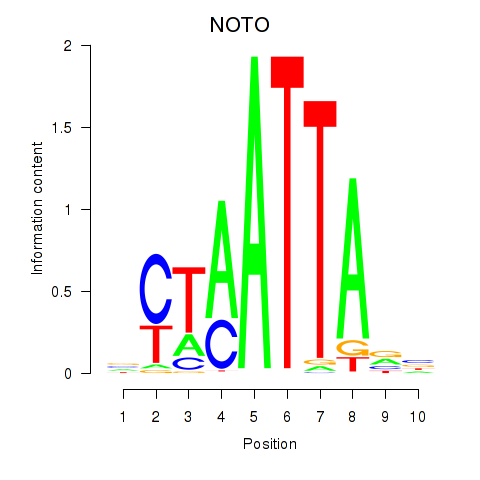
| NOTO_VSX2_DLX2_DLX6_NKX6-1 | 0.31 |
|
|
|
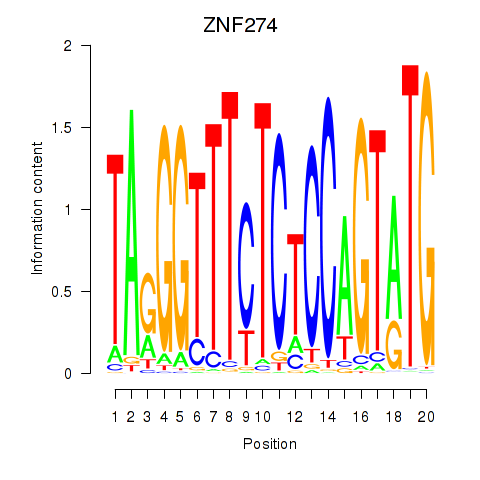
| ZNF274 | 0.31 |
|
|
|
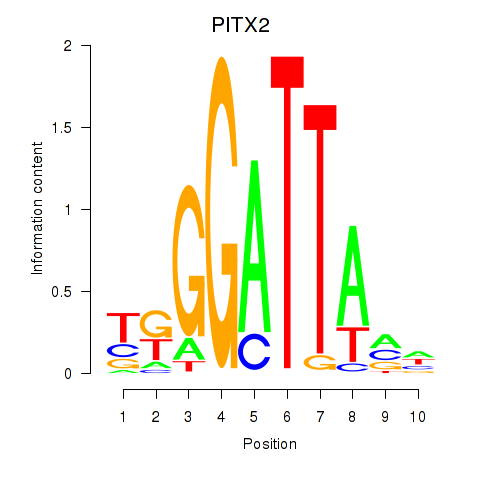
| PITX2 | 0.30 |
|
|
|
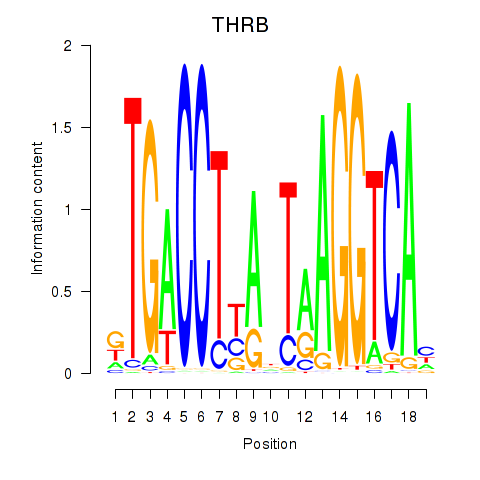
| THRB | 0.30 |
|
|
|
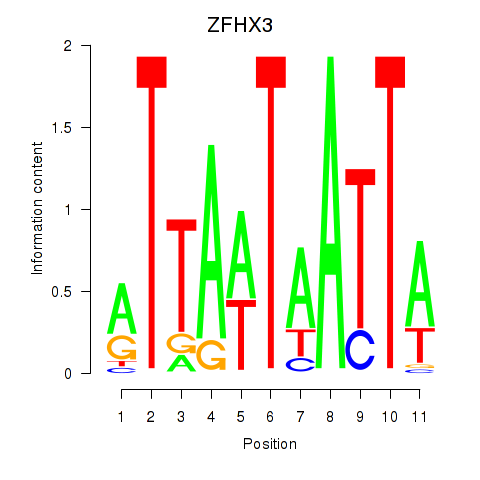
| ZFHX3 | 0.30 |
|
|
|
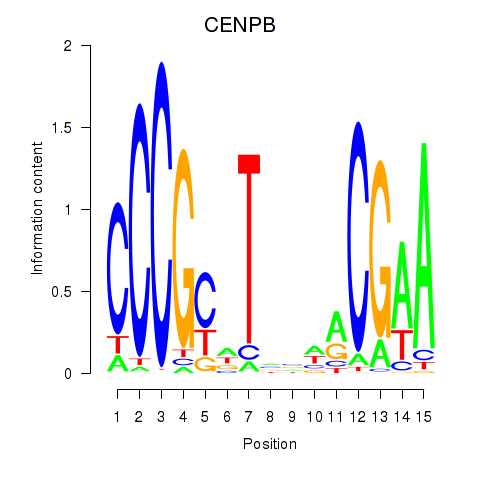
| CENPB | 0.30 |
|
|
|
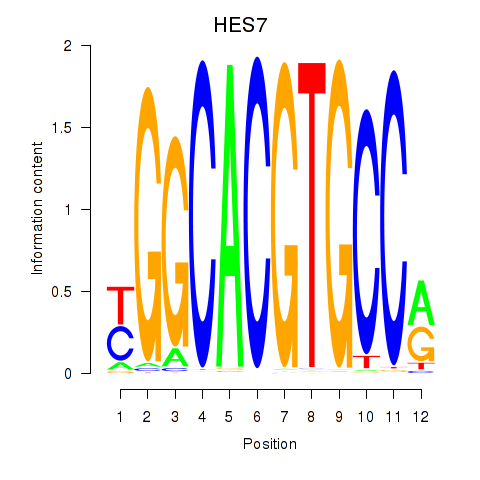
| HES7_HES5 | 0.30 |
|

|
|
| GZF1 | 0.29 |
|
|
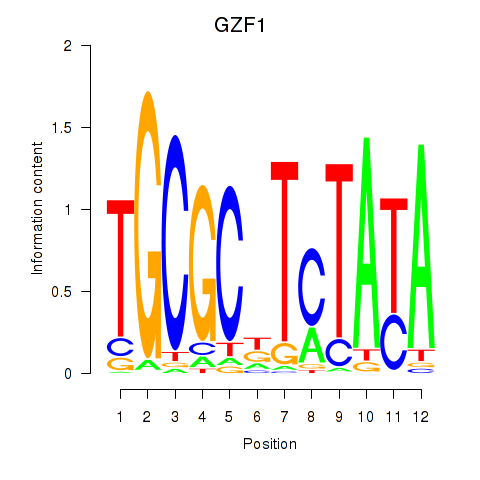
|
| BATF3 | 0.29 |
|

|
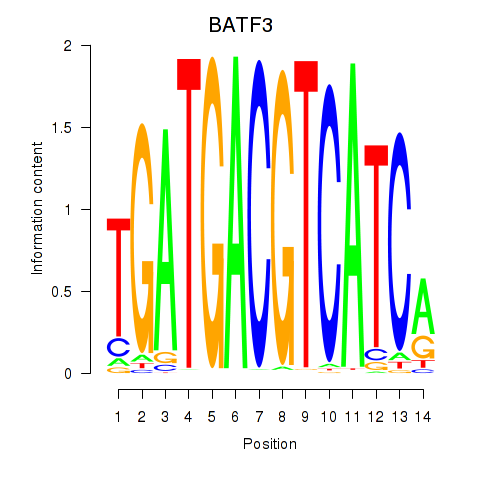
|
| GGCAGUG | 0.29 |
|

|

|
| AUGGCAC | 0.29 |
|
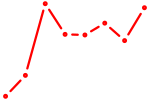
|

|
| AAGUGCU | 0.28 |
|
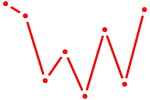
|
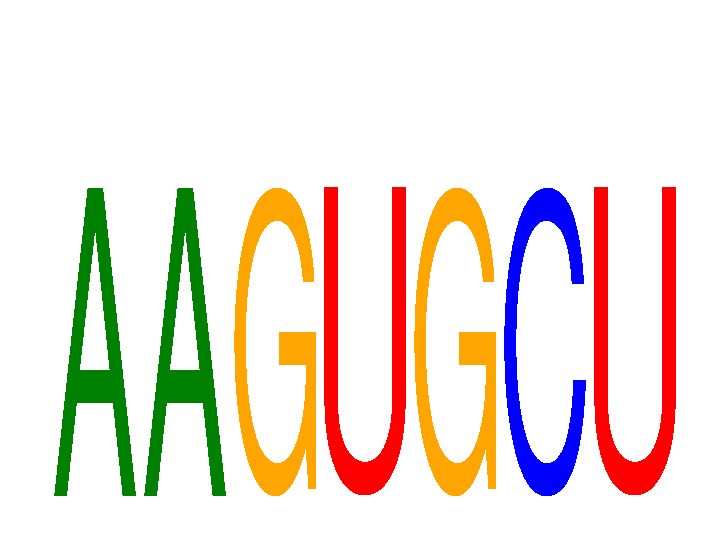
|
| NR4A1 | 0.28 |
|

|
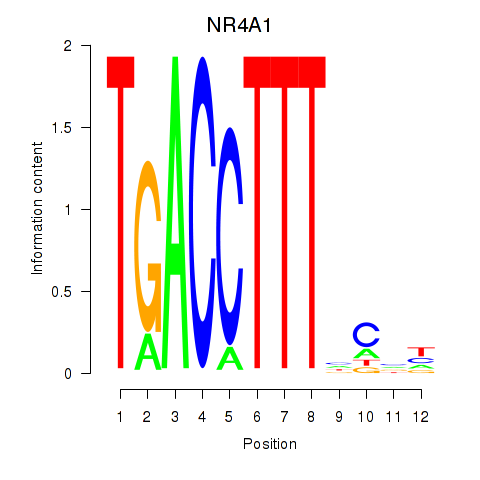
|
| CBFB | 0.28 |
|

|
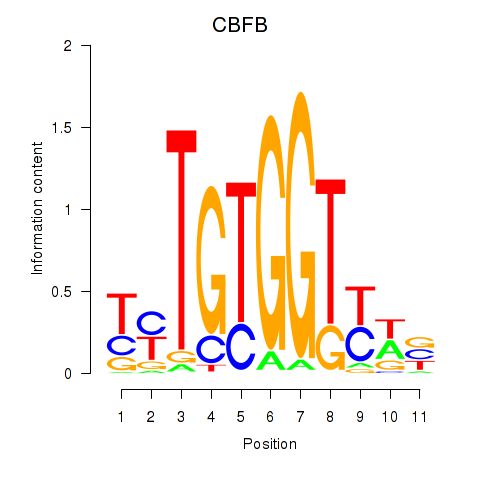
|
| NR2F1 | 0.28 |
|

|
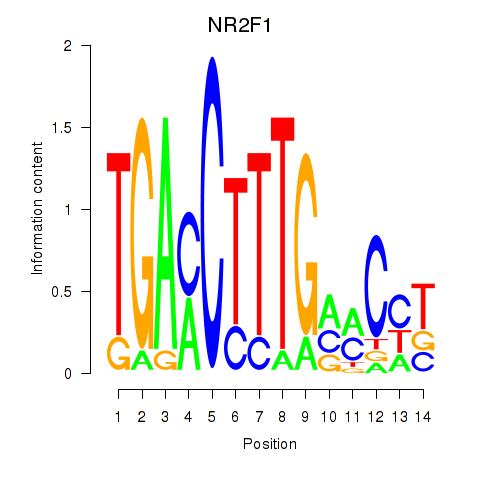
|
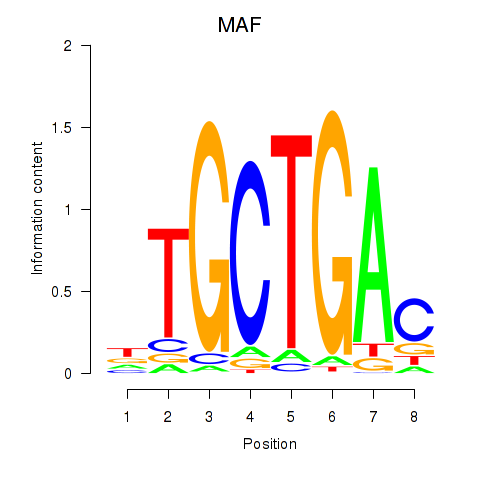
| MAF_NRL | 0.28 |
|
|
|
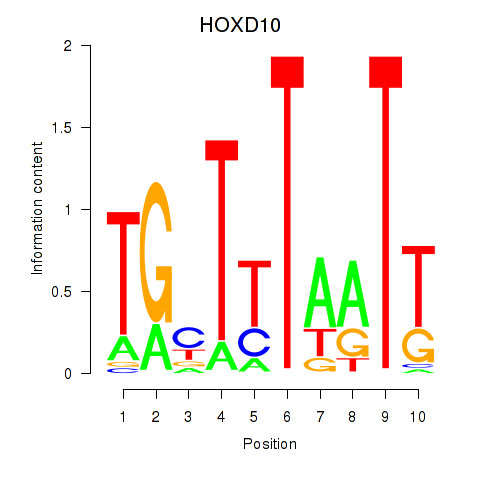
| HOXD10 | 0.27 |
|

|
|
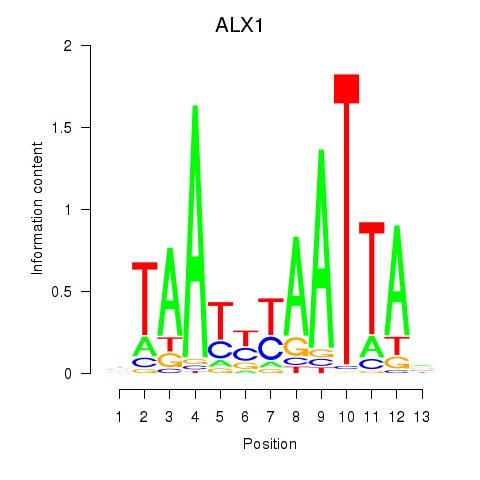
| ALX1_ARX | 0.27 |
|

|
|
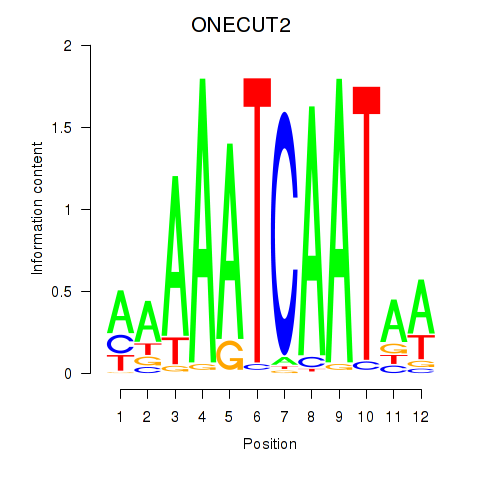
| ONECUT2_ONECUT3 | 0.27 |
|

|
|
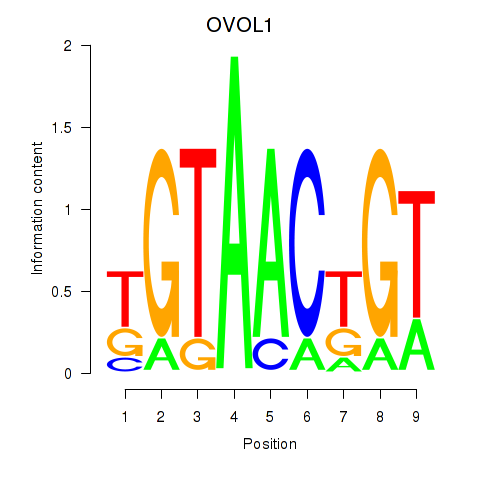
| OVOL1 | 0.27 |
|
|
|
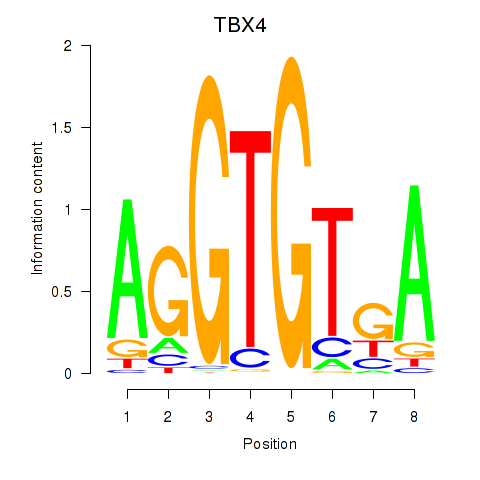
| TBX4 | 0.27 |
|

|
|
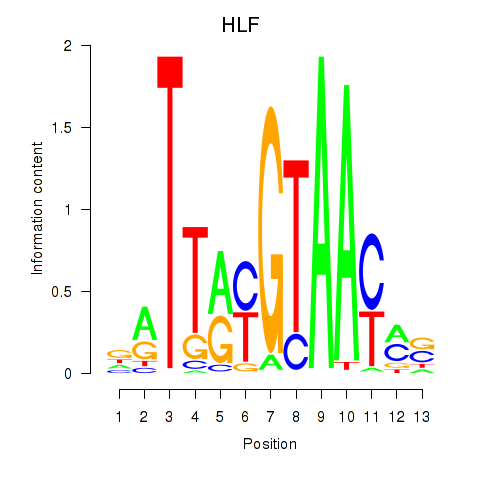
| HLF_TEF | 0.27 |
|
|
|
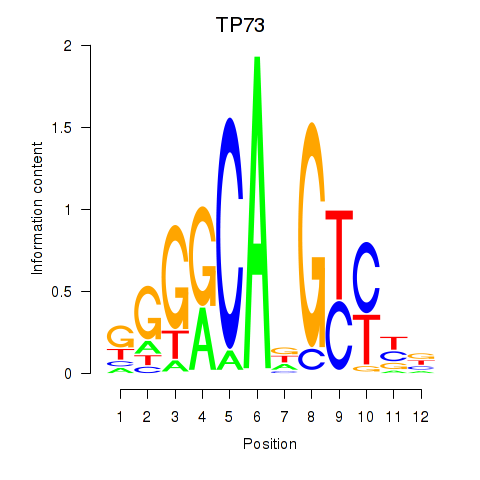
| TP73 | 0.27 |
|

|
|
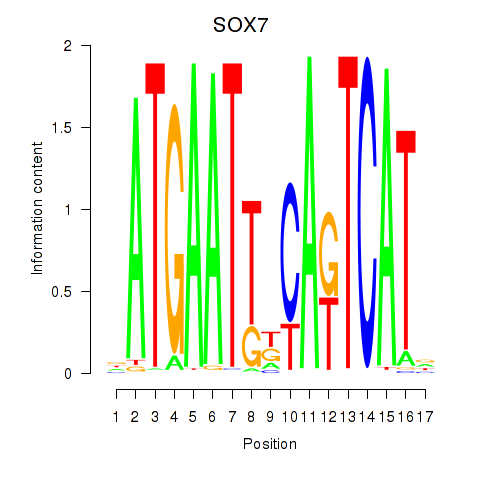
| SOX7 | 0.25 |
|

|
|
| AAGGUGC | 0.25 |
|
|

|
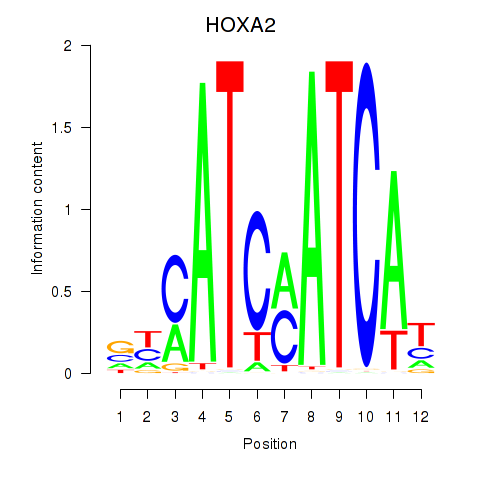
| HOXA2_HOXB1 | 0.25 |
|

|
|
| AAUGCCC | 0.24 |
|
|
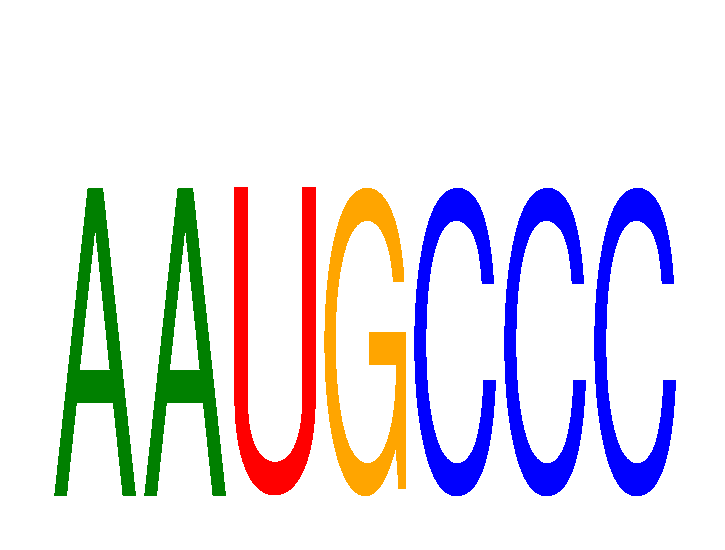
|
| GGCAAGA | 0.24 |
|

|

|
| ZNF282 | 0.24 |
|

|
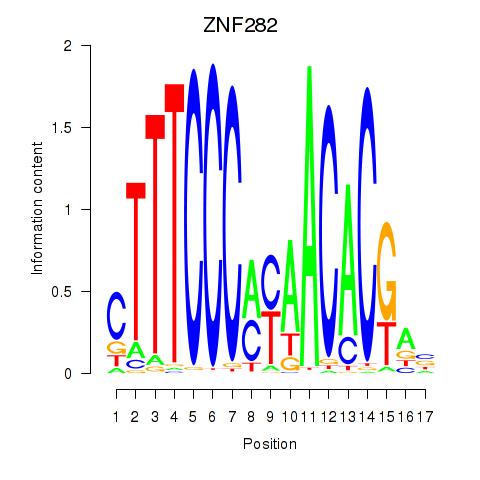
|
| ISL2 | 0.24 |
|

|
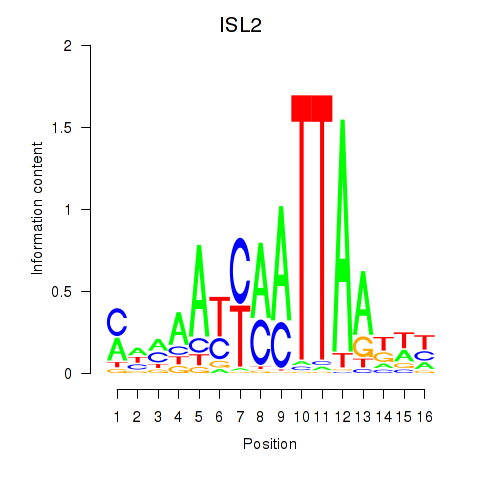
|
| NKX2-8 | 0.24 |
|
|
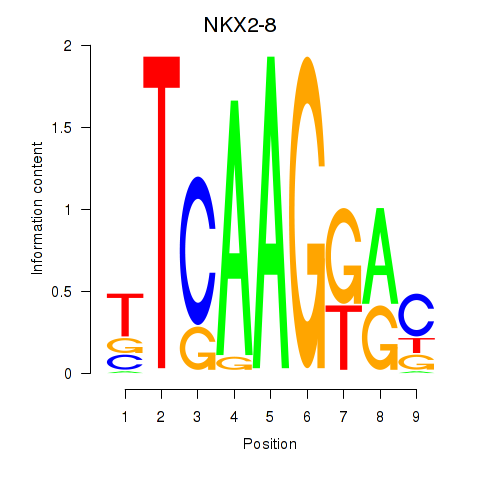
|
| SOX30 | 0.24 |
|

|
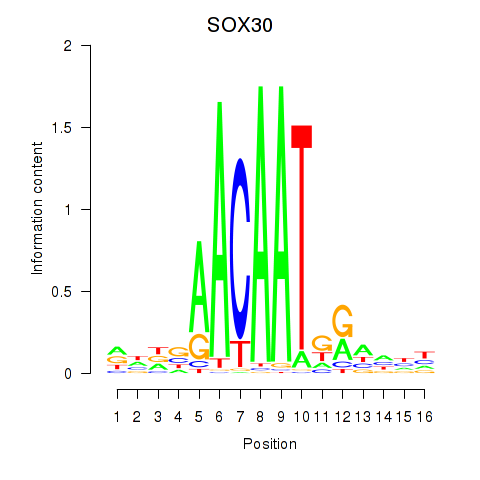
|
| CAGUAGU | 0.23 |
|
|

|
| GAUUGUC | 0.23 |
|

|

|
| UCCCUUU | 0.23 |
|

|
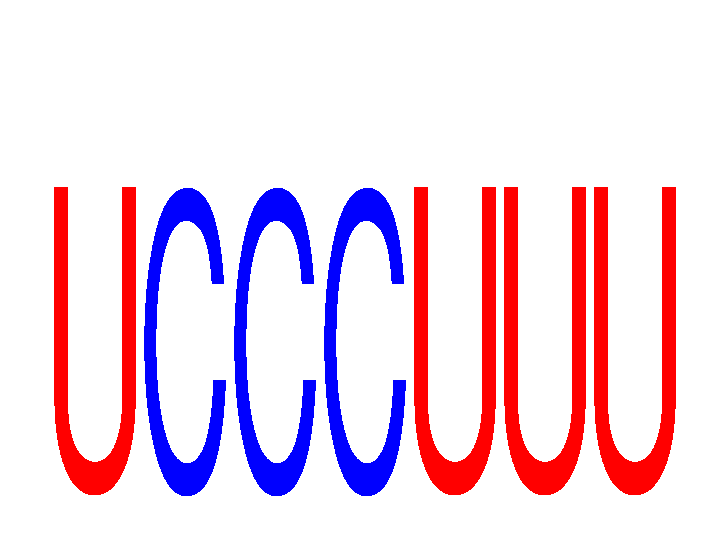
|
| GAGAUGA | 0.23 |
|
|

|
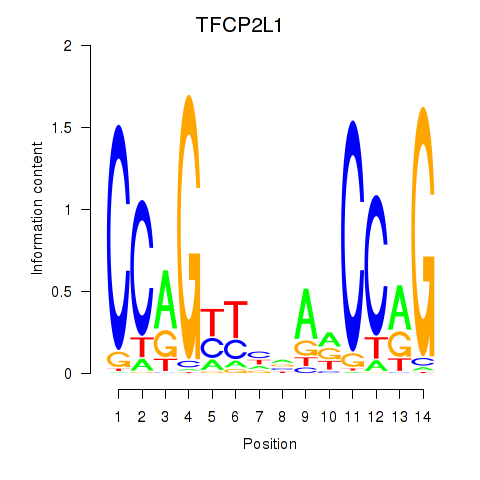
| TFCP2L1 | 0.23 |
|
|
|
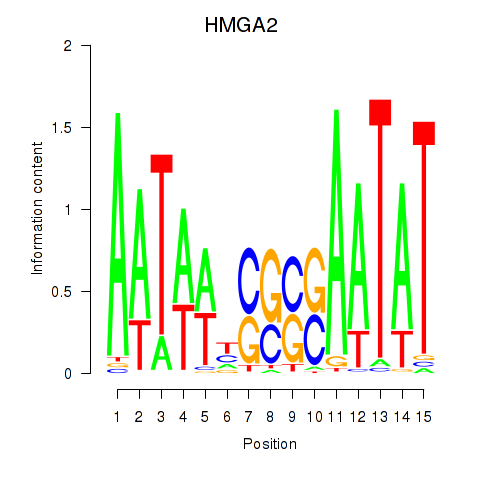
| HMGA2 | 0.22 |
|

|
|
| IRX6_IRX4 | 0.22 |
|
|
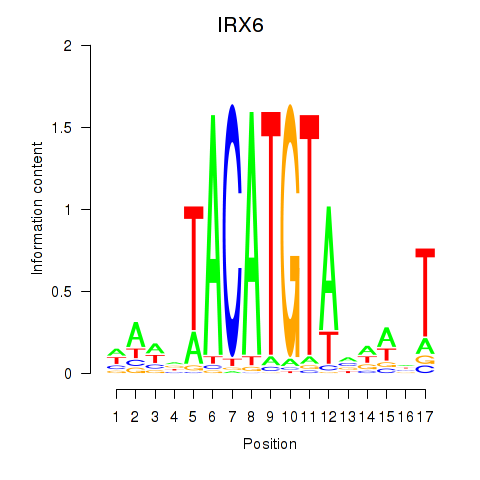
|
| ACAGUAC | 0.22 |
|

|

|
| HOXC6_HOXA7 | 0.22 |
|
|
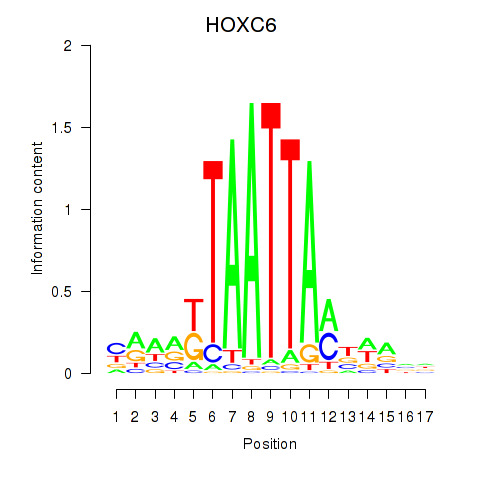
|
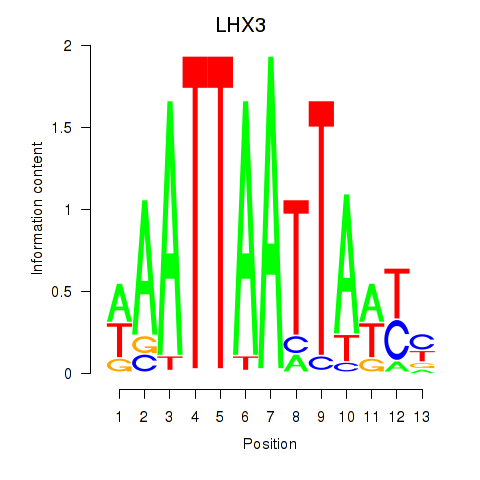
| LHX3 | 0.22 |
|
|
|
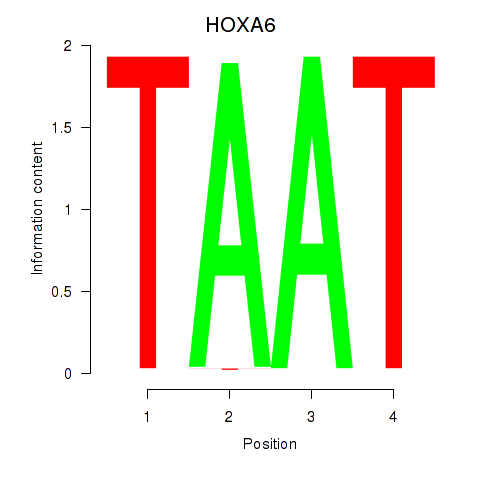
| HOXA6 | 0.22 |
|
|
|
| CACAGUG | 0.21 |
|
|

|
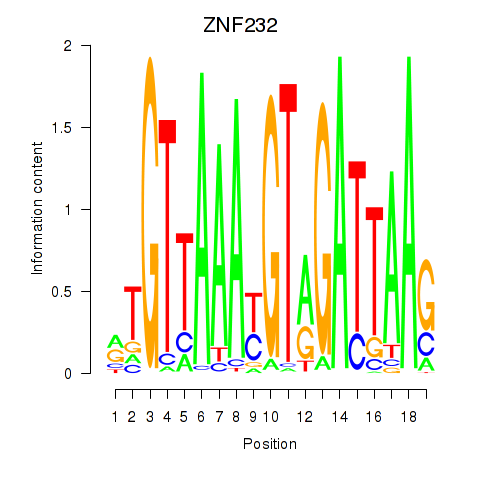
| ZNF232 | 0.21 |
|
|
|
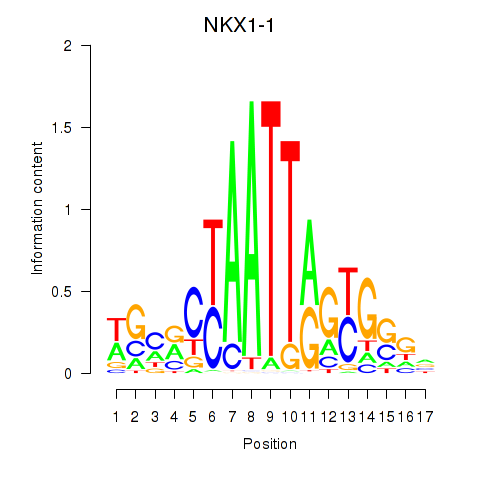
| NKX1-1 | 0.21 |
|
|
|
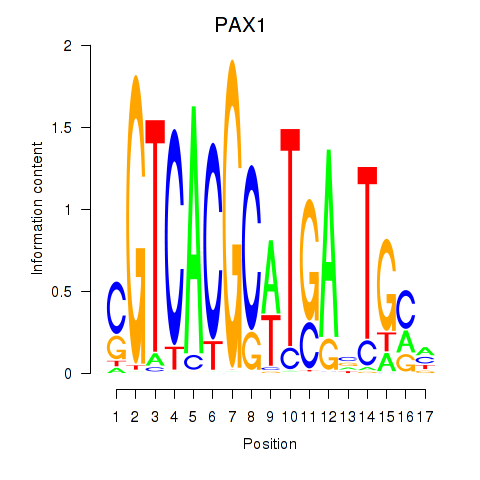
| PAX1_PAX9 | 0.20 |
|
|
|
| UUGGCAA | 0.19 |
|

|

|
| CCAGCAU | 0.18 |
|
|

|
| ACUGGCC | 0.18 |
|

|

|
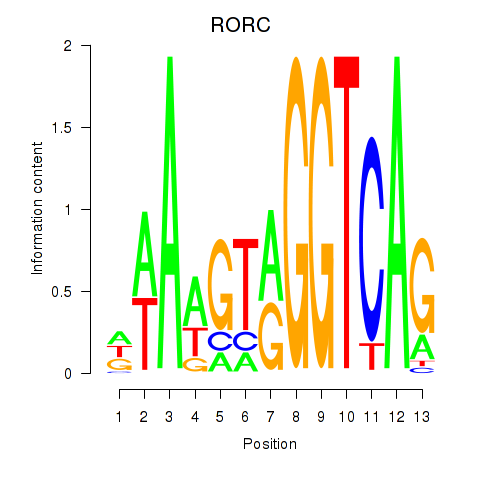
| RORC | 0.18 |
|

|
|
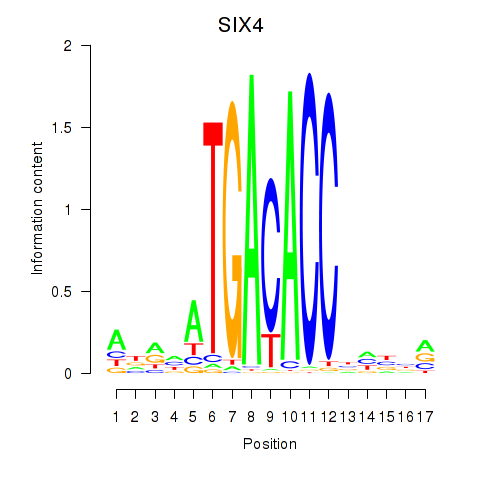
| SIX4 | 0.17 |
|
|
|
| GUAACAG | 0.16 |
|

|

|
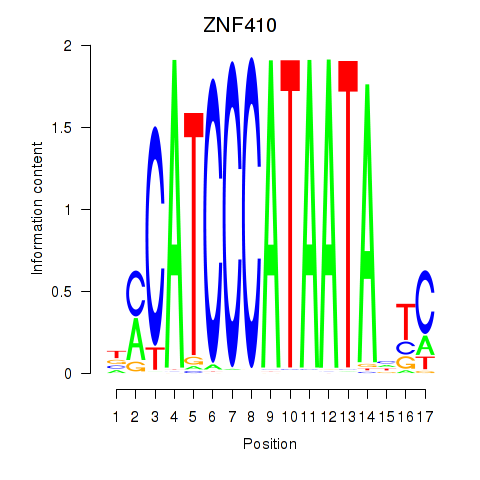
| ZNF410 | 0.16 |
|

|
|
| CCUUCAU | 0.16 |
|
|
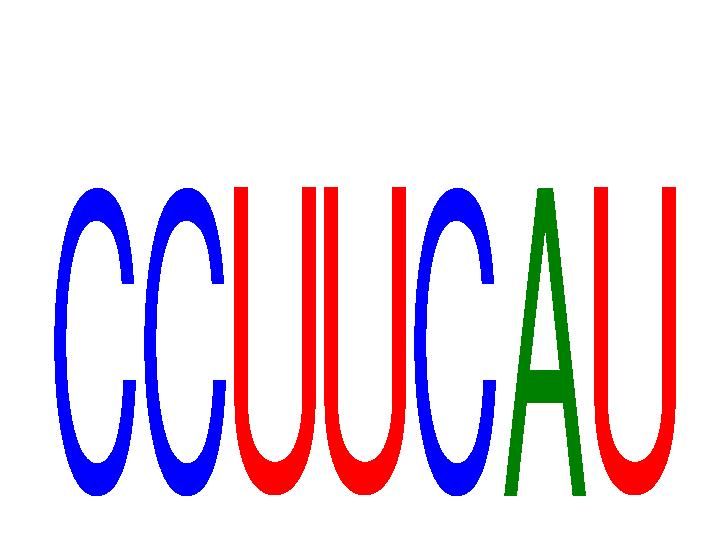
|
| AGCUUAU | 0.16 |
|
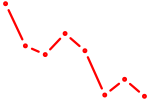
|

|
| ACUGCAU | 0.16 |
|
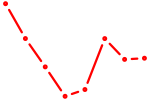
|

|
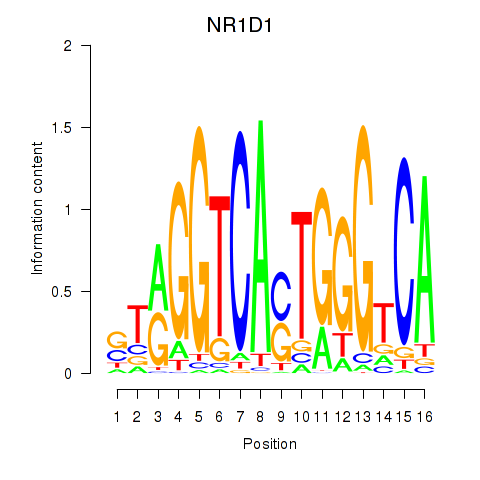
| NR1D1 | 0.16 |
|
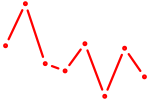
|
|
| UUGUUCG | 0.16 |
|
|

|
| UGCAUUG | 0.15 |
|
|

|
| CUGGCUG | 0.15 |
|
|

|
| CAGUCCA | 0.15 |
|

|

|
| PBX2 | 0.15 |
|
|
|
| UAAGACU | 0.14 |
|
|

|
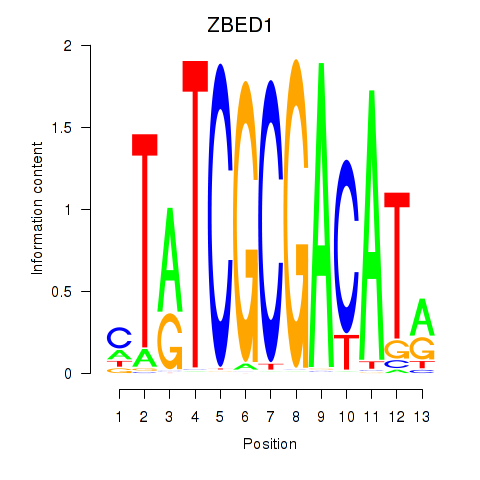
| ZBED1 | 0.14 |
|
|
|
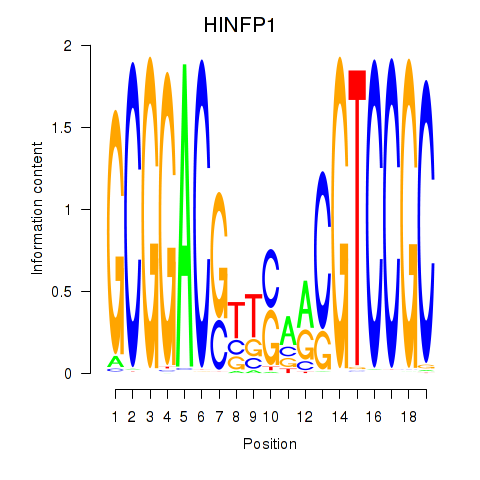
| HINFP1 | 0.14 |
|
|
|
| UGCAGUC | 0.14 |
|
|

|
| AGUGGUU | 0.14 |
|
|

|
| ACCACAG | 0.14 |
|
|

|
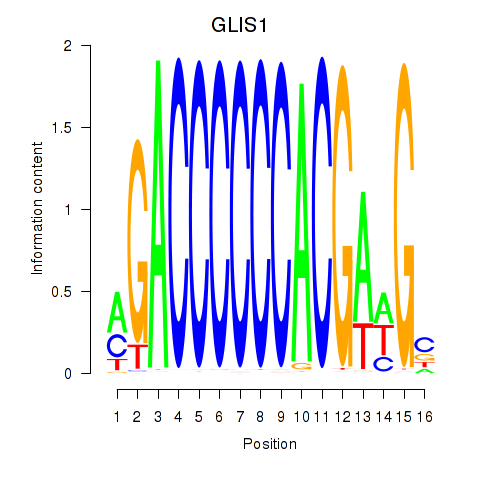
| GLIS1 | 0.12 |
|
|
|
| UAAUGCU | 0.12 |
|

|

|
| ZNF35 | 0.12 |
|
|
|
| CCACAGG | 0.12 |
|

|

|
| UGACCUA | 0.12 |
|
|

|
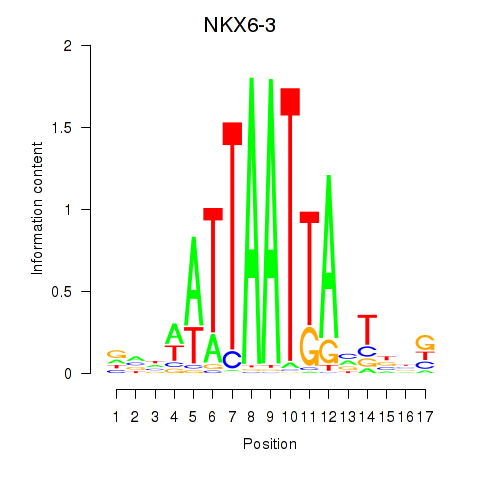
| NKX6-3 | 0.11 |
|
|
|
| CUACAGU | 0.11 |
|

|

|
| GGAGUGU | 0.11 |
|

|

|
| UGACAUC | 0.11 |
|
|

|
| GGACGGA | 0.11 |
|
|

|
| UAAGACG | 0.11 |
|
|

|
| GAUAUGU | 0.10 |
|
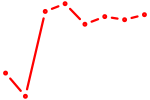
|

|
| AUGUGCC | 0.10 |
|
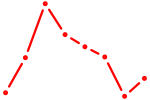
|

|
| AACGGAA | 0.09 |
|

|

|
| AAUCUCA | 0.09 |
|
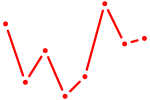
|

|
| AACCUGG | 0.08 |
|
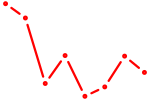
|

|
| CCUUGGC | 0.08 |
|

|

|
| ACCCGUA | 0.07 |
|

|

|
| AAUCUCU | 0.06 |
|

|

|
| GCCUGUC | 0.05 |
|
|

|
| CGUACCG | 0.05 |
|
|

|
| AUGACAC | 0.05 |
|
|

|
| UGUGCGU | 0.04 |
|
|

|
| CACCUCC | 0.04 |
|
|

|
| GAGAACU | 0.04 |
|

|

|
| CGUGUCU | 0.03 |
|

|

|
| UUUUUGC | 0.02 |
|

|

|
| GAUCAGA | 0.02 |
|
|

|
| AGAUCAG | 0.02 |
|

|

|
| AACCGUU | 0.02 |
|

|
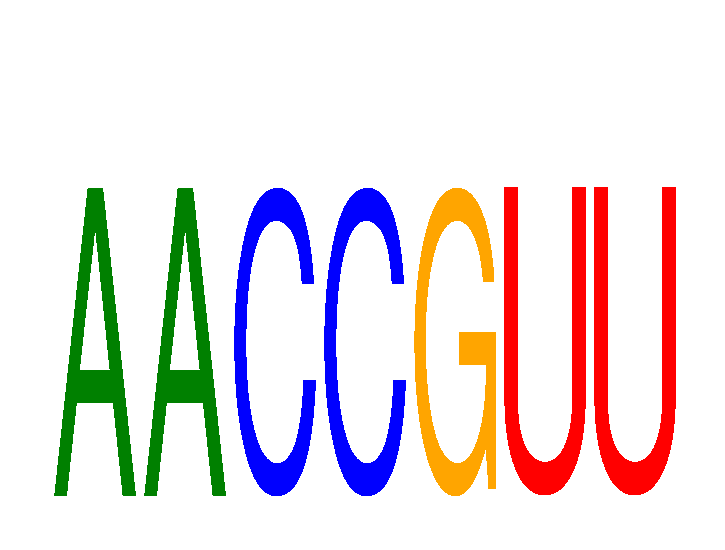
|
| GUACCGU | 0.01 |
|

|

|